Abstract
Heteroduplexes composed of all-DNA and all-2′-OMe RNA strands do not occur in nature, but they have found application in the development of molecular beacons and could also be used as aptamers or elements of nucleic acid-based nanostructures that will contain such structural motifs. The crystallization experiments performed have shown that the introduction of overhangs at the ends of the duplex has a great influence on the success of crystallization, as well as on the DNA:2′-OMe-RNA heteroduplex crystal packing. The molecular and crystal structure of the DNA:2′-O-methyl-RNA heteroduplex in its overhanging and blunt-ended versions was determined at 100 K using synchrotron radiation with a resolution of 1.91 and 1.55 Å, respectively. The Zn-SAD method was used to resolve the original duplex structure when molecular replacement by many existing models of duplex structures failed. Both molecules analyzed adopted a conformation close to the A-RNA double helix. The presented structures provide the first insight into this type of heteroduplexes and allowed a comparative analysis with existing nucleic acid homo- and heteroduplex structures. The results of our research expand the knowledge of the structural properties of new heteroduplexes and may be useful for future applications, such as therapies using this class of compounds.
1. Introduction
2′-O-methylation (2′-OMe) is a common posttranscriptional RNA modification found in all kingdoms of life, but its distribution among archaea, eukaryotes, and bacteria is not uniform [1]. The 2′-OMe decorates the cap portion of messenger RNAs (mRNAs), as well as noncoding and regulatory RNAs, including transfer RNAs (tRNAs), ribosomal RNAs (rRNAs), small nuclear RNAs (snRNAs), and microRNAs (miRNAs) [2,3]. While the chemical properties of this modification do not establish specific functional roles, it is evident that methylation of the 2′-hydroxyl group, which is often involved in contacts that form higher-order RNA structures, can affect RNA secondary structure, its hydrogen-bonding potential, and RNA/nucleic acid and RNA–protein interactions [4]. Although the nucleolytic stability of 2′-OMe RNA is limited [5], the 3′-terminal methylated miRNA molecules have been shown to be more resistant to digestion by 3′→5′ exoribonuclease, and have a higher affinity for Argonaute-2 compared to their unmethylated precursors [6]. To profile the presence of 2′-OMe units in different RNA species, several highly sensitive, quantitative methods have been developed for sequencing the 2′-OMe modifications in RNA chains at one-base resolution [2,4,7].
Re-engineering of nucleic acids into therapeutic molecules requires suitable chemical modifications which can assure their metabolic stability, effective cellular uptake and transport, as well as processing of the target molecule [8]. To date, 2′-O-methyl-RNA (2′-OMe-RNA) along with 2′-deoxy-2′-fluoro-RNA (2′-F-RNA), 2′-O-metoxyethyl-RNA (2′-MOE), S-constrained ethyl-RNA (cET-RNA), locked nucleic acids (LNA) and phosphorothioates (PS-DNA / RNA) have been identified as modifications which are most useful in a variety of RNA backbone biopharmaceuticals including short interfering RNAs (siRNAs), microRNA mimics, anti-miRs, aptamers, riboswitches and CRISPR-Cas9 RNAs that have undergone multiple confirmatory studies and clinical trials [9,10]. Currently, the most spectacular RNA-backbone biopharmaceuticals are those of already the US FDA approved optimized siRNAs, properly equipped with functional groups, with 2′-OMe modifications among others, allowing them to be efficiently stabilized and delivered, including patisiran (Onpattro®, 2018) against polyneuropathy (TTR), givosiran (Givlaari®, 2019) for acute hepatic porphyria (AHP), inclisiran (Leqvio®, 2020), for the treatment of adults with hypercholesterolemia or mixed dyslipidemia, and lumasiran, sold under the brand name Oxlumo® (2020), which is a siRNA medication for the treatment of primary hyperoxaluria type 1 (PH1) [10]. The second class of already the US FDA and EU EMA approved RNA biopharmaceuticals are mRNA vaccines that have been developed for use in combating coronavirus disease 2019 (COVID-19 pandemic) [11,12].
The use of 2′-O-methyl RNAs has been successfully employed in numerous studies with DNA/DNA and RNA/RNA duplexes because they offer several advantages over unmodified models for biopharmaceutical applications. First, the binding affinity for 2′-OMe-containing duplexes is significantly higher than for unmodified duplexes for identical sequences [13,14]. Second, 2′-OMe modification delays the degradation of oligonucleotides by nucleases [15]. Several two-dimensional NMR data on the solution structure of DNA/RNA can be found in the literature, indicating that DNA/RNA hybrid duplexes do not adopt an all-C2′-endo B type conformation in solution; neither do they adopt an all-C3′-endo A type conformation [16]. The single solved NMR structure of a chimeric DNA/2′-OMe-RNA duplex shows that the 2′-O-methylated units create the hydrophobic environment in the minor groove that affects the structure of this duplex [17].
So far, the presence of antiparallel DNA:2′-OMe-RNA heteroduplexes in nature has not been confirmed, but their thermodynamic properties have been studied by Szabat et al. in comparison with unmodified and modified DNA, RNA, and 2′-OMe-RNA homo- and heteroduplexes [18]. This type of molecules seems to be an interesting tool for applications in molecular biology or in structural research of nucleic acids. In the first case, such structures may arise from the interaction of appropriately labeled 2′-OMe-RNA strands with target DNA sequences as an element of molecular beacons [19].
In our research on the crystallographic structure of DNAzyme 10-23, we performed studies on a 52-nucleotide DNA/2′-OMe-RNA oligomer that mimics the deoxyribozyme molecule in complex with its substrate to ensure its sufficient thermodynamic and nucleolytic stability. Although we have succeeded in obtaining a crystal of this oligomer suitable for X-ray diffraction studies [20], the structure of the complex has not yet been solved due to the lack of suitable duplex models. To understand the structure of the DNA:2′-OMe-RNA duplex formed in our model between the 2′-OMe-RNA substrate and the substrate recognition arms of 10-23 DNAzyme, we decided to study the structure of a 9-bp blunt-ended DNA:2′-OMe-RNA duplex and its 8-bp overhang version by X-ray analysis.
2. Materials and Methods
2.1. Chemical Synthesis and Crystallization of Oligonucleotides
The objects for the crystallization experiments were: a duplex with blunt end d(GTCTCCTAG):2′-O-methyl(CUAGGAGAC), and a duplex with overhangs d(TCTCCTAGG):2′-O-methyl(CUAGGAGAC) (Figure 1). Both duplexes have the same 2′-OMe-RNA strand.
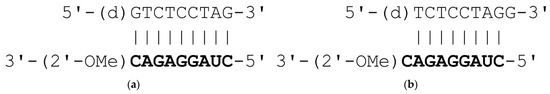
Figure 1.
Sequences of duplexes used in the experiments: (a) with blunt end; (b) with overhanging ends. Characters in normal format mean DNA, bold characters are 2′-O-methyl-RNA.
Oligonucleotides d(GTCTCCTAG), d(TCTCCTAGG), and 2′-O-methyl(CUAGGAGAC) were synthesized on a Gene World H-8 automated DNA/RNA synthesizer (K&A, Schaafheim, Germany) at a 0.2 μmol scale, using commercially available LCA-CPG supports (Biosearch Technologies, Inc., Petaluma, CA, USA) and standard phosphoramidite monomers (Glen Research, Sterling, VA, USA). Oligonucleotides were routinely deprotected and isolated using a Varian binary HPLC system, consisting of two PrepStar 218 pumps and a ProStar 325 UV/VIS detector set at 260 nm. A reversed-phase HPLC column (PRP-1, C18, 7 μm, 305 × 7 mm, Hamilton, Reno, NV, USA) was eluted with a 1% min−1 gradient of CH3CN in 0.1 M TEAB (pH 7.3) at a flow rate 2.5 mL min−1. All oligomers were analyzed by MALDI-TOF MS and polyacrylamide gel electrophoresis (PAGE). Oligonucleotides were dissolved in a buffer containing 5 mM sodium cacodylate pH 7.0 and 10 mM MgCl2 to a final concentration of 2 mM. Duplexes were formed by mixing equal volumes of DNA and 2′-OMe-RNA strands to obtain 1 mM duplex solution, which was heated to 368 K and slowly cooled to room temperature.
Crystallization experiments were performed at room temperature using the hanging drop vapor-diffusion method. An initial search for suitable crystallization conditions was performed using commercially available screens: Nucleic Acid Mini Screen (Hampton Research, Alisa Viejo, CA, USA) and Crystallization Kit for DNA (Sigma-Aldrich Chemie GmbH, Steinheim, Switzerland) for both duplexes, and additionally using Molecular Dimension HELIX (Molecular Dimensions Ltd., Suffolk, UK) for a duplex with overhangs. For the hanging droplet, 1.0–1.5 μL of reservoir solution was added to the same amount of duplex solutions. Each droplet was equilibrated with its individual solution in the wells of 24-well VDX™ plates (Hampton Research, Alisa Viejo, CA, USA). From ten initial hits for a blunt-end duplex and nearly sixty hits for an overhanging-end duplex, the most promising crystallization conditions were selected for an optimization phase. To obtain Zn-derived crystals, ZnCl2 was added to the crystallization solution or used instead of the MgCl2 originally presented. The crystals used for the diffraction experiments are shown in Figure 2.
Figure 2.
Obtained crystals of: (a) an overhanging duplex with Zn2+; (b) a blunt-ended duplex.
2.2. X-ray Data Collection and Processing
Diffraction data were recorded with a single crystal at wavelengths of 0.9184 (for native data) and 1.2831 Å (for anomalous data for Zn) at 100 K on Beamline MX 14.2 at the Berliner Elektronenspeicherring-Gesellschaft für Synchrotronstrahlung (BESSY), Berlin, equipped with a Dectris Pilatus3 S 2M detector [21]. Due to the high concentration of Li2SO4 used for crystallization, the crystals were cryoprotected with the mother liquor. The excess liquid was removed by carefully touching the surface of the plate with the mounting loop. The crystals were then directly immersed in the liquid N2. Complete data sets were collected with a maximum resolution of 1.91 Å for a blunt-ended duplex and up to 1.55 Å for the overhanging duplex. Diffraction data were processed, integrated, and scaled with XDS [22] using the XDSAPP2.0 GUI interface [23]. Data acquisition and processing statistics are shown in Table 1.

Table 1.
Crystallographic parameters and data-collection statistics. Values in parentheses correspond to the highest resolution shell.
2.3. Solution, Refinement and Analysis of the Structures
The first crystal structure of the overhanging duplex with Zn2+ was solved by the SAD method [24,25]. The locations of the Zn2+ ions sites were determined based on the anomalous signal using SHELXD [26]. These Zn2+ substructures were used as input to SHELXE [27], which performed the calculation and modification of the electron density map. Models of the DNA and 2′-OMe-RNA strands were manually inserted into the obtained electron density maps using COOT [28] and improved by alternating restrained refinement with phenix. refine from the PHENIX suite [29] and model building. Stereochemical restrains for refinement were adopted from RestraintLib (http://achesym.ibch.poznan.pl/restraintlib; access date 3 September 2020) [30,31,32] and Grade Web Server (http://grade.globalphasing.org; access date 20 October 2020) for DNA and 2′-O-methyl-RNA, respectively. The overhanging duplex structural model, modified by manual reconstruction in COOT, was used as a search model for molecular replacement with MOLREP [33] to determine the crystal structure of the blunt-ended duplex. The final refinement statistics of the described duplex structures are listed in Table 2. The crystal structures and the amplitudes of the structure factors were deposited in the Protein Data Bank under the accession codes given in Table 2. Detailed analysis of the structural parameters of the duplexes was performed using the Web 3DNA 2.0 server [34,35]. All figures were generated using PyMOL v.2.5.0 [36].

Table 2.
Refinement statistics of the studied structures. The values in parentheses correspond to the highest resolution shell.
3. Results and Discussion
We report here the first crystal structures of DNA:2′-O-methyl-RNA duplexes in two variants: both blunt-ended and overhang-ended. The sequences of the duplexes analyzed in this work correspond to one of the recognition arms of the deoxyribozyme 10–23 model used in our previous work [20]. Initial crystallization experiments of the blunt-ended duplex variant yielded trigonal crystals suitable for diffraction experiments, and data were collected to 1.91 Å resolution in the 321 point group, which was identified as the P3221 space group after analysis of systematic absences. Unfortunately, many attempts at phasing by molecular replacement using various models derived from existing nucleic acid homoduplexes and heteroduplexes or generated in silico with ideal geometry parameters failed. In a next step, we attempted to obtain duplex derivatives for SAD/MAD experiments that contained 5-bromouridine instead of thymidine in the DNA strand, and by soaking native crystals with compounds commonly used in macromolecular crystallography for anomalous phasing, such as [Co(NH6)]3+ or [Ta6Br12]2+. The presence of 5-BrU in the DNA strand altered the crystallization properties of the duplex studied and the crystals obtained were smaller and diffracted only to a resolution of 4–5 Å. On the other hand, we observed cracking of the tested native crystals after a short period of soaking with phasing compounds. To improve crystallization properties and obtain crystals that diffract to higher resolution limits, we decided to design a duplex consisting of eight complete base pairs with single overhangs on each strand by simple moving the first guanine in the previously used sequence from the 5′ end to the 3′ end position of the DNA strand. This small change significantly improved the crystallization properties and resulted in a higher number of screening hits, as well as improved diffraction properties of the obtained crystals. Among the nearly sixty initial crystallization hits, we observed the formation of two main crystal forms: well-diffracted tetragonal crystals belonging to the 422 point group (identified as the P43212 space group after analysis of systematic absences) and hexagonal crystals belonging to the 622 point group with weak diffraction properties. The conditions under which tetragonal crystals were obtained were further used for crystallization experiments in the presence of Zn2+ ions instead of Mg2+ as the source of the anomalous signal. Finally, the Zn-SAD methodology enabled the solution of the structure of the overhanging duplex (Figure 3), which, after manual rebuilding, was used as a search model for the molecular replacement of the data of the blunt-ended duplex.
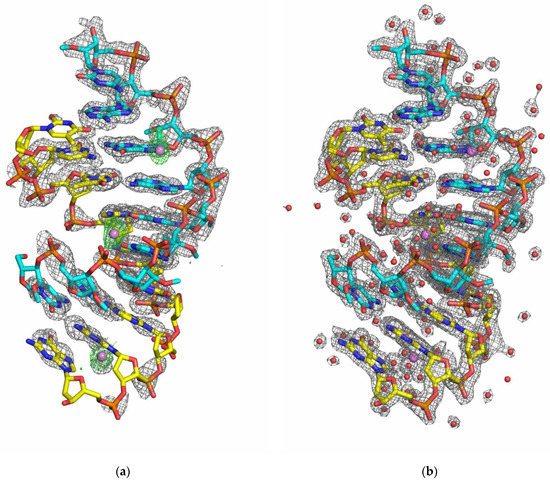
Figure 3.
The crystal structure of the overhanging DNA:2′-OMe-RNA duplex. (a) The initial density-modified electron density map contoured at the 1.0σ level (SHELXE output; grey mesh) with an anomalous electron density map contoured at the 3.0σ level (green mesh) and the refined structure of the overhanging duplex with Zn2+. (b) The final 2FO-FC electron density map (grey mesh) contoured at 1.0 σ and the refined structure of the overhanging duplex with Zn2+. The DNA and 2′-OMe-RNA strands are shown as sticks in yellow and cyan color schemes, respectively. Zn2+ ions and water molecules are shown as purple and red spheres, respectively.
3.1. The Refined Models and Crystal Packing
The high quality of the crystals and the use of synchrotron radiation allowed the refinement of the models of DNA:2′-OMe-RNA duplex structures with a good accuracy. The obtained models of the duplexes are complete and form a right-handed helix. The high-resolution model structure of the overhanging duplex (1.55 Å resolution) was refined to a final R/Rfree of 0.221/0.251. It also contains 92 water molecules and three Zn2+ ions identified by strong peaks on the anomalous difference map. No additional sites were found for Mg2+ ions, spermine, or compounds presented in folding buffers and used for crystallization experiments. The overhanging DNA:2′-OMe-RNA duplex molecule has been solved in the tetragonal space group P43212 with one duplex per asymmetric unit and eight duplexes in each unit cell (Figure 4a). The crystal lattice consists of infinite columns of overhanging duplex helices stacked head-to-tail. The overhang dG9 folds out of the duplex to form a base pair with the overhang Cm1 of a symmetrical counterpart. The eight complete base pairs and an additional base pair from the overhangs form a pseudo-nonamer.
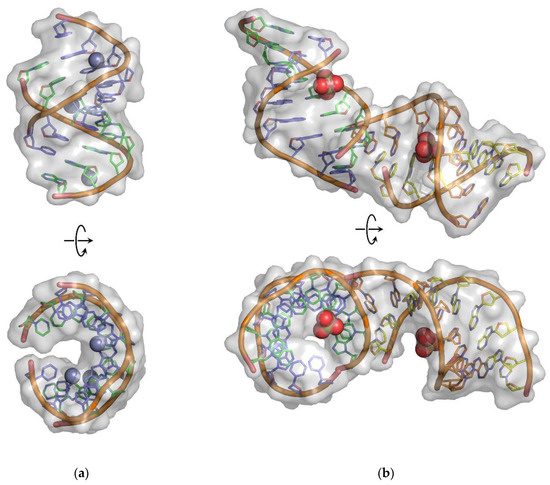
Figure 4.
Molecular structures of (a) overhanging and (b) blunt-ended DNA:2′-OMe-RNA duplexes. The DNA strands are colored as green and purple chains, whereas the 2′-OMe-RNA strands are shown in the cyan and yellow color schemes. The positions of the Zn2+ and SO42− ions are shown as spheres.
Each molecule of the overhanging duplex forms contacts with eight symmetrically related molecules through stacking interactions between terminal bases and through water-mediated intermolecular contacts. The most important interactions are localized at the junction between three molecules surrounding the Cm9 base (Figure 5a). The Cm9 overhang of the reference molecule forms a typical Watson–Crick base pair with dG9’ from a symmetry-related counterpart, SYM1. Moreover, the interaction of the overhanging ends of the symmetry-related molecules is stabilized by stacking interactions of Cm9 with Cm1′ and dT1 with dG9′, resulting in a continuous double helix structure in the crystal lattice. An additional direct hydrogen bond is formed between the O2′ atom of the 2′-O-methyl group of Cm9 and the N2 atom of Gm5′′ of the second symmetry-related molecule, SYM2.
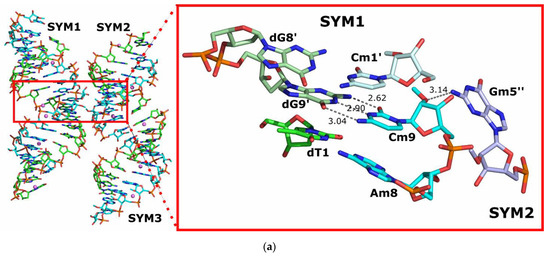
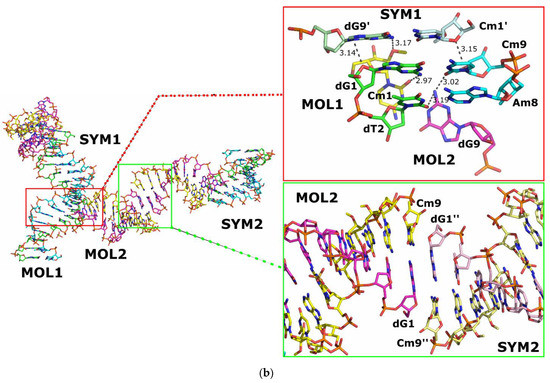
Figure 5.
Intermolecular interactions: (a) between an overhanging reference DNA:2′-OMe-RNA duplex and a symmetry-related object SYM1; (b) between blunt-ended reference DNA:2′-OMe-RNA duplexes and symmetry related objects SYM1 and SYM2. Hydrogen bonds are indicated by dark grey dashed lines.
For the blunt-ended duplex crystal data, the model was refined to a final R/Rfree of 0.184/0.195. The molecule has been solved in the trigonal space group P3221 with two duplexes per asymmetric unit and a total number of twelve duplexes in each unit cell (Figure 4b). Pairs of A:B and C:D chains form the duplexes referred to as MOL1 and MOL2, respectively. Additionally, 77 water molecules and two SO42− ions (one per duplex molecule) were also included in the final model structure. Again, no other chemicals used for folding and crystallization were indicated in the electron density maps obtained.
Analysis of the obtained crystal structure of the blunt-end duplex showed that three interfaces of interactions between the component molecules can be distinguished (Figure 5b). The MOL1 molecule and its symmetry-related counterparts create pseudo-infinite helices parallel to the c-axis of the crystal by stacking the terminal bases in “head-to-tail” order. Another interaction occurs between MOL1 and MOL2 molecules within the same asymmetric unit, with the first two “head” base pairs dG1-Cm9 and dT2-Am8 of the MOL1 molecule interacting with the “tail” base pair dG9-Cm1 of the MOL2 duplex via hydrogen bonds. Finally, the last interaction occurs between the MOL2 molecule and its symmetric counterpart SYM2 by stacking the terminal bases in “head-to-head” order, with the initial “head” base pairs dG1-Cm9 forming a series of hydrogen bonds with the “head” base pairs dG1′′-Cm9′′ of a symmetrically related MOL2 molecule belonging to the SYM2 object.
The structure of the motif formed by the first two interactions listed is based on three RNA fragments that form a unique set of tertiary interactions (Figure 6a). Analysis of the secondary structure pattern shows that two helices (H1 and H2) are stacked on top of each other due to the stacking interaction, but are shifted counterclockwise by about 25°. The third helix (H3) is located in the shallow groove of the stacking helices due to the positioning of its top base pair. This creates the possibility for a tertiary interaction (SE/SE) [37,38] between the 2-NH2 amino group of dG9MOL1,SYM1 and the 2′-OMe of Cm1MOL2 (H3). Another 2-NH2 group of dG1MOL1 (H1) forms a strong 3.0 Å hydrogen bond with the 2-O of the same Cm1MOL2. In addition, interesting double H-bonds (SE/SE) with a distances of 3.0 and 3.2 Å, are found between dG9MOL2 (H3) and 2-O’s of dT2MOL1 and Cm9MOL1, both from H1 (Figure 6c). The 2′-OH of this guanine dG9MOL2 (H3) is also involved in an interesting interaction with the sugar oxygen of the distant thymine dT4MOL1. In general, the sequence of the motif consists of twelve nucleotides arranged in six Watson–Crick base pairs, with seven nucleotides being essential (in red, Figure 6b). Of these nucleotides, five appear to be conservative and three could be random pyrimidines (Y) that interact with G or dG to form a 3-way junction interaction (3wj-YdG). The whole structure looks like a stable compact motif with some possible modifications (Figure 6d).
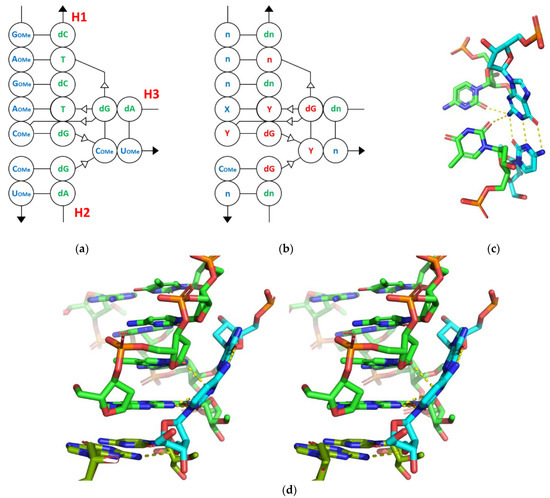
Figure 6.
3wj-YdG interaction: (a) original motif sequence; (b) secondary structure pattern; (c) 3D representation of 3wj-YdG interaction derived from pdb:7osx; (d) tertiary structure in stereoscopic 3D view (parallel). Hydrogen bonds are indicated by yellow dashed lines.
3.2. Structural Features of DNA:2′-O-Me-RNA Duplexes
Failures in molecular replacement experiments suggested that the geometry of the DNA:2′-O-methyl-RNA heteroduplex might differ from the geometries of used search duplex models, e.g., DNA:RNA [39], RNA:RNA [40], or 2′-OMe-RNA:2′-OMe-RNA [41]. The helix conformation parameters of the analyzed overhanging and blunt-ended duplexes are close to the values described for the double helix in A-form [42] (see Table 3). In this duplex, there are 10.93 residues per helical turn with an average helical twist of 32.9°, which corresponds to the typical A-RNA structure. The average rise per residue is 3.3 Å, compared to the range of 2.6–3.0 Å observed for A-form duplexes. The base-pair inclination (17.3°) is in the upper range of values observed for A-form duplexes (10°–20°). The average propeller twist is −11.5 Å, and the displacement of the base pairs towards the minor groove is 4.8 Å. The width of the minor and major grooves is 15.1 and 12.4 Å, respectively. All deoxyribose and ribose rings of the presented structures are in the strict C3′-endo puckering conformation with an average sugar pseudorotation phase angle of 16.6° and 12.8° [43] and with a corresponding intrastrand phosphate–phosphate distance of 5.77 and 5.90 Å on average, for DNA and 2′-O-methyl-RNA strands, respectively. All residues are characterized by anti glycosidic bond angles. The α-, β-, γ-, δ-, ε-, ζ-backbone torsion angles (Table S1) also fall within the range assumed for the A-family of right-handed helices. As can be seen in Table S1, the exocyclic C4′-C5′ bonds are all in the +g conformation, except for those at the Gm4 residue, which adopt the trans conformation. This observation correlates with the O5′-P bonds being in the −g conformation, again with the exception of the Gm4 residue, where the +ac conformation is observed.

Table 3.
Average base-pair and local base-pair helical parameters.
In discussing the data obtained for the blunt-end duplex, one should note some differences between two molecules present in the asymmetric unit of the crystal. Obviously, both duplexes belong to the A-form double helix family, with 11.5 and 10.8 residues per helical turn and with an average helical twist of 31.4° and 33.3° for MOL1 and MOL2, respectively. Comparison of the average base-pair and local base-pair helix parameters (Table 3) shows that the structure of the MOL1 molecule is similar to the overhanging DNA:2′-OMe-RNA duplex discussed previously, while the MOL2 duplex parameters are similar to those calculated for a DNA:RNA duplex [39]. The average rise per residue is 2.9 and 2.7 Å, compared to the range of 2.6–3.0 Å observed for A-form duplexes. The base pair inclination is different for the MOL1 and MOL2 duplexes, with values of 10.5 and 17.3°, respectively, but is still within the range observed for A-form duplexes. The average propeller twist is −8.8 and −10.6 Å, and the displacement of base pairs toward the minor groove is 4.2 and 3.9 Å, measured for the MOL1 and MOL2 duplexes, respectively.
The widths of the minor and major grooves measured for MOL1 average 15.3 and 17.9 Å, respectively, while for MOL2 they assume values of 15.6 and 13.6 Å. In the MOL1 duplex, the 2′-O-methyl-RNA strand is purely A-form, but there appears to be C2′-endo on the DNA strand at the 3′-end (residues dA8 and dG9). Analysis of the MOL2 duplex showed that almost all the deoxyribose and ribose rings of this molecule are in the strict C3′-endo puckering, with the exception of residues dC3 and Am3, where a C2′-oxo conformation is observed [43]. All residues of the two duplex molecules are characterized by anti glycosidic bond angles. Again, the α-, β-, γ-, δ-, ε-, ζ-backbone torsion angles (Table S1) fall within the range accepted for the A-family of right-handed helices. Detailed data analysis (Table S1) showed that the exocyclic C4′-C5′ bonds are all in the +g conformation, except for those at residues dC6, Gm4 and Gm7 of MOL1, and dC3, Am3, and Gm7 of MOL2, which adopt the trans conformation, correlating with the −g and +ac conformations of the O5′-P bonds, respectively.
The conformation of the 2′-O-methyl residues was also analyzed for all 2′-OMe-RNA chains, and the values of the C1′-C2′-O2′-CM2 dihedral angles were measured. Almost all 2′-O-methyl residues in the two structures discussed were in the +g conformation, with some outliers belonging to the +ac conformation. These results are consistent with those previously observed for two known structures with continuous 2′-OMe-RNA strands (PDB IDs: 310d, 1i7j) [41,46]. The largest deviation was observed for the Am6 residue of the MOL1 molecule from data of the blunt-ended duplex, where the discussed value of the dihedral angle is almost 150°, compared to the typical range of 60–85° observed for the rest of the 2′-O-methyl residues. However, a detailed analysis of this structural region does not explain the reason for this disorder.
To highlight the helical differences between DNA:2′-OMe-RNA, DNA:RNA, RNA:RNA, and 2′-OMe-RNA:2′-OMe-RNA duplexes, we extended the helices to a maximum of 40 bp using the regular helical structure of nucleic acids based on their average helical parameters using the Web 3DNA 2.0 server [34,35]. We present the overall geometry in side view and top view in the space-filling models (Figure 7). Surprisingly, the structure of the DNA:2′-OMe-RNA duplex was found to be very similar to the RNA:RNA and not to the DNA:RNA duplex, including the width of the minor and major grooves of these helices. This observation could be attributed to the “compressing” effect of the presence of the 2′-O-methyl RNA strand compared to the DNA:RNA helix, whose structure is more “stretched”. However, some differences in buckle, opening, and tilt parameters render the RNA:RNA model useless for solving the structure of the DNA:2′-OMe-RNA duplex.
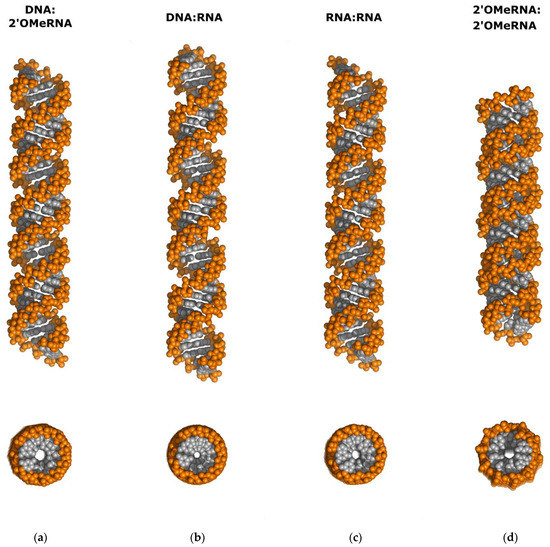
Figure 7.
Comparison of idealized duplexes of (a) DNA:2′-OMe-RNA (derived from 7ow0.pdb), (b) DNA:RNA (derived from 479d.pdb), (c) RNA:RNA (derived from 1sdr.pdb), and (d) 2′-OMe-RNA:2′-OMe-RNA (derived from 1i7j.pdb). All helices were expanded to 40 bp per duplex and represented as space-filling models. Side view (top) and top view (bottom). Base pairs are shown in gray, while phosphates and sugar units are colored orange. Details are described in the text.
3.3. Hydration of the Duplexes
The overhanging DNA:2′-OMe-RNA duplex is hydrated by 92 uniquely identified water molecules. Among them, four waters are located at specific positions with an occupation of one half; 74 of the hydration sites belong to the first hydration shell and 18 to the second. An equal number of water molecules (37 per chain) are involved in the interactions with DNA and 2′-OMe-RNA strands. A total of 38 water molecules hydrate the backbone phosphates, 22 hydrate the sugar atoms O2′, O3′, O4′ and O5′, and 22 hydrate the base units, while 15 are found in the coordination spheres of the zinc ions. The pyrimidine bases have one to two hydrogen-bonded water molecules, with the exception of the guanines dG8, Gm4 and Gm7, which are involved in the coordination of the zinc ions, where the number of bonded water molecules varies from four to seven. Water molecules bridging adjacent phosphate groups in the same strand are also observed. No single water molecule was found bound to the O2′ atom of the 2′-O-methyl group of the 2′-OMe-RNA strand, with the exception of Am8, which distinguishes it from other A-form RNA duplexes. Detailed information on the hydration of the overhanging DNA:2′-OMe-RNA hybrid duplex is provided in Table S3.
The asymmetric unit of the blunt-ended duplex crystal structure contains two molecules of DNA:2′-OMe-RNA duplexes hydrated by 77 well-defined water molecules, 53 of which are associated with the first duplex (A:B chains), and only 24 with the second (C:D chains). In total, 68 of the hydration sites belong to the first hydration shell and 9 to the second. In total, 23 water molecules hydrate the backbone phosphates, 20 hydrate the sugar atoms O2′, O3′, O4′ or O5′, and 38 hydrate the base units, while 4 interact with sulfate ions. The purine bases have between two and three associated waters, with the exception of residues Gm5 and Am6 of chain A, where four and six waters were identified, respectively. The pyrimidine bases generally have one to two hydrogen-bonded water molecules, but no water was found for some residues. The lower number of water molecules observed is related to both the lower resolution of the data collected and differences in crystal packing. Detailed information on the hydration of these hybrid duplexes with blunt ends can be found in Table S3.
3.4. Zn2+ Ions Coordination
Due to problems with phasing by the molecular replacement method and with the use of incorporated Br-U or soaking with [Co(NH6)]3+ or [Ta6Br12]2+ for SAD/MAD experiments, we decided to test the Zn-SAD method [24,25]. Zinc ions were incorporated into the overhanging duplex structure by simply using ZnCl2 instead of MgCl2 in the optimized crystallization conditions of Crystallization Kit for DNA no. 30 (Sigma-Aldrich Chemie GmbH, Steinheim, Switzerland). The final crystallization conditions contained 1.8 M Li2SO4, 50 mM sodium cacodylate pH 6.5, 5 mM spermine, and 50 mM ZnCl2, and allowed to obtain crystals with good X-ray diffraction properties. The anomalous signal from the stably bound Zn2+ was sufficient to determine three Zn2+ sites (per asymmetric unit). The heavy atom search method implemented in SHELXD, followed by density modification with SHELXE, resulted in interpretable experimental density maps (Figure 3a).
All three Zn2+ ions were coordinated to the N7 atoms of three of the five guanine bases presented: dG8, Gm4 and Gm7, while there is no evidence of Zn2+ ion binding for dG9 and Gm5. Moreover, Zn2+ is bound to the first guanine from the 5′-end in a series of guanine double strands in both DNA and 2′-O-methyl RNA strand. There are no zinc ions bound to the backbone phosphate groups as observed by Hou and Tsodikov [25]. All three Zn2+ ions were in an octahedral coordination with the N7 atom of the guanines and the water molecules (Figure 8a). The bond lengths were in the range of 2.04–2.62 Å for Zn2+…N7 and Zn2+…O of the water molecules, as expected for an octahedral coordination (see Table S3). All three Zn2+ ions had a complete coordination shell with five water ligands, and were refined with an occupancy of 1.0, resulting in B-factors in the range of 33–45 Å2.
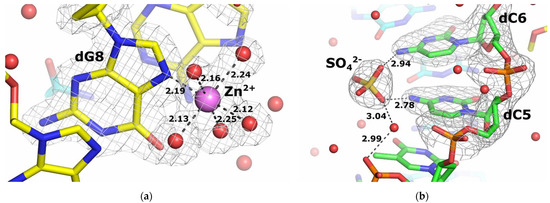
Figure 8.
Ions bonding in duplex structures under study. (a) An enlarged view of a representative Zn2+ coordination shell in the structure of the overhanging DNA:2′-OMe-RNA duplex. (b) A view of the SO42− binding site in the structure of the blunt-ended duplex A:B.
3.5. SO42− Ions Binding
The binding of sulfate ions by RNA duplexes has already been observed and described [47,48]. Here we have demonstrated the presence of SO42− ions bound to the blunt-ended duplex at the same site of the two duplex molecules in the asymmetric unit. The sulfate was used as a precipitant in the form of 2.0 M lithium sulfate. The SO42− ion binding pocket is formed by two deoxycytidines dC5 and dC6, with the oxygen atoms of the sulfate ion forming hydrogen bonds with the N4 atoms of the cytidine rings (Figure 8b). The SO42− ion bound to chain A is stabilized by an additional interaction with the phosphate group of dT4 via a water molecule. Analysis of the sulfate-binding pocket of chain C revealed two additional interactions formed via water molecules with the O4 atom of dT7 of the same strand, as well as with the O4 atom of Um2 and the N6 and N7 atoms of the Am3 residues of chain D. Detailed information on all interactions between SO42− ions and oligonucleotide strands is given in Table S4.
4. Conclusions
In this work, we have studied the structure of DNA:2′-O-methyl RNA hybrid duplexes in two forms: with and without overhangs. So far, the three-dimensional structure of this type of molecule has not been described in the literature. We found that the presence of overhangs significantly increases the number of hits when screening suitable crystallization conditions, and also improved the crystallization process of the hybrid DNA:2′-OMe-RNA duplex. In the next step, the application of the methodology based on the detection of an anomalous signal from zinc atoms introduced into the hybrid duplex structure enabled the solution of the structure using the SAD technique. The crystal structures of the DNA:2′-O-methyl RNA duplexes presented here show a nucleic acid conformation belonging to the canonical A-type nucleic acid geometry, similar to the RNA:RNA duplexes, but with some significant differences that explain the problems in phase determination by molecular substitution in this type of model. Finally, both duplexes were refined to high resolution, allowing detailed structural analysis, including binding of zinc or sulfate ions and identification of the 3wj-YdG interaction between two helices within the blunt-ended duplex model.
Knowledge of the molecular structure of the DNA:2′-O-methyl-RNA duplex could be useful not only for using these models to solve future structures with this motif, such as molecular beacons or complexes with target proteins, but also for designing nanostructures containing these types of duplexes or for developing therapies based on the use of this class of compounds.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cryst12060760/s1, Table S1: Backbone and glycosyl torsion angles; Table S2: Dihedral angles C1′-C2′-O2′-CM2 calculated for all 2′-OMe-RNA residues; Table S3: Distances between water molecules and non-hydrogen nucleic acid atoms or Zn2+ ions for overhanging DNA:2′-OMe-RNA duplex (PDB ID: 7ow0); Table S4: Distances between water molecules and non-hydrogen nucleic acid atoms or SO42− ions for blunt-ended DNA:2′-OMe-RNA duplex (PDB ID: 7oxs).
Author Contributions
Synthesis and purification of oligonucleotides, A.M. and B.M.; crystallization, X-ray data-collection, structure solution and refinement, data visualization, validation, deposition and analysis, writing—original draft, R.D.; writing—review and editing, supervision, B.N. All authors have read and agreed to the published version of the manuscript.
Funding
This research was financially supported by CMMS PAS Statutory Funds, Poland.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Atomic coordinates and structure factor amplitudes for blunt-ended and overhanging DNA:2′-OMe-RNA heteroduplexes are available from the Protein Data Bank as entries 7oxs and 7ow0, respectively.
Acknowledgments
Diffraction data were collected at the BL14.2 beamline at the BESSY II electron storage ring at Helmholtz-Zentrum Berlin (HZB), Germany. We would especially like to thank Martin Gerlach and Manfred S. Weiss for their help and support during the experiments. The authors are grateful to Piotr Guga and Arkadiusz Chworoś for constructive suggestions and discussions.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Motorin, Y.; Helm, M. RNA nucleotide methylation. Wiley Interdiscip. Rev. RNA 2011, 2, 611–631. [Google Scholar] [CrossRef] [PubMed]
- Motorin, Y.; Marchand, V. Detection and analysis of RNA ribose 2′-O-methylations: Challenges and solutions. Genes 2018, 9, 642. [Google Scholar] [CrossRef]
- Roundtree, I.A.; Evans, M.E.; Pan, T.; He, C. Dynamic RNA modifications in gene expression regulation. Cell 2017, 169, 1187–1200. [Google Scholar] [CrossRef]
- Incarnato, D.; Anselmi, F.; Morandi, E.; Neri, F.; Maldotti, M.; Rapelli, S.; Parlato, C.; Basile, G.; Oliviero, S. High-throughput single-base resolution mapping of RNA 2΄-O-methylated residues. Nucleic Acids Res. 2017, 45, 1433–1441. [Google Scholar] [CrossRef]
- Kratschmer, C.; Levy, M. Effect of chemical modifications on aptamer stability in serum. Nucleic Acids Ther. 2017, 27, 335–344. [Google Scholar] [CrossRef]
- Liang, H.; Jiao, Z.; Rong, W.; Qu, S.; Liao, Z.; Sun, X.; Wei, Y.; Zhao, Q.; Wang, J.; Liu, Y.; et al. 3′-Terminal 2′-O-methylation of lung cancer miR-21-5p enhances its stability and association with Argonaute 2. Nucleic Acids Res. 2020, 48, 7027–7040. [Google Scholar] [CrossRef]
- Hsu, P.J.; Fei, Q.; Dai, Q.; Shi, H.; Dominissini, D.; Ma, L.; He, C. Single base resolution mapping of 2′-O-methylation sites in human mRNA and in 3′ terminal ends of small RNAs. Methods 2019, 156, 85–90. [Google Scholar] [CrossRef]
- Egli, M.; Manoharan, M. Re-engineering RNA molecules into therapeutic agents. Acc. Chem. Res. 2019, 52, 1036–1047. [Google Scholar] [CrossRef]
- Moumné, L.; Marie, A.-C.; Crouvezier, N. Oligonucleotide therapeutics: From discovery and development to patentability. Pharmaceutics 2022, 14, 260. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Zhong, L.; Weng, Y.; Peng, L.; Huang, Y.; Zhao, Y.; Liang, X.J. Therapeutic siRNA: State of the art. Signal Transduct. Target. Ther. 2020, 5, 101. [Google Scholar] [CrossRef] [PubMed]
- Sandbrink, J.B.; Shattock, R.J. RNA Vaccines: A suitable platform for tackling emerging pandemics? Front. Immunol. 2020, 11, 608460. [Google Scholar] [CrossRef] [PubMed]
- Wright, J.T. COVID-19 vaccination: Science, politics and public health. J. Am. Dent. Assoc. 2021, 152, 181–183. [Google Scholar] [CrossRef] [PubMed]
- Freier, S.M.; Altmann, K.H. The ups and downs of nucleic acid duplex stability: Structure-stability studies on chemically-modified DNA:RNA duplexes. Nucleic Acids Res. 1997, 25, 4429–4443. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, A.S.; Sousa, I.M.; Fernandes, R.M.; Azevedo, N.F.; Almeida, C. Optimizing locked nucleic acid/2′-O-methyl-RNA fluorescence in situ hybridization (LNA/2′OMe-FISH) procedure for bacterial detection. PLoS ONE 2019, 14, e0217689. [Google Scholar] [CrossRef]
- Cummins, L.L.; Owens, S.R.; Risen, L.M.; Lesnik, E.A.; Freier, S.M.; McGee, D.; Guinosso, C.J.; Cook, P.D. Characterization of fully 2′-modified oligoribonucleotide hetero- and homoduplex hybridization and nuclease sensitivity. Nucleic Acids Res. 1995, 23, 2019–2024. [Google Scholar] [CrossRef]
- Salazar, M.; Fedoroff, O.Y.; Miller, J.M.; Ribeiro, N.S.; Reid, B.R. The DNA strand in DNA:RNA hybrid duplexes is neither B-form nor A-form in solution. Biochemistry 1993, 32, 4207–4215. [Google Scholar] [CrossRef]
- Tsao, Y.P.; Wang, L.Y.; Hsu, S.T.; Jain, M.L.; Chou, S.H.; Huang, C.; Cheng, J.W. The solution structure of [d(CGC)r(amamam)d(TTTGCG)]2. J. Biomol. NMR 2001, 21, 209–220. [Google Scholar] [CrossRef]
- Szabat, M.; Pędziński, T.; Czapik, T.; Kierzek, E.; Kierzek, R. Structural aspects of the antiparallel and parallel duplexes formed by DNA, 2′-O-methyl RNA and RNA oligonucleotides. PLoS ONE 2015, 10, e0143354. [Google Scholar] [CrossRef]
- Gao, J.; Li, Y.; Li, W.; Zeng, C.; Xi, F.; Huang, J.; Cui, L. 2′-O-methyl molecular beacon: A promising molecular tool that permits elimination of sticky-end pairing and improvement of detection sensitivity. RSC Adv. 2020, 10, 41618–41624. [Google Scholar] [CrossRef]
- Dolot, R.; Sobczak, M.; Mikołajczyk, B.; Nawrot, B. Synthesis, crystallization and preliminary crystallographic analysis of a 52-nucleotide DNA/2′-OMe-RNA oligomer mimicking 10-23 DNAzyme in the complex with a substrate. Nucleosides Nucleotides Nucleic Acids 2017, 36, 292–301. [Google Scholar] [CrossRef]
- Gerlach, M.; Mueller, U.; Weiss, M.S. The MX beamlines BL14.1-3 at BESSY II. J. Large-Scale Res. Facil. 2016, 2, A47. [Google Scholar] [CrossRef]
- Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Sparta, K.M.; Krug, M.; Heinemann, U.; Mueller, U.; Weiss, M.S. XDSAPP2.0. J. Appl. Cryst. 2016, 49, 1085–1092. [Google Scholar] [CrossRef]
- Kim, M.K.; Lee, S.; An, Y.J.; Jeong, C.S.; Ji, C.J.; Lee, J.W.; Cha, S.S. In-house zinc SAD phasing at Cu Kα edge. Mol. Cells 2013, 36, 74–81. [Google Scholar] [CrossRef] [PubMed]
- Hou, C.; Tsodikov, O.V. Utilizing guanine-coordinated Zn2+ ions to determine DNA crystal structures by single-wavelength anomalous diffraction. Acta Crystallogr. D Biol. Crystallogr. 2019, 75, 32–40. [Google Scholar] [CrossRef]
- Schneider, T.R.; Sheldrick, G.M. Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 2002, 58, 1772–1779. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Macromolecular phasing with SHELXE. Z. Kristallogr. 2002, 217, 644–650. [Google Scholar] [CrossRef]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef]
- Liebschner, D.; Afonine, P.V.; Baker, M.L.; Buknóczi, G.; Chen, V.B.; Croll, T.I.; Hintze, B.; Hung, L.-W.; Jain, S.; McCoy, A.J.; et al. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Biol. Crystallogr. 2019, 75, 861–877. [Google Scholar] [CrossRef]
- Kowiel, M.; Brzeziński, D.; Jaskólski, M. Conformation-dependant restrains for polynucleotides: I. clustering of the geometry of the phosphodiester group. Nucleic Acids Res. 2016, 44, 8479–8489. [Google Scholar] [CrossRef][Green Version]
- Gilski, M.; Zhao, J.; Kowiel, M.; Brzeziński, D.; Turner, D.H.; Jaskólski, M. Accurate geometrical restrains for Watson-Crick base pairs. Acta Crystallogr. B Struct. Sci. Cryst. Eng. Mater. 2019, 75, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Kowiel, M.; Brzeziński, D.; Gilski, M.; Jaskólski, M. Conformation-dependent restraints for polynucleotides: The sugar moiety. Nucleic Acids Res. 2020, 48, 962–973. [Google Scholar] [CrossRef] [PubMed]
- Vagin, A.; Teplyakov, A. MOLREP: An automated program for molecular replacement. J. Appl. Crystallogr. 1997, 30, 1022–1025. [Google Scholar] [CrossRef]
- Li, S.; Olson, W.K.; Lu, X.-J. Web 3DNA 2.0 for the analysis, visualization, and modeling of 3D nucleic acid structures. Nucleic Acids Res. 2019, 47, W26–W34. [Google Scholar] [CrossRef] [PubMed]
- Olson, W.K.; Bansal, M.; Burley, S.K.; Dickerson, R.E.; Gerstein, M.; Harvey, S.C.; Heinemann, U.; Lu, X.J.; Neidle, S.; Shakked, Z.; et al. A standard reference frame for the description of nucleic acid base-pair geometry. J. Mol. Biol. 2001, 313, 229–237. [Google Scholar] [CrossRef]
- The PyMOL. Molecular Graphics System, Version 2.0; Schrödinger, LLC: New York, NY, USA, 2018. [Google Scholar]
- Leontis, N.B.; Stombaugh, J.; Westhof, E. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 2002, 30, 3497–3531. [Google Scholar] [CrossRef]
- Leontis, N.B.; Westhof, E. Analysis of RNA motifs. Curr. Opin. Struct. Biol. 2003, 13, 300–308. [Google Scholar] [CrossRef]
- Xiong, Y.; Sundaralingam, M. Crystal structure of a DNA·RNA hybrid duplex with a polypurine RNA r(gaagaagag) and complementary polypyrimidine DNA d(CTCTTCTTC). Nucleic Acids Res. 2000, 28, 2171–2176. [Google Scholar] [CrossRef]
- Shindelin, H.; Zhang, M.; Bald, R.; Furste, J.P.; Erdmann, V.A.; Heinemann, U. Crystal structure of and RNA dodecamer containing the Escherichia coli Shine-Dalgarno sequence. J. Mol. Biol. 1995, 249, 595–603. [Google Scholar] [CrossRef]
- Adamiak, D.A.; Rypniewski, W.R.; Milecki, J.; Adamiak, R.W. The 1.19 Å X-ray structure of 2′-O-Me(CGCGCG)2 duplex shows dehydrated RNA with 2-methyl-2,4-pentanediol in the minor groove. Nucleic Acids Res. 2001, 29, 4144–4153. [Google Scholar] [CrossRef]
- Saenger, W. Principles of Nucleic Acids Structure; Springer Publishers: New York, NY, USA, 1984. [Google Scholar]
- Altona, C.; Sundaralingam, M. Conformational analysis of the sugar ring in nucleosides and nucleotides. New description using the concept of pseudorotation. J. Am. Chem. Soc. 1972, 94, 8205–8212. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, R.; Arnott, S. The structures of DNA and RNA helices in oriented fibers. In Landolt-Börnstein Numerical Data and Functional Relationships in Science and Technology, Group VII/1b, Nucleic Acids; Saenger, W., Ed.; Springer: Berlin, Germany, 1989; pp. 31–170. [Google Scholar]
- Arnott, S. Polynucleotide secondary structures: An historical perspective. In Oxford Handbook of Nucleic Acid Structure; Neidle, S., Ed.; Oxford University Press: Oxford, UK, 1999; pp. 1–38. [Google Scholar]
- Adamiak, D.A.; Milecki, J.; Popenda, M.; Adamiak, W.A.; Dauter, Z.; Rypniewski, W.R. Crystal structure of 2′-O-Me(CGCGCG)2, and RNA duplex at 1.30 Å resolution. Hydration pattern of 2′-O-methylated RNA. Nucleic Acids Res. 1997, 25, 4599–4607. [Google Scholar] [CrossRef] [PubMed]
- Rypniewski, W.; Adamiak, D.A.; Milecki, J.; Adamiak, R.W. Noncanonical G(syn)-G(anti) base pairs stabilized by sulphate anions in two X-ray structures of the (GUGGUCUGAUGAGGCC) RNA duplex. RNA 2008, 14, 1845–1851. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Masquida, B.; Sauter, C.; Westhof, E. A sulfate pocket formed by three GoU pairs in the 0.97 Å resolution X-ray structure of a nonameric RNA. RNA 1999, 5, 1384–1395. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).