Abstract
Effective biocatalysts for the synthesis of optically pure amines from keto precursors are highly required in organic synthesis. Transaminases are a large group of PLP-dependent enzymes, which can be utilized for production of chiral amines or amino acids. The bioinformatic approach previously made to search for promising transaminases with unusual characteristics surprisingly revealed mysterious genes in some Gram-negative bacteria, which products were annotated as aminotransferases, but they lacked the key catalytic lysine residue required for covalent binding of the PLP-cofactor. To address the question of which products these genes encode, we obtained the first structure of such a type of protein from the bacterium Variovorax paradoxus (VP5454) and provided its comprehensive analysis. We demonstrated that VP5454 has a typical aminotransferase fold and architecture of the active site, where substitution of the catalytic lysine with asparagine was observed. Despite that no covalent adduct can be formed between PLP and asparagine residue, using X-ray analysis and molecular dynamic (MD) simulation, we demonstrated that VP5454 is able to bind the PLP molecule in the transaminase in a specific manner, with PLP coordinated via its phosphate moiety. Taking into account a number of sequences homologous to VP5454 with a substituted catalytic lysine found in the genomes of various bacteria, we speculate that the proteins encoded by these sequences may have hidden functional roles.
1. Introduction
The development of biocatalysts for the synthesis of optically pure amines from keto precursors, cascade processes, and whole-cell catalysis is in great demand in organic synthesis [1,2,3]. Transaminases (TAs; aminotransferases; EC 2.6.1.X) are a large group of pyridoxal-5′-phosphate (PLP)-dependent enzymes that catalyze the stereoselective reversible transfer of an amino group from an amino substrate to a keto acid or ketone to form a chiral amine or amino acid and a new keto compound [4,5].
Despite noticeable progress, the engineering of a tailormade biocatalyst is still an intuitive task, which is hindered by a lack of data on sequence–structure–function relationships. A search for the enzymes with unusual characteristics with subsequent structural and functional characteristics is highly promising for expanding our knowledge in this field. Since the amino acid sequences of TAs contain well-known conserved motifs discriminating their substrate specificity [6], substitutions in such motifs can be used to search for TAs with altered catalytic properties.
TAs belong to I and IV fold types of PLP-dependent enzymes [7]. Here, we focus on TAs of PLP fold type IV (class IV), which are represented by three distinct families: (R)-selective D-amino acid aminotransferases (DAATs), (R)-amine TAs (R-TAs), and (S)-selective branched-chain L-amino acid TAs (BCATs) [8,9]. The minimal catalytically active unit of TAs of PLP fold type IV is a homodimer, in which the active site is formed by residues of two adjacent subunits [4]. TAs can be tetramers or hexamers in a crystal and solution [10,11,12]. Each TA’s subunit consists of two domains connected by an interdomain loop, a small N-terminal domain with an α/β-structure, and a large C-terminal domain with a pseudobarrel structure. The active site is located at the bottom of the cleft formed by the residues of both domains of one subunit and a small domain of the adjacent subunit [8]. A cofactor, PLP molecule, binds to the catalytic lysine residue in the active site via Schiff base linkage, producing a so-called holo-form of the enzyme. Other highly conserved among TAs residues are involved in noncovalent interactions with PLP [13,14]: glutamate or aspartate residue form hydrogen bonds with the protonated N1 atom of the PLP pyridine ring, family-specific residues form hydrogen bonds with the phenolic group of the PLP molecule, and arginine and threonine residues form hydrogen bonds with the phosphate moiety of PLP [13,14,15,16]. The active site of TAs is a combination of two pockets: P-pocket on the phosphate side of the PLP and O-pocket on the opposite side with the PLP molecule positioned in between the pockets [17]. A number of conserved secondary structure elements are involved in the formation of the pockets, including an interdomain loop, an O-loop, βX- and βY-strands, and two β-turns [8]. The size and amino acid composition of the pockets vary between different families of TAs of fold type IV, which cause differences in catalytic properties of these enzymes.
We revealed a puzzling aminotransferase class IV sequence in the genome of Gram-negative bacteria Variovorax paradoxus. This sequence has a substitution in a key catalytic lysine residue, which is known to form a covalent adduct with the cofactor PLP. To shed light on properties of this puzzling protein, called VP5454, we purified and examined it biochemically. We also succeeded in VP5454 crystallization and obtained its crystal structure at 2.3 Å resolution using X-ray analysis. We performed a comparative analysis of our results using structural data of known TAs with special focus on the structure of the active site. Finally, we performed molecular dynamic simulations to further support our conclusions. To sum up, we demonstrated that VP5454 has a typical architecture of PLP fold type IV TAs, that it shares some BCAT features in the active site, and that it is able to non-covalently bind PLP. Moreover, we suppose that the substitution of the active site N174 residue for lysine might result in the appearance of BCAT activity. We also speculate that the VP5454 gene falls in a special group of “pseudo TA” genes, which might be a result of a special evolution mechanism for deactivating these type of enzymes, aimed at obtaining of a novel obscured function.
2. Materials and Methods
2.1. VP5454 Expression, Purification and Characterization
Enzyme expression and purification were performed as described in [18]. Briefly, the gene VP5454_1c54540 (WP_021013128.1) encoding transaminase of PLP fold type IV was identified in the complete genome sequence of V. paradoxus. The nucleotide sequence of VP5454_1c54540 was optimized for expression in E. coli using server Optimizer (http://genomes.urv.es/OPTIMIZER/; accessed on 27 February 2019) and was synthesized by ATG Service Gene (St. Petersburg, Russia). The synthetic gene was cloned into the pET-21d vector (Novagen, Darmstadt, Germany) modified as described by [19]. Recombinant VP5454 (317 a.a., 33.89 kDa) fused at the N-terminus to a His6TEV-tag was expressed in E. coli Rosetta (DE3) (Novagen, Darmstadt, Germany). After incubation for 18 h at 24 °C, the cells were harvested by centrifugation, resuspended in 50 mM Tris-HCl buffer, pH 8.0, supplemented with 500 mM NaCl, 20 mM imidazole, 1 mM b-mercaptoethanol, 0.1% (v/v) Triton X-100, 100 µM PLP and 1 mM phenylmethylsulfonyl fluoride (PMSF), and disrupted by sonication. The crude cell extract was centrifuged for 25 min at 18,500× g. The supernatant was applied to a HisTrap HP column (Cytiva, Marlborough, MA, USA). The (His)6-tagged recombinant VP5454 was eluted with a linear gradient from 20 to 500 mM imidazole in the same buffer without Triton X-100. The collective fraction was transferred into 50 mM Tris-HCl buffer, pH 8.0, supplemented with 100 mM NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA), 1 mM β-mercaptoethanol, 5% (v/v) glycerol and tobacco etch virus (TEV) protease (1 mg per 10 mg of the protein). The solution was incubated overnight at +4 °C, dialyzed against the binding buffer, and applied to a HisTrap HP column. The flow-through was collected, concentrated and transferred into 50 mM pyrophosphate buffer, pH 9.0. The VP5454 solution was applied to MonoQ column (Cytiva, Marlborough, MA, USA) equilibrated in 50 mM pyrophosphate buffer, pH 9.0. The main fraction was collected using a linear gradient of NaCl from 0 to 500 mM in pyrophosphate buffer. Finally, the eluted VP5454 fraction was adjusted to 50 mM NaCl by dilution and was further concentrated to 20 mg/mL. To prevent cysteine residues oxidation, 5 mM DTT was added. For storage and crystallization, 1mM PLP was added to purified VP5454 solution. The protein purity was analyzed by SDS-PAGE (12%). The protein concentration was determined spectrophotometrically [20]. The amino acid sequences were checked by MALDI-TOF MS analysis (UltraFlex-treme Bruker Daltonik, Bremen, Germany).
2.2. Protein Crystallization
An initial crystallization screening of VP5454 was performed with a robotic crystallization system (Rigaku, The Woodlands, TX, USA) and commercially available 96-well crystallization screens (Hampton Research and Anatrace, Aliso Viejo, CA, USA) at 20 °C using the sitting drop vapor diffusion method. The protein concentration was 20 mg/ml in the following buffer: 50 mM pyrophosphate buffer pH 9.0, supplemented with 50 mM NaCl, 5 mM dithiothreitol (DTT). Diffraction quality crystals were obtained by the counter-diffusion method in capillaries in microgravity conditions (aboard the ISS). Rod-shaped crystals of 100 × 50 × 50 µm3 size were obtained within 2 weeks in the following conditions: 0.1 M citric acid BIS-TRIS propane pH 4.1; 20% PEG3350.
2.3. Data Collection, Processing, Structure Solution and Refinement
VP5454 crystals were extracted from the capillaries, briefly soaked in a mother liquor containing 20% glycerol immediately prior to diffraction data collection and flash-frozen in liquid nitrogen. The data were collected at 100K at BL41XU beamline (SPring8, Harima, Japan). The data were indexed, integrated and scaled using the Dials program [21] (Table 1). The program Pointless [22] suggested the P3221 space group.

Table 1.
Data collection, processing and refinement.
The structure was solved by the molecular replacement method using the MOLREP program [23] and the structure of BCAT TA from Geoglobus acetiviorans (PDB ID-5CM0, [24]) as an initial model. The refinement of the structure at 2.3 Å using the isotropic individual atom B-factors was carried out using the REFMAC5 program of the CCP4 suite [25]. The visual inspection of electron density maps and the manual rebuilding of the model were carried out using the COOT interactive graphics program [26]. NCS was used during the refinement, as well as TLS, which were introduced during the final refinement cycles.
In the final model, an asymmetric unit contained six copies of the protein (1794 visible residues), 611 water molecules, one PLP molecule, 5 phosphate moieties of partially modelled PLP, 6 phosphate molecules, 6 PEG molecules and 9 glycerol molecules from the cryo-solution. Fourteen N-terminal amino acids in chain A, 8 N-terminal amino acids in chain B, 17 N-terminal amino acids in chains C, D and E, 16 N-terminal amino acids in chain F as well as three C-terminal amino acids in chains A, B, C, D and F have no electron density, possibly due to their high mobility. In D and F subunits, residues 148–150 and 149–150, respectively, have no electron density, while in other chains, this region is clearly visible.
2.4. Structure Analysis and Validation
The search for VP5454 homologous sequences and multiple sequence alignments was made with BLAST service. The visual inspection of the structure was carried out using the COOT program and the PyMOL Molecular Graphics System, Version 1.9.0.0 Schrödinger, LLC. The structure comparison and superposition were made using the PDBeFold program [27], while contacts were analyzed using the PDBePISA [28]. Omit and polder maps were calculated with PHENIX [29].
2.5. Molecular Modeling
Molecular dynamics simulations were performed using NAMD3 software [30]. A crystal structure obtained in this study was utilized as a source of coordinates of non-hydrogen atoms. Protonation states of amino acid residues were as follows: positively charged arginines and lysines, negatively charged aspartates and glutamates, and neutral histidines except H70. The latter is located close to the negatively charged E55 and likely exists in the protonated state.
In VP5454, the position of the PLP phosphate group is determined by the P-pocket; however, due to the absence of a covalent bond between PLP and the protein, two possible orientations of the pyridine moiety may exist. The aldehyde group might be oriented toward N174 or in the opposite direction. Thus, we prepared two systems composed of the VP5454 dimer with the PLP in both binding sites that differ in PLP orientation. Additionally, we obtained in silico point mutation N174K and reconstructed the covalent bond between the PLP and the side chain of L174 as it should be in TAs.
All systems were solvated in rectangular water boxes such that the distances from the protein surface to the border of the cell exceeded 12 Å. Two chloride ions were added to neutralize the charge of the entire system. A protein macromolecule was described using CHARMM36 force field [31,32] parameters, and TIP3P force field parameters were utilized for water molecules [33]. Force field parameters for the PLP molecules were obtained in Ref [18] using the CGenFF force field [34]. The NPT ensemble was utilized with p = 1 atm and T = 300 K with the 1 fs integration time step; the first 50 ns of trajectories were not considered in analysis. Production runs were three 100 ns trajectories for each model system. Cutoff distances for all non-covalent interactions were 12 Å with the switch to the smoothing potential at 10 Å. Additionally, we performed MM-PBSA [35] calculations of the PLP binding energies at different orientations in the active site. For each system, energies were averaged over 50 frames equally distributed along MD trajectories. For MM-PBSA calculations, we utilized a café plugin to VMD [36].
3. Results
3.1. VP5454 Purification and Biochemical Analysis
A bioinformatic screen for TAs genes (unpublished data) was previously performed to identify a puzzling aminotransferase class IV (VP5454) sequence in a genome of Gram-negative bacteria Variovorax paradoxus (NCBI accession number: WP_021013128.1).
VP5454 was cloned and expressed in a soluble form in E. coli and was purified to homogeneity. The molecular weight of the purified VP5454 was approximately 170 kDa, as determined by gel filtration (Figure 1), indicating VP5454 as a homotetramer in solution. However, the purified VP5454 solution was colorless, indicating an absence of a bound cofactor. The absorption spectrum of the purified VP5454 had only one peak at 280 nm. No characteristic peak at 410–420 nm (Shiff base band) was observed after the addition of the equimolar amount of PLP to the protein solution, while the absorbance at 390 nm indicated a free form of PLP in the protein solution.
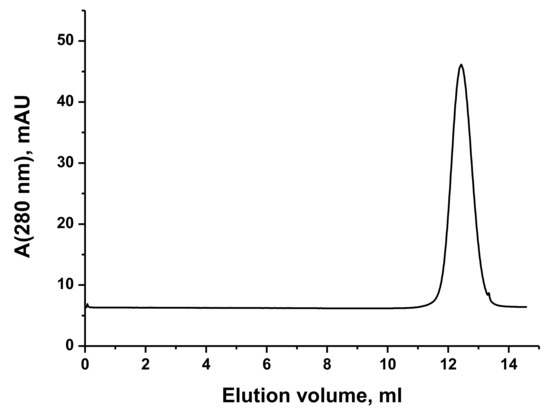
Figure 1.
VP5454 gel filtration elution profile. Column size and specifications—a calibrated Superdex 200 column (Cytiva, Marlborough, MA, USA) bed volume of 24 mL equilibrated in 50 mM pyrophosphate buffer pH 9.0 supplemented with 50 mM NaCl. The peak corresponds to a tetramer for the native enzyme. The column was calibrated using Gel Filtration calibration kit (Cytiva).
3.2. VP5454 Has Typical TAs Fold and Is Mostly Similar to BCAT TAs
To figure out whether VP5454 has a common TA fold, we determined its crystal structure using X-ray at 2.3 Å resolution. There were six almost identical protein subunits (RMSD between subunits do not exceed 0.14 Å) in the asymmetric unit of the crystal belonging to the P3221 space group, and contact analysis revealed that the protein is a hexamer in a crystal (Figure 2), which is similar to those found for some other TAs [12,13,24]. The hexamer can be represented as a trimer of dimers. Similar to the subunit organization of classical PLP fold type IV TAs, each VP5454 subunit consists of a small N-terminal domain with an α/β-structure (residues 1–141 and 310–314) and a large C-terminal domain (residues 154–309) connected by an interdomain loop (residues 142–153). The latter has relatively poor electron density and high B-factors in all the subunits, reflecting its flexibility also found in other TA structures [14,37]. A relative domain orientation in VP5454 is also supported by a C-terminal helix 290PLTQRLDALYRAYIA304, which interacts with the N-terminal domain. Despite a moderate amino acid identity, structure superposition demonstrated that the VP5454 structure had the most similarity to those of BCAT TAs (Table 2, Figure 2).
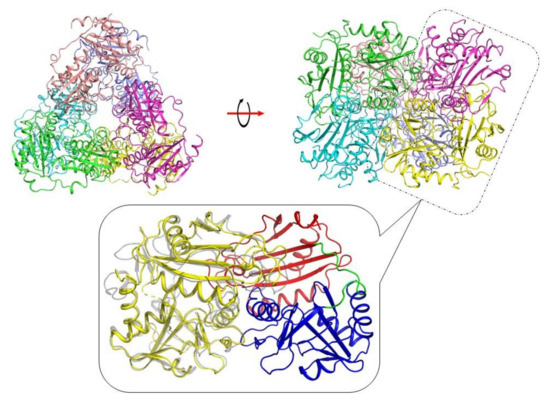
Figure 2.
X-ray structure of the VP5454. (top) Hexamer from the asymmetric unit is colored by chains and is depicted in two projections related by a 90° rotation (red arrow). (bottom) A zoomed view of the functional VP5454 dimer highlighted by the dotted box in hexameric structure. Color codes are: blue—C-terminal domain, red—N-terminal domain, green—interdomain loop, and yellow—adjacent subunit of the dimer. Superposed eBCAT (PDB ID-1IYE) subunit is colored in gray and is depicted as semitransparent for clarity.

Table 2.
Superposition of the VP5454 with the closest homologs. Superposition was made for one protein subunit.
A dimer of VP5454, found within the crystal hexamer, contains two identical active site cavities formed by the large and small domains of one subunit and the small domain of the second subunit of the dimer. VP5454 active site cavity has a bi-lobed structure, which represents canonical P- and O-pockets. In the TA of PLP fold type IV, the size of the active site and its entry are regulated by the length and amino acid composition of both the flexible interdomain loop (142HKVVTPEAKRNG153—hereinafter VP5454 numbering, unless otherwise indicated) and the O-pocket loop (118GVKKTPNQDPRFII131) as well as the length of βX-strand, βY-strand and two beta-turns. In the VP5454 structure, the interdomain loop is partially disordered and is oriented far from the active site, leaving its entrance open. The O-pocket of VP5454 is formed by residues W54, R112, R178, L181, region 208CNST211, as well as residues from the adjacent subunit C49*, T122*, P123*, N124* and Q125*. The P-pocket includes residues belonging to the β-turn 269GTLA272, as well as G56, R58, H70, R73, H110, G232, I233, T234 and R235. Amino acid residues F230 and N231 are involved in the formation of both pockets (Figure 3).

Figure 3.
Amino acid alignment between VP5454 and eBCAT. Canonical TA motifs are highlighted, and conserved amino acids are marked with asterisk. Digital labels are following: 1—characteristic motif 1, 2—βX-strand, 3—characteristic motif 2, 4—O-pocket loop, 5—βY-strand, 6—interdomain loop, 7—catalytic lysine residue (in eBCAT), 8—GAGE-region, 9—230FNGITR235 region, 10—β turn, 11—C-terminal helix.
Comparison of both VP5454 pockets to the ones in the model of the canonical BCAT from E.coli in the complex with L-glutamate bound to PLP (eBCAT, PDB ID-1IYE) revealed some common features (Figure 4a,b). First, the γ-carboxylic group in the O-pocket in eBCAT is coordinated by R97, Y31*, Y129 and the N atom of V109* (eBCAT numbering). In VP5454, the side chain of R112 and the N atom of Q125* form similar binding sites for a negatively charged group of substrates. There is a peak of electron density in this position in VP5454 (Figure 4a), which clearly corresponds to a phosphate molecule, which might be a phosphate group of a pyrophosphate ion from the protein buffer or phosphate originating from dissociation of the pyrophosphate [38]. At the same time, important for eBCAT substrate binding, Y31* is substituted for C49* in VP5454. A thiolate group of C49 is located far from the O-pocket and does not participate in phosphate ion coordination. Contrary to the interdomain loop of eBCAT, the interdomain loop of VP5454 is oriented out of the O-pocket, and residues from this loop do not participate in the phosphate group coordination. The same is for the O-pocket loop of VP5454, indicating that the O-pocket of VP5454 has more open conformation compared to that in the structure of eBCAT complexed with L-glutamate (PDB ID-1IYE). The peculiar feature of the active site of VP5454 is tryptophan W54 (F36 in eBCAT) at the bottom of the O-pocket. Together with R178, the side chain of W54 makes a shape of the bottom of the O-pocket different from that of eBCAT.
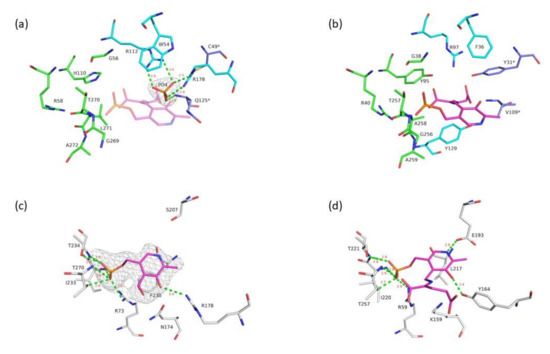
Figure 4.
Active site comparison between VP5454 and eBCAT. (a) VP5454 active site. For clarity, only the residues described in the text are shown. P-pocket residues are colored in green, O-pocket residues in cyan, and residues of the O-pocket related to the adjacent subunit of the dimer are in blue. Omit Fo-Fc electron density map (gray mesh) for the phosphate molecule is depicted at 4σ level. Hydrogen bonds fixing phosphate are shown with green lines, and corresponding distances are labeled in red. A PLP molecule with bound L-glutamate from superimposed eBCAT (PDB ID-1IYE) structure is shown in pink and is semitransparent for clarity. (b) eBCAT active site. Color codes and orientation is the same as in panel (a). Cofactor covalently bound to L-glutamate is shown in pink. (c) A PLP molecule bound in the VP5454 active site. Hydrogen bonds fixing PLP are shown with green lines, and corresponding distances are labeled in red. Polder map for PLP is shown as gray mesh at 4σ level. Only one conformation of the F230 side chain is shown for clarity. (d) PLP coordination in eBCAT. PLP molecule covalently bound to L-glutamate is shown in pink. Orientation and color scheme is similar to that in panel (c).
P-pocket in VP5454 also has some similarities to those of eBCAT, including composition of β-turn 269GTLA272 (256GTAA259 in eBCAT), as well as residues G56 (G38 in eBCAT) and R58 (R40 in eBCAT) (Figure 3). The latter in eBCAT forms one of two α-carboxylic group-binding sites through hydrogen bonding to O atoms of the main chain of β-turn 256GTAA259. The similar coordination site in VP5454 is formed by R58 and 269GTLA272 residues. The second coordination site of the α-carboxylic group in the P-pocket of eBCAT is formed by Y95 polarized by R97 (both residues are a part of the specificity-determining motives in canonical BCATs). The similar site is absent in VP5454 because H110, which occupies the position of Y95, does not interact with R112 in a similar mode. To sum up, the canonical specificity-determining sites of BCAT are partially disturbed in VP5454. At the same time, two keys for BCAT activity residues R112 (R97 in eBCAT) and R58 (R40 in eBCAT) are in their typical places.
3.3. Active Site of VP5454 Can Accommodate PLP Molecule
In TAs a cofactor, PLP molecule, forms a Schiff base linkage with the catalytic lysine situated in between the pockets and is stabilized by a number of hydrogen bonds, presumably via its phosphate moiety. In VP5454, such lysine residue is replaced by N174, clearly visible in electron density. Despite this substitution, there were two blobs of electron density found in the active site of VP5454, which is larger than that for solvent molecules. The first peak, located near to R112 and Q125, was attributed to a phosphate molecule (see above). Comparison of the VP5454 and the eBCAT structures demonstrated that the second peak corresponds to the phosphate group of the PLP molecule, bound in its typical TA of PLP fold type IV position. Taking into account that an excess of PLP was added to the protein prior to crystallization, a PLP molecule was placed in the corresponding electron density peak of VP5454. Being placed as such, a phosphate group of PLP forms polar interactions with conserved residues R73 (R59 in eBCAT), I233 (I220), T234 (T221) and T270 (T257) (Figure 4c,d). In the VP5454 structure, a side chain of F230 (L217 in eBCAT) residue has a double conformation. A pyridine ring of PLP (modeled in VP5454 with partial occupancy) is in place of one of F230 conformations. However, the second conformation allows for binding of the cofactor in the active site and could stabilize it via formation of stacking interaction (Figure 4c). The mobility of the side chain of F230 may be caused by the absence of a bound cofactor in some subunits due to its inability to form a covalent adduct without the active site lysine.
Compared to eBCAT, where the nitrogen atom of the PLP ring forms a hydrogen bond with the side group of glutamic acid E193, in VP5454, similar residue is replaced by S207, which, due to the shorter side chain, is not involved in PLP coordination in the VP5454 crystal structure. Finally, the O3 oxygen atom of PLP in VP5454 can be coordinated by the side chain of the R178 (OH-group of Y164 in eBCAT). A known feature of TAs is a proper binding of the PMP molecule in the active site, which does not require Schiff base linkage [12,39]. Thus, the fact that PLP can be bound in the VP5454 active site even without catalytic lysine seems to be a result of tight fixation of its phosphate group together with pyridoxal moiety orientation via F230.
3.4. Molecular Modeling Supports the Ability of VP5454 to Non-Covalently Bind PLP
To further study the details of PLP binding in VP5454, we performed classical MD simulations. We placed PLP in two different ways with respect to the pyridine ring of PLP. Both of them have the same typical eBCAT location of the phosphate group in the P-pocket (Figure 5). First, we discuss the PLP orientation that mimics the typical orientation in eBCAT (PLP-I conformation on Figure 5). In eBCAT, the positively charged NH fragment of the pyridine moiety forms a hydrogen bond with the negatively charged carboxylate group of the glutamate residue E193. In VP5454, the same position is occupied by serine S207, which is not involved in PLP coordination due to its short side chain. However, an OD atom of nearby N212 (N198 in eBCAT) forms a stable hydrogen bond with the PLP NH fragment during the entire MD trajectory (Figure 5). In eBCAT, the conservative Y164 residue is responsible for the stabilization of the negative charge on the O3 atom. In VP5454, O3 forms two stable hydrogen bonds with the NH groups of the R178 side chain. Thus, even in the lack of covalent linkage, the PLP molecule is fixed in the active site due to the stable interactions with the residues of the P-pocket as well as R178 and N212 residues. Another PLP conformation might exist (PLP-II conformation of Figure 5), which differs in the orientation of the pyridine moiety being rotated 180°. In this case, interactions between the phosphate group and P-pocket as well as between the O3 atom and R178 are strong, and therefore, these parts of PLP are firmly fixed. An alternative hydrogen bond partner was found for the NH group of PLP. In second conformation, the stable hydrogen bond was found in the MD simulations with the E55 residue (Figure 5). The MM-PBSA was utilized to calculate binding energies of the PLP in both orientations. For each state, binding energies were averaged over 50 frames. The values were found to be similar for both orientations: −22.5 ± 7.9 and −25.2 ± 7.1 kcal/mol for PLP-I and PLP-II, respectively. We also examined the SASA for both complexes and obtained 369 ± 5 and 370 ± 5 Å2 for PLP-I and PLP-II, respectively. Thus, we cannot discriminate which conformation is more preferable without Schiff base linkage; moreover, lack of the well-resolved electron density in this region in the crystal structure might be an indication of the presence of both of these conformations.
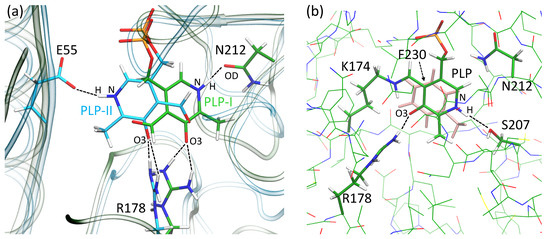
Figure 5.
(a) Alignment of the binding sites of VP5454 with two different orientations of PLP, PLP-I and PLP-II. The protein backbone is shown with transparent ribbons, green for VP5454 with PLP-I and cyan for VP5454 with PLP-II. (b) The VP5454 N174K variant with the Schiff base between the side chain of K174 and PLP; the side chain of F230 that forms π–stacking interactions with the pyridine ring of PLP is shown in pink. Color code: carbon—green (cyan for the model with PLP-II), oxygen—red, nitrogen—blue, phosphorus—orange, hydrogen—white. Hydrogen bonds are shown with black dashed lines.
We also obtained in silico N174K substitution in VP5454 and introduced a covalent bond between K174 and PLP, restoring the Schiff base. This results in the formation of a more rigid binding site with the stable π–stacking interactions between the pyridine moiety of the PLP and the side chain of F230. In the N174K variant, the dihedral angle CA–CB–CG–CD, which determines F230 benzene ring rotation, fluctuates mainly between 70 and 100 degrees. To compare, in wild-type VP5454 with the PLP-I orientation, the fluctuations of this angle are twice as large. In complex with the PLP-II orientation, F230 does not interact with the cofactor at all. Another difference between the VP5454 N174K and wild-type VP5454 with PLP-I orientation is the hydrogen bond partner of the NH group. In the former case, it changes from N212 to the S207 (Figure 5). This occurs because, upon formation of the Schiff base, the orientation of the pyridine moiety changes slightly; that is, the pyridine ring rotates clockwise around the axis orthogonal to the pyridine ring. Remarkably, the S207 position corresponds to the E193 in eBCAT, indicating that upon formation of the Schiff base in VP5454 N174K, a PLP is bound in the same way as a PLP in the active site of BCAT. The interactions between the PLP, the P-pocket residues and R178 remain the same in both the wild-type VP5454 and N174K variant.
4. Discussion
A conserved lysine residue in the active site of all known TAs plays a vital role via covalent binding of a cofactor, PLP, and allows for an aminotransferase reaction to proceed. In the genetic alphabet, a lysine residue is coded by two codons—AAA or AAG. However, mutation of the only last nucleotide to U or C converts a coding amino acid from lysine to asparagine and thus must destroy all enzymatic activity. A BLAST search for homologous VP5454 amino acid sequences, where the catalytic lysine is substituted for asparagine, surprisingly revealed a number of sequences annotated as TAs from different Gram-negative bacteria. Noteworthy, no structural data for such proteins are known to date.
The VP5454 structure revealed that substitution of the catalytic lysine does not alter the classical aminotransferase fold as well as the architecture of the protein’s active site. Based on structural comparison, we can argue that VP5454 originates from the BCAT family of TA, despite some notable differences in their active site (Figure 4). Among these differences, the following residues worth mentioning are C49, W54, H110, N125, R178, S207 and F230. F230 (L217 in eBCAT) and W54 (F36) limit the active site cavity. The bulky side chains of W54 reduce the cavity size compared to those of eBCAT, while big F230, being located at the re-side of the PLP, limits the cofactor mobility in the absence of the catalytic lysine and can stabilize PLP via stacking interactions. From the other side, a small side chain of S207 (E193) does able to form a stabilizing hydrogen bond to the cofactor nitrogen atom as is revealed by molecular dynamic simulations. Finally, O3 oxygen of PLP is hydrogen bonded to Y164 in eBCAT, while in VP5454, this oxygen is coordinated by the side chain of R178. The latter, together with N125 (V109), could also stabilize a substrate γ-COOH group. In contrast to eBCAT, where the γ-COOH group is additionally coordinated by a conserved Y31*, in VP5454, a short side chain of corresponding C49 does not participate in substrate fixation, possibly allowing for binding of more extended substrates. Coordination of the α-COOH group of substrates is more conserved in VP5454 with the only notable difference, H100, corresponding to a conserved Y95 in eBCAT.
Single nucleotide mutations are frequently occurring process, which can cause amino acid substitution in a protein-coding gene (missense mutations). Such mutations, if they negatively alter protein properties, will not be inherited in the population. However, this evolutionary mechanism can in theory be used to deactivate the whole enzyme if its activity is no longer required. The advantage of this process is that only one amino acid substitution will turn off the enzyme activity. Taking into account that a single nucleotide mutation of the lysine coding triplet is required to completely deactivate TA and the fact that there are a number of sequences annotated as TA with lysine substituted for asparagine, we speculate that a corresponding substitution in VP5454 has a functional role rather than a full-genome sequencing fault. Another question arises—why did the VP5454 gene remain in the genome if it is not no longer required by the organism more. One of the possible answers could be that the VP5454 protein still has an obscured function. For example, one can suppose that for a PLP carrier/transfer role as it is known, an excess of PLP affects different cellular processes and can be toxic to the cell [40,41]. However, PLP was identified as one of the most damage-prone molecules [42]. However, to address the functional role of VP5454 and proteins with similar lysine substitutions in detail, a complicated gene knockout together with transcription level experiments is worth performing.
5. Conclusions
In conclusion, we successfully elucidate the 3D structure of an uncharacterized protein from the bacterium Variovorax paradoxus, VP5454, annotated as class IV aminotransferase and lacking a conservative catalytic lysine residue, required for covalent cofactor binding. We demonstrated that VP5454 possesses a canonical BCATs fold and a common architecture of the active site consisting of two pockets, which however differ from those of BCATs in some detail.
Despite the absence of the catalytic lysine, crystal structure analysis indicated a conformation of the PLP molecule bound at the VP5454 active site, resembling those in canonical BCATs. Cofactor binding is mostly determined by hydrogen bond interactions of its phosphate moiety conserved within TAs of PLP fold IV residues of the active site. This allowed for PLP binding even in the absence of the catalytic lysine, but it tolerates an uncertainty of binding mode of the cofactor’s pyridine ring. Molecular dynamic simulations also supported our findings.
Taking into account the fact that most of the genomes encode at least a few different TAs, we hypothesize that the catalytic lysine substitutions found in some of the annotated TAs sequences can be a result of a specific evolutionary mechanism for the deactivation of enzymes of this type, possibly to switch their function to another that is more necessary for the organism.
Author Contributions
Conceptualization, K.M.B., E.Y.B. and M.G.K.; Data curation, K.M.B.; Funding acquisition, K.M.B., M.G.K. and V.O.P.; Investigation, K.M.B., I.O.M., A.Y.N., T.V.R. and E.Y.B.; Methodology, K.M.B. and M.G.K.; Supervision, V.O.P.; Visualization, K.M.B. and M.G.K.; Writing—original draft, K.M.B., I.O.M., E.Y.B. and M.G.K.; Writing—review and editing, K.M.B., E.Y.B. and M.G.K. All authors reviewed the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded in part by the Ministry of Science and Higher Education of the Russian Federation in the framework of agreement no. 075-15-2021-1354 (07.10.2021) (biochemical and structural studies), the Russian Federal Space Agency, grant 8418.20.05 (crystallization in microgravity), and the grant of the President of the Russian Federation (MD-1390.2022.1.4) (molecular modeling).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The article contains the data. Structural data are deposited in the RCSB data bank with accession code 7Z79.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript or in the decision to publish the results.
References
- Gu, X.; Zhao, J.; Chen, L.; Li, Y.; Yu, B.; Tian, X.; Min, Z.; Xu, S.; Gu, H.; Sun, J.; et al. Application of Transition-Metal Catalysis, Biocatalysis, and Flow Chemistry as State-of-the-Art Technologies in the Synthesis of LCZ696. J. Org. Chem. 2020, 85, 6844–6853. [Google Scholar] [CrossRef]
- Savile, C.K.; Janey, J.M.; Mundorff, E.C.; Moore, J.C.; Tam, S.; Jarvis, W.R.; Colbeck, J.C.; Krebber, A.; Fleitz, F.J.; Brands, J.; et al. Biocatalytic Asymmetric Synthesis of Chiral Amines from Ketones Applied to Sitagliptin Manufacture. Science 2010, 329, 305–309. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.; Snajdrova, R.; Moore, J.C.; Baldenius, K.; Bornscheuer, U.T. Biocatalysis: Enzymatic Synthesis for Industrial Applications. Angew. Chem. Int. Ed. 2020, 60, 88–119. [Google Scholar] [CrossRef] [PubMed]
- Eliot, A.C.; Kirsch, J.F. Pyridoxal Phosphate Enzymes: Mechanistic, Structural, and Evolutionary Considerations. Annu. Rev. Biochem. 2004, 73, 383–415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toney, M.D. Controlling reaction specificity in pyridoxal phosphate enzymes. Biochim. et Biophys. Acta (BBA) Proteins Proteom. 2011, 1814, 1407–1418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Höhne, M.; Schätzle, S.; Jochens, H.; Robins, K.; Bornscheuer, U.T. Rational assignment of key motifs for function guides in silico enzyme identification. Nat. Chem. Biol. 2010, 6, 807–813. [Google Scholar] [CrossRef] [PubMed]
- Grishin, N.V.; Phillips, M.A.; Goldsmith, E.J. Modeling of the spatial structure of eukaryotic ornithine decarboxylases. Protein Sci. 1995, 4, 1291–1304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bezsudnova, E.Y.; Popov, V.; Boyko, K.M. Structural insight into the substrate specificity of PLP fold type IV transaminases. Appl. Microbiol. Biotechnol. 2020, 104, 2343–2357. [Google Scholar] [CrossRef]
- Steffen-Munsberg, F.; Vickers, C.; Kohls, H.; Land, H.; Mallin, H.; Nobili, A.; Skalden, L.; Bergh, T.V.D.; Joosten, H.-J.; Berglund, P.; et al. Bioinformatic analysis of a PLP-dependent enzyme superfamily suitable for biocatalytic applications. Biotechnol. Adv. 2015, 33, 566–604. [Google Scholar] [CrossRef]
- Iwasaki, A.; Matsumoto, K.; Hasegawa, J.; Yasohara, Y. A novel transaminase, (R)-amine:pyruvate aminotransferase, from Arthrobacter sp. KNK168 (FERM BP-5228): Purification, characterization, and gene cloning. Appl. Microbiol. Biotechnol. 2011, 93, 1563–1573. [Google Scholar] [CrossRef]
- Stekhanova, T.N.; Rakitin, A.L.; Mardanov, A.V.; Bezsudnova, E.Y.; Popov, V.O. A Novel highly thermostable branched-chain amino acid aminotransferase from the crenarchaeon Vulcanisaeta moutnovskia. Enzym. Microb. Technol. 2017, 96, 127–134. [Google Scholar] [CrossRef]
- Bezsudnova, E.Y.; Boyko, K.M.; Nikolaeva, A.Y.; Zeifman, Y.S.; Rakitina, T.V.; Suplatov, D.A.; Popov, V.O. Biochemical and structural insights into PLP fold type IV transaminase from Thermobaculum terrenum. Biochimie 2018, 158, 130–138. [Google Scholar] [CrossRef]
- Boyko, K.M.; Stekhanova, T.N.; Nikolaeva, A.Y.; Mardanov, A.V.; Rakitin, A.L.; Ravin, N.V.; Bezsudnova, E.Y.; Popov, V.O. First structure of archaeal branched-chain amino acid aminotransferase from Thermoproteus uzoniensis specific for l-amino acids and R-amines. Extremophiles 2016, 20, 215–225. [Google Scholar] [CrossRef]
- Okada, K.; Hirotsu, K.; Hayashi, H.; Kagamiyama, H. Structures of Escherichia coli Branched-Chain Amino Acid Aminotransferase and Its Complexes with 4-Methylvalerate and 2-Methylleucine: Induced Fit and Substrate Recognition of the Enzyme. Biochemistry 2001, 40, 7453–7463. [Google Scholar] [CrossRef]
- Sugio, S.; Petsko, G.A.; Manning, J.M.; Soda, K.; Ringe, D. Crystal Structure of a D-Amino Acid Aminotransferase: How the Protein Controls Stereoselectivity. Biochemistry 1995, 34, 9661–9669. [Google Scholar] [CrossRef]
- Thomsen, M.; Skalden, L.; Palm, G.J.; Höhne, M.; Bornscheuer, U.T.; Hinrichs, W. Crystallographic characterization of the (R)-selective amine transaminase fromAspergillus fumigatus. Acta Crystallogr. Sect. D Biol. Crystallogr. 2014, 70, 1086–1093. [Google Scholar] [CrossRef]
- Wybenga, G.G.; Crismaru, C.G.; Janssen, D.B.; Dijkstra, B.W. Structural Determinants of the β-Selectivity of a Bacterial Aminotransferase. J. Biol. Chem. 2012, 287, 28495–28502. [Google Scholar] [CrossRef] [Green Version]
- Bakunova, A.K.; Nikolaeva, A.Y.; Rakitina, T.V.; Isaikina, T.Y.; Khrenova, M.G.; Boyko, K.M.; Popov, V.O.; Bezsudnova, E.Y. The Uncommon Active Site of D-Amino Acid Transaminase from Haliscomenobacter hydrossis: Biochemical and Structural Insights into the New Enzyme. Molecules 2021, 26, 5053. [Google Scholar] [CrossRef]
- Boyko, K.; Gorbacheva, M.; Rakitina, T.; Korzhenevskiy, D.; Vanyushkina, A.; Kamashev, D.; Lipkin, A.; Popov, V. Expression, purification, crystallization and preliminary X-ray crystallographic analysis of the histone-like HU protein fromSpiroplasma melliferumKC3. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2015, 71, 24–27. [Google Scholar] [CrossRef] [Green Version]
- Pace, C.N.; Vajdos, F.; Fee, L.; Grimsley, G.; Gray, T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995, 4, 2411–2423. [Google Scholar] [CrossRef] [Green Version]
- Winter, G.; Waterman, D.G.; Parkhurst, J.M.; Brewster, A.S.; Gildea, R.J.; Gerstel, M.; Fuentes-Montero, L.; Vollmar, M.; Michels-Clark, T.; Young, I.D.; et al. DIALS: Implementation and evaluation of a new integration package. Acta Crystallogr. Sect. D Struct. Biol. 2018, 74, 85–97. [Google Scholar] [CrossRef] [Green Version]
- Evans, P. Scaling and assessment of data quality. Acta Crystallogr. Sect. D Biol. Crystallogr. 2005, 62, 72–82. [Google Scholar] [CrossRef]
- Vagin, A.A.; Teplyakov, A. MOLREP: An Automated Program for Molecular Replacement. J. Appl. Crystallogr. 1997, 30, 1022–1025. [Google Scholar] [CrossRef]
- Isupov, M.N.; Boyko, K.M.; Sutter, J.-M.; James, P.; Sayer, C.; Schmidt, M.; Schönheit, P.; Nikolaeva, A.Y.; Stekhanova, T.N.; Mardanov, A.V.; et al. Thermostable Branched-Chain Amino Acid Transaminases From the Archaea Geoglobus acetivorans and Archaeoglobus fulgidus: Biochemical and Structural Characterization. Front. Bioeng. Biotechnol. 2019, 7, 7. [Google Scholar] [CrossRef] [PubMed]
- Collaborative Computational Project, Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 1994, 50, 760–763. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D Struct. Biol. Crystallogr. 2004, D60, 2126–2132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krissinel, E.; Henrick, K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, 60, 2256–2268. [Google Scholar] [CrossRef] [PubMed]
- Krissinel, E.; Henrick, K. Inference of Macromolecular Assemblies from Crystalline State. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef]
- Liebschner, D.; Afonine, P.V.; Baker, M.L.; Bunkóczi, G.; Chen, V.B.; Croll, T.I.; Hintze, B.; Hung, L.-W.; Jain, S.; McCoy, A.J.; et al. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. Sect. D Struct. Biol. 2019, 75, 861–877. [Google Scholar] [CrossRef] [Green Version]
- Phillips, J.C.; Hardy, D.J.; Maia, J.D.C.; Stone, J.E.; Ribeiro, J.V.; Bernardi, R.C.; Buch, R.; Fiorin, G.; Hénin, J.; Jiang, W.; et al. Scalable molecular dynamics on CPU and GPU architectures with NAMD. J. Chem. Phys. 2020, 153, 044130. [Google Scholar] [CrossRef]
- Best, R.B.; Zhu, X.; Shim, J.; Lopes, P.E.M.; Mittal, J.; Feig, M.; MacKerell, J.A.D. Optimization of the Additive CHARMM All-Atom Protein Force Field Targeting Improved Sampling of the Backbone ϕ, ψ and Side-Chain χ1 and χ2 Dihedral Angles. J. Chem. Theory Comput. 2012, 8, 3257–3273. [Google Scholar] [CrossRef] [Green Version]
- Denning, E.J.; Priyakumar, U.D.; Nilsson, L.; Mackerell, A.D.M. Impact of 2′-hydroxyl sampling on the conformational properties of RNA: Update of the CHARMM all-atom additive force field for RNA. J. Comput. Chem. 2011, 32, 1929–1943. [Google Scholar] [CrossRef] [Green Version]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Vanommeslaeghe, K.; Hatcher, E.; Acharya, C.; Kundu, S.; Zhong, S.; Shim, J.; Darian, E.; Guvench, O.; Lopes, P.; Vorobyov, I.; et al. CHARMM General Force Field: A Force Field for Drug-Like Molecules Compatible with the CHARMM All-Atom Additive Biological Force Fields. J. Comput. Chem. 2010, 31, 671–690. [Google Scholar] [CrossRef] [Green Version]
- Wang, E.; Sun, H.; Wang, J.; Wang, Z.; Liu, H.; Zhang, J.Z.H.; Hou, T. End-Point Binding Free Energy Calculation with MM/PBSA and MM/GBSA: Strategies and Applications in Drug Design. Chem. Rev. 2019, 119, 9478–9508. [Google Scholar] [CrossRef]
- Liu, H.; Hou, T. CaFE: A tool for binding affinity prediction using end-point free energy methods. Bioinformatics 2016, 32, 2216–2218. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.-D.; Lin, C.-H.; Chuankhayan, P.; Huang, Y.-C.; Hsieh, Y.-C.; Huang, T.-F.; Guan, H.-H.; Liu, M.-Y.; Chang, W.-C.; Chen, C.-J. Crystal Structures of Complexes of the Branched-Chain Aminotransferase from Deinococcus radiodurans with α-Ketoisocaproate and l- Glutamate Suggest the Radiation Resistance of This Enzyme for Catalysis. J. Bacteriol. 2012, 194, 6206–6216. [Google Scholar] [CrossRef] [Green Version]
- Mistry, D.; Powles, N. The relative hydrolytic reactivities of pyrophosphites and pyrophosphates. Org. Biomol. Chem. 2013, 11, 5727–5733. [Google Scholar] [CrossRef] [Green Version]
- Goto, M.; Miyahara, I.; Hayashi, H.; Kagamiyama, H.; Hirotsu, K. Crystal Structures of Branched-Chain Amino Acid Aminotransferase Complexed with Glutamate and Glutarate: True Reaction Intermediate and Double Substrate Recognition of the Enzyme. Biochemistry 2003, 42, 3725–3733. [Google Scholar] [CrossRef]
- Mizushina, Y.; Xu, X.; Matsubara, K.; Murakami, C.; Kuriyama, I.; Oshige, M.; Takemura, M.; Kato, N.; Yoshida, H.; Sakaguchi, K. Pyridoxal 5′-phosphate is a selective inhibitor in vivo of DNA polymerase α and ε. Biochem. Biophys. Res. Commun. 2003, 312, 1025–1032. [Google Scholar] [CrossRef]
- Vega, D.E.; Margolin, W. Suppression of a Thermosensitive zipA Cell Division Mutant by Altering Amino Acid Metabolism. J. Bacteriol. 2018, 200, e00535-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lerma-Ortiz, C.; Jeffryes, J.G.; Cooper, A.J.; Niehaus, T.D.; Thamm, A.M.; Frelin, O.; Aunins, T.; Fiehn, O.; de Crécy-Lagard, V.; Henry, C.S.; et al. ‘Nothing of chemistry disappears in biology’: The Top 30 damage-prone endogenous metabolites. Biochem. Soc. Trans. 2016, 44, 961–971. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).