Effect of Hydrated Deep Eutectic Solvents on the Thermal Stability of DNA
Abstract
1. Introduction
2. Materials and Methods
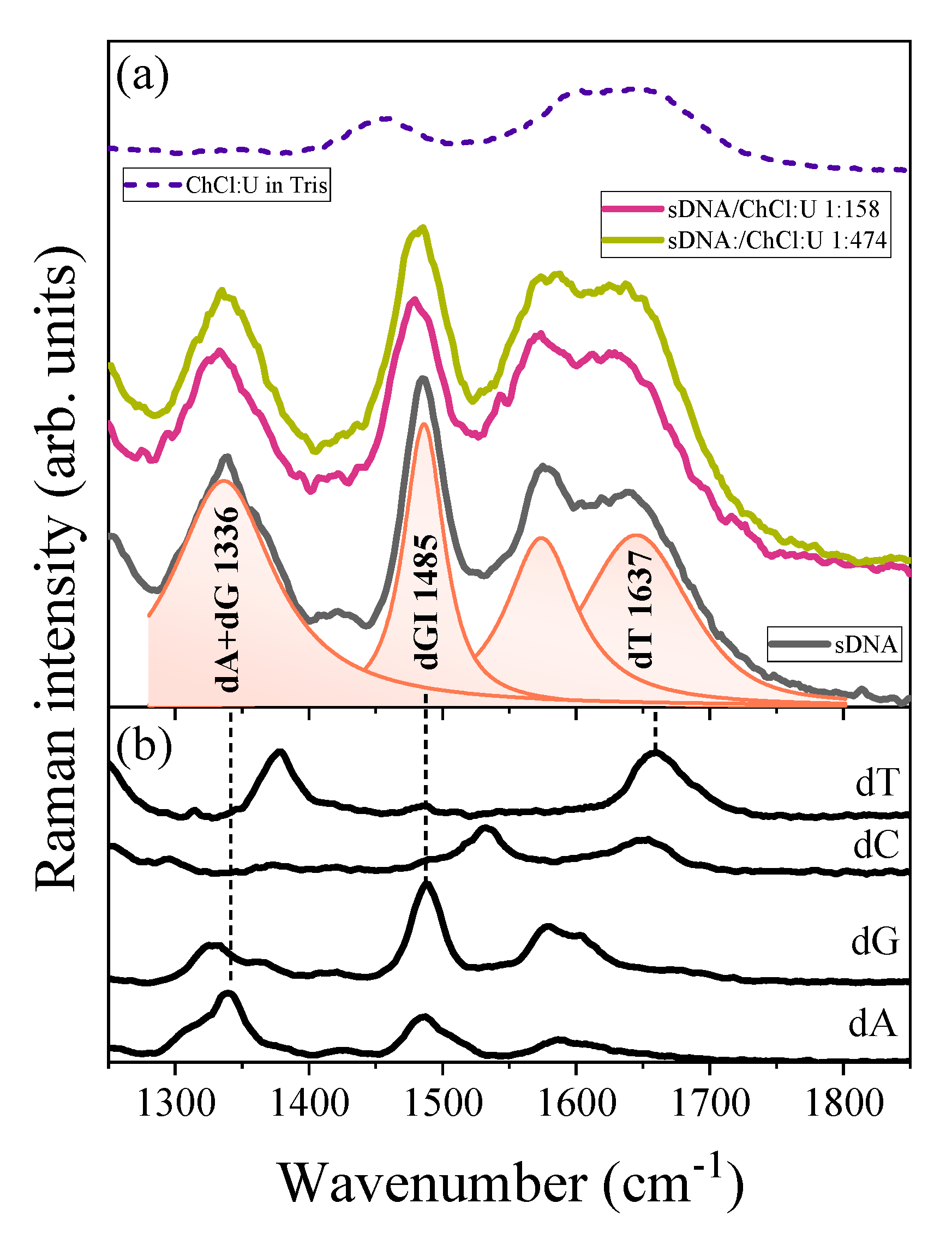
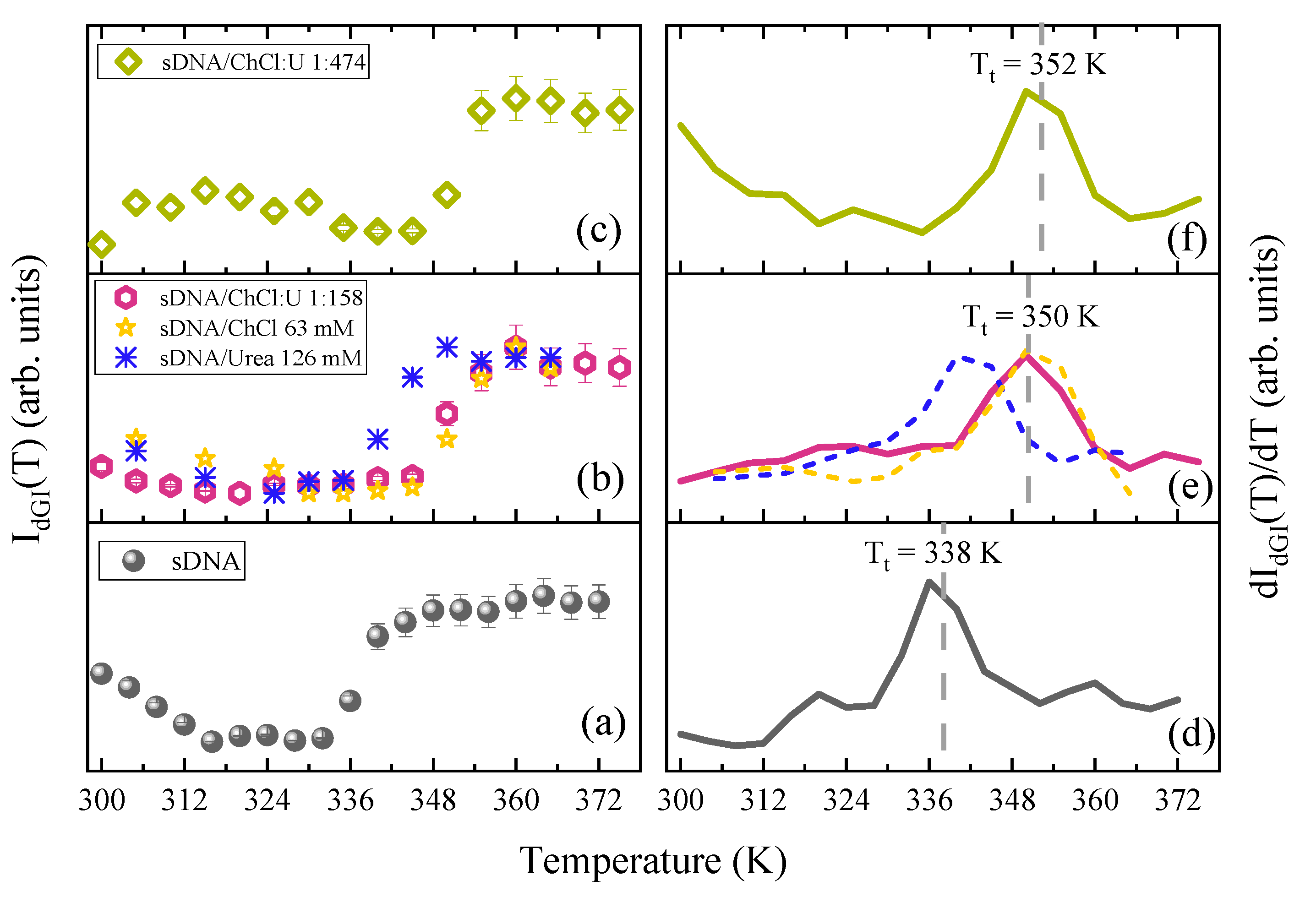
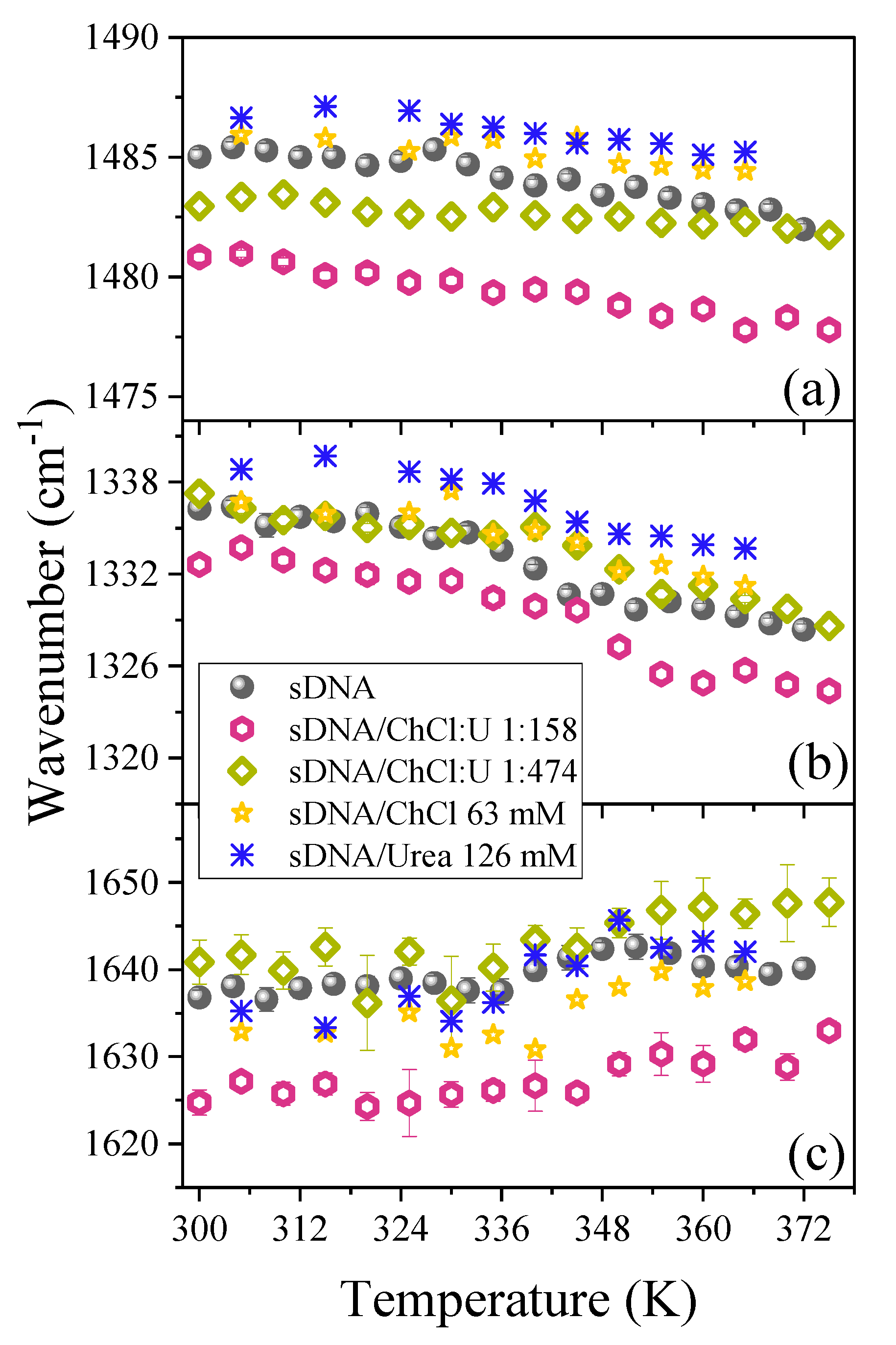
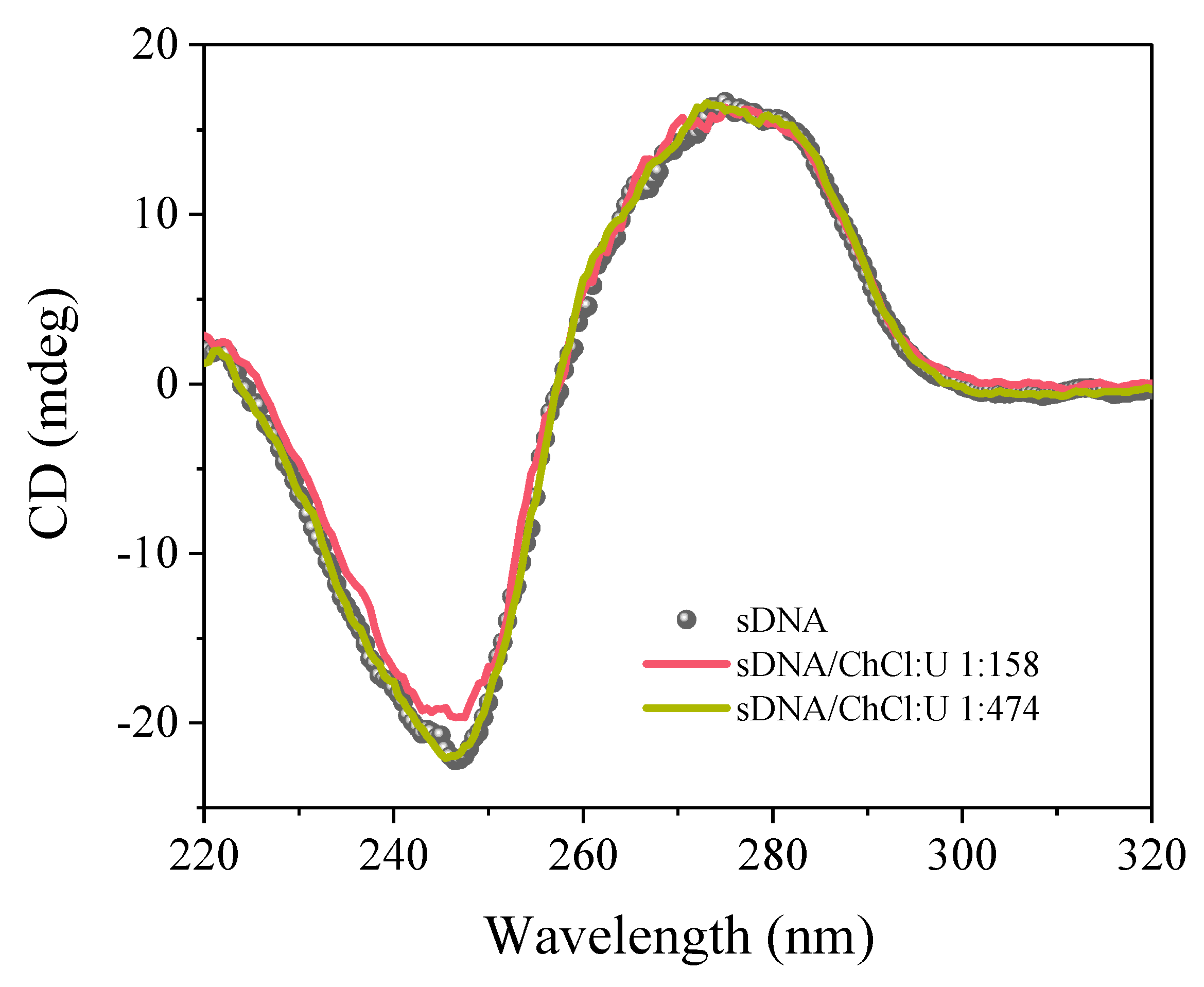
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Martins, M.A.R.; Pinho, S.P.; Coutinho, J.A.P. Insights into the Nature of Eutectic and Deep Eutectic Mixtures. J. Solut. Chem. 2019, 48, 962–982. [Google Scholar] [CrossRef]
- Zhang, Q.; De Oliveira Vigier, K.; Royer, S.; Jérôme, F. Deep Eutectic Solvents: Syntheses, Properties and Applications. Chem. Soc. Rev. 2012, 41, 7108–7146. [Google Scholar] [CrossRef]
- Smith, E.L.; Abbott, A.P.; Ryder, K.S. Deep Eutectic Solvents (DESs) and Their Applications. Chem. Rev. 2014, 114, 11060–11082. [Google Scholar] [CrossRef]
- Kist, J.A.; Zhao, H.; Mitchell-Koch, K.R.; Baker, G.A. The study and application of biomolecules in deep eutectic solvents. J. Mater. Chem. B 2021, 9, 536–566. [Google Scholar] [CrossRef]
- Vilková, M.; Płotka-Wasylka, J.; Andruch, V. The role of water in deep eutectic solvent-base extraction. J. Mol. Liq. 2020, 304, 112747. [Google Scholar] [CrossRef]
- Oliveira, G.; Wojeicchowski, J.P.; Oliveira Farias, F.; Igarashi-Mafra, L.; de Pelegrini Soares, R.; Mafra, M.R. Enhancement of biomolecules solubility in aqueous media using designer solvents as additives: An experimental and COSMO-based models’ approach. J. Mol. Liq. 2020, 318, 114266. [Google Scholar] [CrossRef]
- Cao, C.; Nian, B.; Li, Y.; Wu, S.; Liu, Y. Multiple Hydrogen-Bonding Interactions Enhance the Solubility of Starch in Natural Deep Eutectic Solvents: Molecule and Macroscopic Scale Insights. J. Agric. Food Chem. 2019, 67, 12366–12373. [Google Scholar] [CrossRef]
- Pogozelski, W.K.; Tullius, T.D. Oxidative strand scission of nucleic acids: Routes initiated by hydrogen abstraction from the sugar moiety. Chem. Rev. 1998, 98, 1089–1108. [Google Scholar] [CrossRef]
- Cadet, J.; Delatour, T.; Douki, T.; Gasparutto, D.; Pouget, J.P.; Ravanat, J.L.; Sauvaigo, S. Hydroxyl radicals and DNA base damage. Mutat. Res. Fundam. Mol. Mech. Mutagen. 1999, 424, 9–21. [Google Scholar] [CrossRef]
- Pal, S.; Paul, S. Understanding the Role of Reline, a Natural DES, on Temperature-Induced Conformational Changes of C-Kit G-Quadruplex DNA: A Molecular Dynamics Study. J. Phys. Chem. B 2020, 124, 3123–3136. [Google Scholar] [CrossRef]
- Nordstrom, L.J.; Clark, C.A.; Andersen, B.; Champlin, S.M.; Schwinefus, J.J. Effect of Ethylene Glycol, Urea, and N-Methylated Glycines on DNA Thermal Stability: The Role of DNA Base Pair Composition and Hydration. Biochemistry 2006, 45, 9604–9614. [Google Scholar] [CrossRef] [PubMed]
- Mondal, D.; Mukesh, S.; Chandrakant, M.; Vishal, G.; Kamalesh, P. Improved solubility of DNA in recyclable and reusable bio-based deep eutectic solvents with long-term structural and chemical stability. Chem. Commun. 2013, 49, 9606–9608. [Google Scholar] [CrossRef] [PubMed]
- de La Harpe, K.; Kohl, F.R.; Zhang, Y.; Kohler, B. Excited-State Dynamics of a DNA Duplex in a Deep Eutectic Solvent Probed by Femtosecond Time-Resolved IR Spectroscopy. J. Phys. Chem. A 2018, 122, 2437–2444. [Google Scholar] [CrossRef] [PubMed]
- Mamajanov, I.; Engelhart, A.; Bean, H.; Hud, N. DNA and RNA in Anhydrous Media: Duplex, Triplex, and G-Quadruplex Secondary Structures in a Deep Eutectic Solvent. Angew. Chem. Int. Ed. 2010, 49, 6310–6314. [Google Scholar] [CrossRef] [PubMed]
- Nakano, M.; Tateishi-Karimata, H.; Tanaka, S.; Sugimoto, N. Choline Ion Interactions with DNA Atoms Explain Unique Stabilization of A–T Base Pairs in DNA Duplexes: A Microscopic View. J. Phys. Chem. B 2014, 118, 379–389. [Google Scholar] [CrossRef]
- Sequeira, R.A.; Bhatt, J.; Prasad, K. Recent Trends in Processing of Proteins and DNA in Alternative Solvents: A Sustainable Approach. Sustain. Chem. 2020, 1, 10. [Google Scholar] [CrossRef]
- Abbott, A.P.; Capper, G.; Davies, D.L.; Rasheed, R.K.; Tambyrajah, V. Novel Solvent Properties of Choline Chloride/Urea Mixtures. Chem. Commun. 2003, 1, 70–71. [Google Scholar] [CrossRef]
- Bottari, C.; Catalini, S.; Foggi, P.; Mancini, I.; Mele, A.; Perinelli, D.R.; Paciaroni, A.; Gessini, A.; Masciovecchio, C.; Rossi, B. Base-specific pre-melting and melting transitions of DNA in presence of ionic liquids probed by synchrotron-based UV resonance raman scattering. J. Mol. Liq. 2021, 330, 115433. [Google Scholar] [CrossRef]
- Bottari, C.; Mancini, I.; Mele, A.; Gessini, A.; Masciovecchio, C.; Rossi, B. Conformational stability of DNA in hydrated ionic liquid by synchrotron-based UV resonance raman. In UV and Higher Energy Photonics: From Materials to Applications 2019; International Society for Optics and Photonics: Bellingham, WA, USA, 2019. [Google Scholar]
- Rossi, B.; Bottari, C.; Catalini, S.; Gessini, A.; D’Amico, F.; Masciovecchio, C. Synchrotron based UV Resonant Raman scattering for material science. In Molecular and Laser Spectroscopy, 1st ed.; Gupta, V.P., Ozaki, Y., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; Chapter 13; Volume 2, pp. 447–478. [Google Scholar]
- Rossi, B.; Tortora, M.; Catalini, S.; Vigna, J.; Mancini, I.; Gessini, A.; Masciovecchio, C.; Mele, A. Insight into the thermal stability of DNA in hydrated ionic liquids from multi-wavelength UV resonance raman experiments. Phys. Chem. Chem. Phys. 2021, 23, 15980–15988. [Google Scholar] [CrossRef]
- Fodor, S.P.A.; Rava, R.P.; Hays, T.R.; Spiro, T.G. Ultraviolet resonance Raman spectroscopy of the nucleotides with 266-, 240-, 218-, and 200-nm pulsed laser excitation. J. Am. Chem. Soc. 1985, 107, 1520–1529. [Google Scholar] [CrossRef]
- Fodor, S.P.A.; Spiro, T.G. Ultraviolet resonance Raman spectroscopy of DNA with 200-266-nm laser excitation. J. Am. Chem. Soc. 1986, 108, 3198–3205. [Google Scholar] [CrossRef]
- Bianchi, F.; Comez, L.; Biehl, R.; D’Amico, F.; Gessini, A.; Longo, A.M.; Masciovecchio, C.; Petrillo, C.; Radulescu, A.; Rossi, B.; et al. Structure of human telomere G-quadruplex in the presence of a model drug along the thermal unfolding pathway. Nucleic Acids Res. 2018, 46, 11927–11938. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.Q.; Thomas, G.J., Jr. UV resonance raman spectroscopy of DNA and protein constituents of viruses: Assignments and cross sections for excitations at 257, 244, 238, and 229 nm. Biopolymers 1998, 45, 247–256. [Google Scholar] [CrossRef]
- Duguid, J.G.; Bloomfield, V.A.; Benevides, J.M.; Thomas, G.J. DNA melting investigated by differential scanning calorimetry and Raman spectroscopy. Biophys. J. 1996, 71, 3350–3360. [Google Scholar] [CrossRef]
- Duguid, J.G.; Bloomfield, V.A.; Benevides, J.M.; Thomas, G.J., Jr. Raman Spectroscopy of DNA-Metal Complexes. I. The Thermal Denaturation of DNA in the Presence of Sr2+, Ba2, Mg2+, Ca2+, Mn2+, C02+, Ni2+, and Cd2+. Biophys. J. 1995, 69, 2623–2641. [Google Scholar] [CrossRef]
- Toyama, A.; Takeuchi, H.; Harada, I. Ultraviolet resonance Raman spectra of adenine, uracil and thymine derivatives in several solvents. Correlation between band frequencies and hydrogen-bonding states of the nucleic acid bases. J. Mol. Struct. 1991, 242, 87–98. [Google Scholar] [CrossRef]
- Turpin, P.Y.; Chinsky, L.; Laigle, A.; Jollès, B. DNA structure studies by resonance Raman spectroscopy. J. Mol. Struct. 1989, 214, 43–70. [Google Scholar] [CrossRef]
- Erfurth, S.C.; Peticolas, W.I.S. Melting and premelting phenomenon in DNA by laser Raman scattering. Biopolymers 1975, 14, 247–264. [Google Scholar] [CrossRef]
- Tsuboi, M.; Komatsu, M.; Hoshi, J.; Kawashima, E.; Sekine, T.; Ishido, Y.; Russell, M.P.; Benevides, J.M.; Thomas, G.J. Raman and Infrared Spectra of (2′S)-[2′-2H] Thymidine: Vibrational Coupling between Deoxyribosyl and Thymine Moieties and Structural Implications. J. Am. Chem. Soc. 1997, 119, 2025–2032. [Google Scholar] [CrossRef]
- Jirasek, A.; Schulze, H.G.; Hughesman, C.; Creagh, A.L.; Haynes, C.A.; Blades, M.W.; Turner, R.F.B. Discrimination between UV radiation-induced and thermally induced spectral changes in AT-paired DNA oligomers using UV resonance raman spectroscopy. J. Raman Spectrosc. 2006, 37, 1368–1380. [Google Scholar] [CrossRef]
- Mukerji, I.; Williams, A.P. UV resonance raman and circular dichroism studies of a DNA duplex containing an A(3)T(3) tract: Evidence for a premelting transition and three-centered H-bonds. Biochemistry 2002, 41, 69–77. [Google Scholar] [CrossRef][Green Version]
- Toyama, A.; Hanada, N.; Ono, J.; Yoshimitsu, E.; Takeuchi, H. Assignments of guanosine UV resonance raman bands on the basis of 13C, 15N and 18O substitution effects. J. Raman Spectrosc. 1999, 30, 623–630. [Google Scholar] [CrossRef]
- Movileanu, L.; Benevides, J.M.; Thomas, G.J., Jr. Temperature Dependence of the Raman Spectrum of DNA. Part I—Raman Signatures of Premelting and Melting Transitions of Poly(dA–dT)·Poly(dA–dT). J. Raman Spectrosc. 1999, 30, 637–649. [Google Scholar] [CrossRef]
- Gállego, I.; Grover, M.A.; Hud, N.V. Folding and Imaging of DNA Nanostructures in Anhydrous and Hydrated Deep-Eutectic Solvents. Angew. Chem. Int. Ed. 2015, 54, 6765–6769. [Google Scholar] [CrossRef]
- Oprzeska-Zingrebe, E.A.; Smiatek, J. Preferential Binding of Urea to Single-Stranded DNA Structures: A Molecular Dynamics Study. Biophys. J. 2018, 114, 1551–1562. [Google Scholar] [CrossRef]
- Hong, J.; Capp, M.W.; Anderson, C.F.; Saecker, R.M.; Felitsky, D.J.; Anderson, M.W.; Record, M.T., Jr. Preferential interactions of glycine betaine and of urea with DNA: Implications for DNA hydration and for effects of these solutes on DNA stability. Biochemistry 2004, 43, 14744–14758. [Google Scholar] [CrossRef]
- Herskovits, T.T. Nonaqueous Solutions of DNA; Denaturation by Urea and Its Methyl Derivatives. Biochemistry 1963, 2, 335–340. [Google Scholar] [CrossRef] [PubMed]
- Tateishi-Karimata, H.; Sugimoto, N. Structure, stability and behaviour of nucleic acids in ionic liquids. Nucleic Acids Res. 2014, 42, 8831–8844. [Google Scholar] [CrossRef]
- Tateishi-Karimata, H.; Sugimoto, N. A–T Base Pairs are More Stable Than G–C Base Pairs in a Hydrated Ionic Liquid. Angew. Chem. Int. Ed. 2012, 51, 1416–1419. [Google Scholar] [CrossRef]
- Portella, G.; Germann, M.W.; Hud, N.V.; Orozco, M. MD and NMR Analyses of Choline and TMA Binding to Duplex DNA: On the Origins of Aberrant Sequence-Dependent Stability by Alkyl Cations in Aqueous and Water-Free Solvents. J. Am. Chem. Soc. 2014, 136, 3075–3086. [Google Scholar] [CrossRef]
- Shobhna, K.P.; Kaur, S.; Kashyap, H.K. Influence of Hydration on the Structure of Reline Deep Eutectic Solvent: A Molecular Dynamics Study. ACS Omega 2018, 3, 15246–15255. [Google Scholar]
- Di Pietro, M.E.; Hammond, O.; van den Bruinhorst, A.; Mannu, A.; Padua, A.; Mele, A.; Costa Gomes, M. Connecting chloride solvation with hydration in deep eutectic systems. Phys. Chem. Chem. Phys 2021, 21, 107–111. [Google Scholar] [CrossRef] [PubMed]
- Shah, D.; Mjalli, F.S. Effect of water on the thermo-physical properties of Reline: An experimental and molecular simulation based approach. Phys. Chem. Chem. Phys 2014, 16, 23900–23907. [Google Scholar] [CrossRef] [PubMed]
- Duguid, J.G.; Bloomfield, V.A.; Benevides, J.M.; Thomas, G.J. Raman Spectroscopy of DNA-Metal Complexes. 1. Interactions and Conformational Effects of the Divalent Cations: Mg, Ca, Sr, Ba, Mn, Co, Ni, Cu, Pd, and Cd. Biophys. J. 1993, 65, 1916–1928. [Google Scholar] [PubMed]
- Chandran, A.; Ghoshdastidar, D.; Senapati, S. Groove Binding Mechanism of Ionic Liquids: A Key Factor in Long-Term Stability of DNA in Hydrated Ionic Liquids. J. Am. Chem. Soc. 2012, 134, 20330–20339. [Google Scholar] [CrossRef]
- Jumbri, K.; Abdul Rahman, M.B.; Abdulmalek, E.; Ahmada, H.; Micaelo, N.M. An insight into structure and stability of DNA in ionic liquids from molecular dynamics simulation and experimental studies. Phys. Chem. Chem. Phys. 2014, 16, 14036–14046. [Google Scholar] [CrossRef]
- Zhao, H. DNA stability in ionic liquids and deep eutectic solvents. J. Chem. Technol. Biotechnol. 2015, 90, 19–25. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tortora, M.; Vigna, J.; Mancini, I.; Mele, A.; Gessini, A.; Masciovecchio, C.; Rossi, B. Effect of Hydrated Deep Eutectic Solvents on the Thermal Stability of DNA. Crystals 2021, 11, 1057. https://doi.org/10.3390/cryst11091057
Tortora M, Vigna J, Mancini I, Mele A, Gessini A, Masciovecchio C, Rossi B. Effect of Hydrated Deep Eutectic Solvents on the Thermal Stability of DNA. Crystals. 2021; 11(9):1057. https://doi.org/10.3390/cryst11091057
Chicago/Turabian StyleTortora, Mariagrazia, Jacopo Vigna, Ines Mancini, Andrea Mele, Alessandro Gessini, Claudio Masciovecchio, and Barbara Rossi. 2021. "Effect of Hydrated Deep Eutectic Solvents on the Thermal Stability of DNA" Crystals 11, no. 9: 1057. https://doi.org/10.3390/cryst11091057
APA StyleTortora, M., Vigna, J., Mancini, I., Mele, A., Gessini, A., Masciovecchio, C., & Rossi, B. (2021). Effect of Hydrated Deep Eutectic Solvents on the Thermal Stability of DNA. Crystals, 11(9), 1057. https://doi.org/10.3390/cryst11091057








