Synthesis and Characterization of New Guanine Complexes of Pt(IV) and Pd(II) by X-ray Diffraction and Hirshfeld Surface Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Synthesis Procedure of [PdCl2HGua2]Cl2·H2O (1)
2.3. Synthesis Procedure of PtCl5HGua·2H2O (2)
2.4. Single-Crystal XRD Analysis
3. Results and Discussions
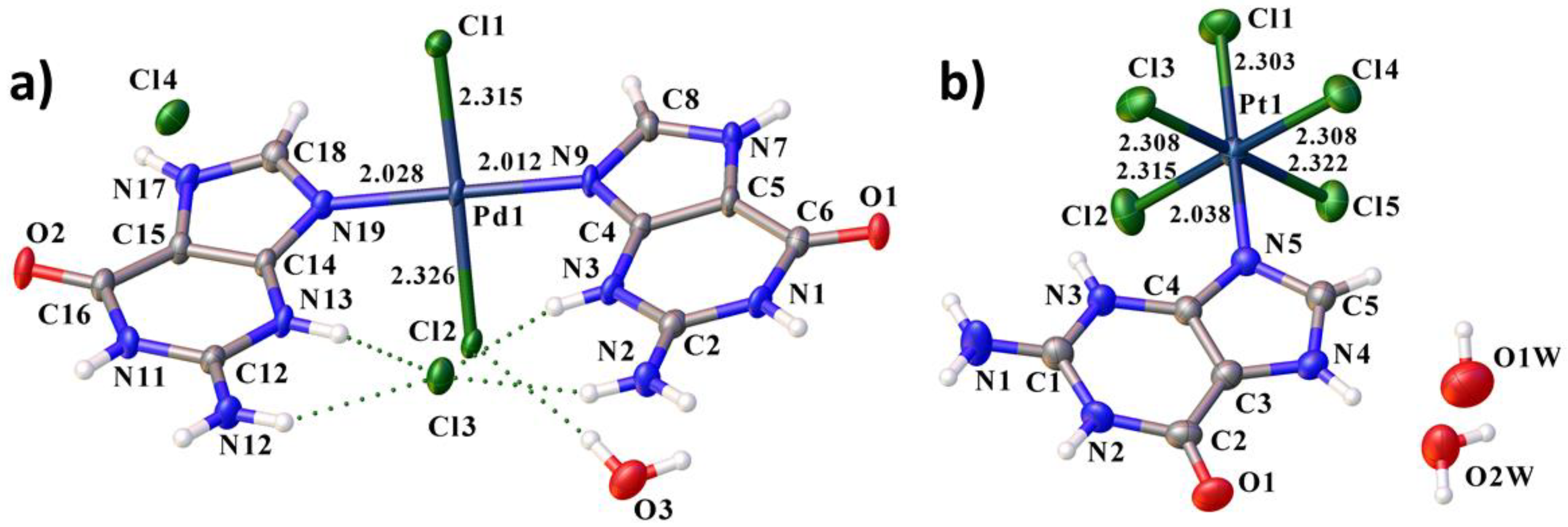
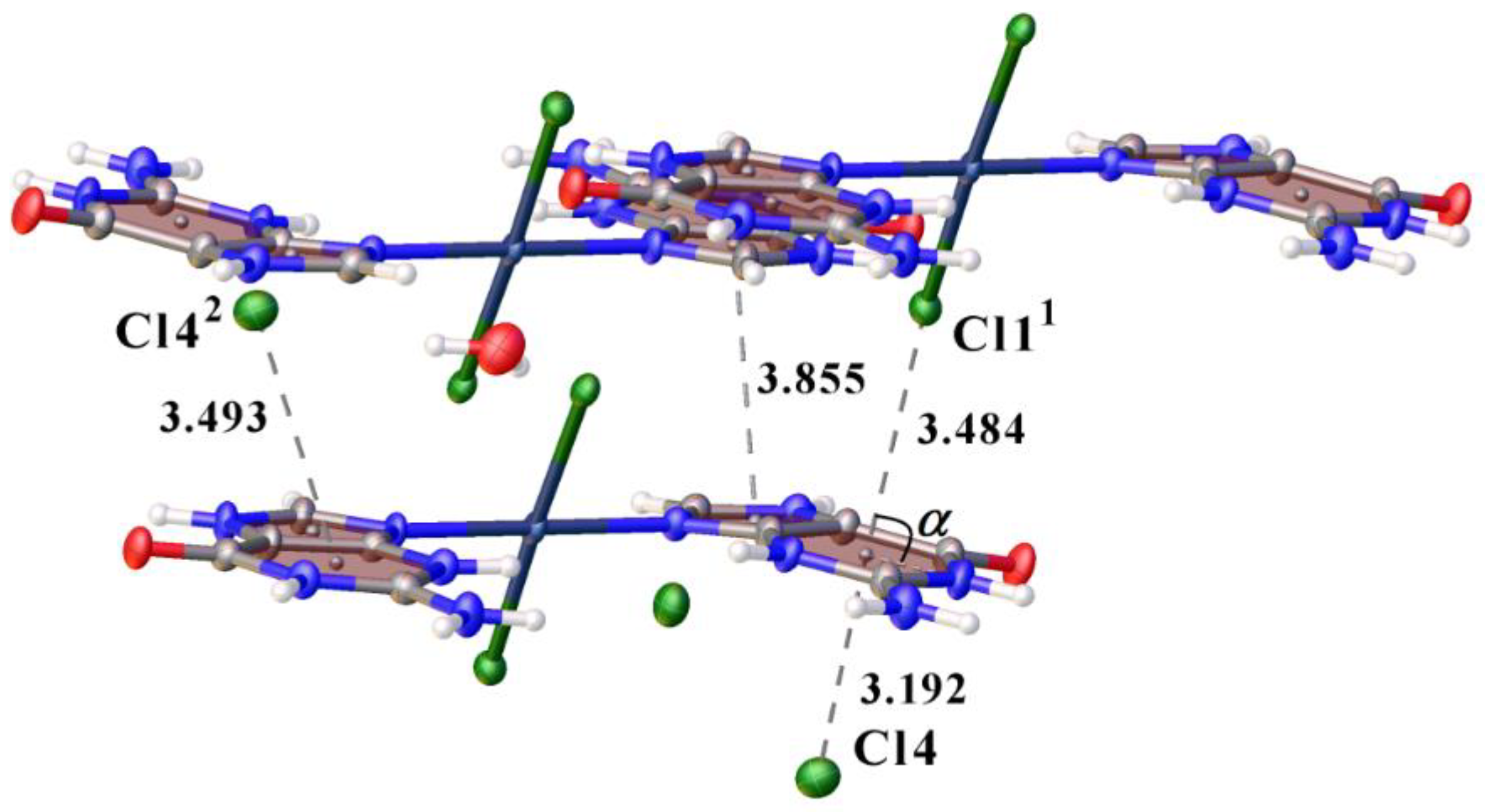
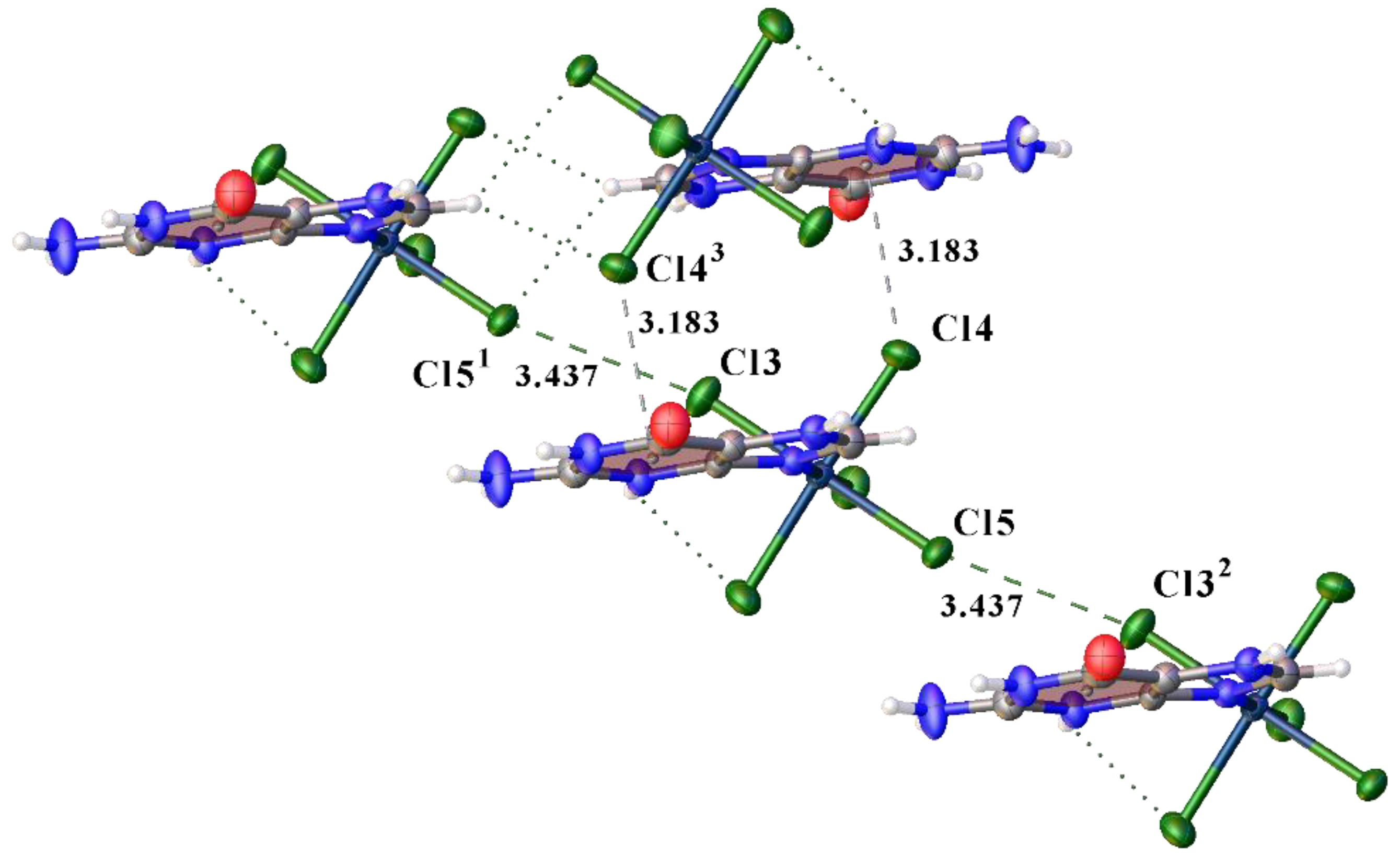
3.1. Structural Description
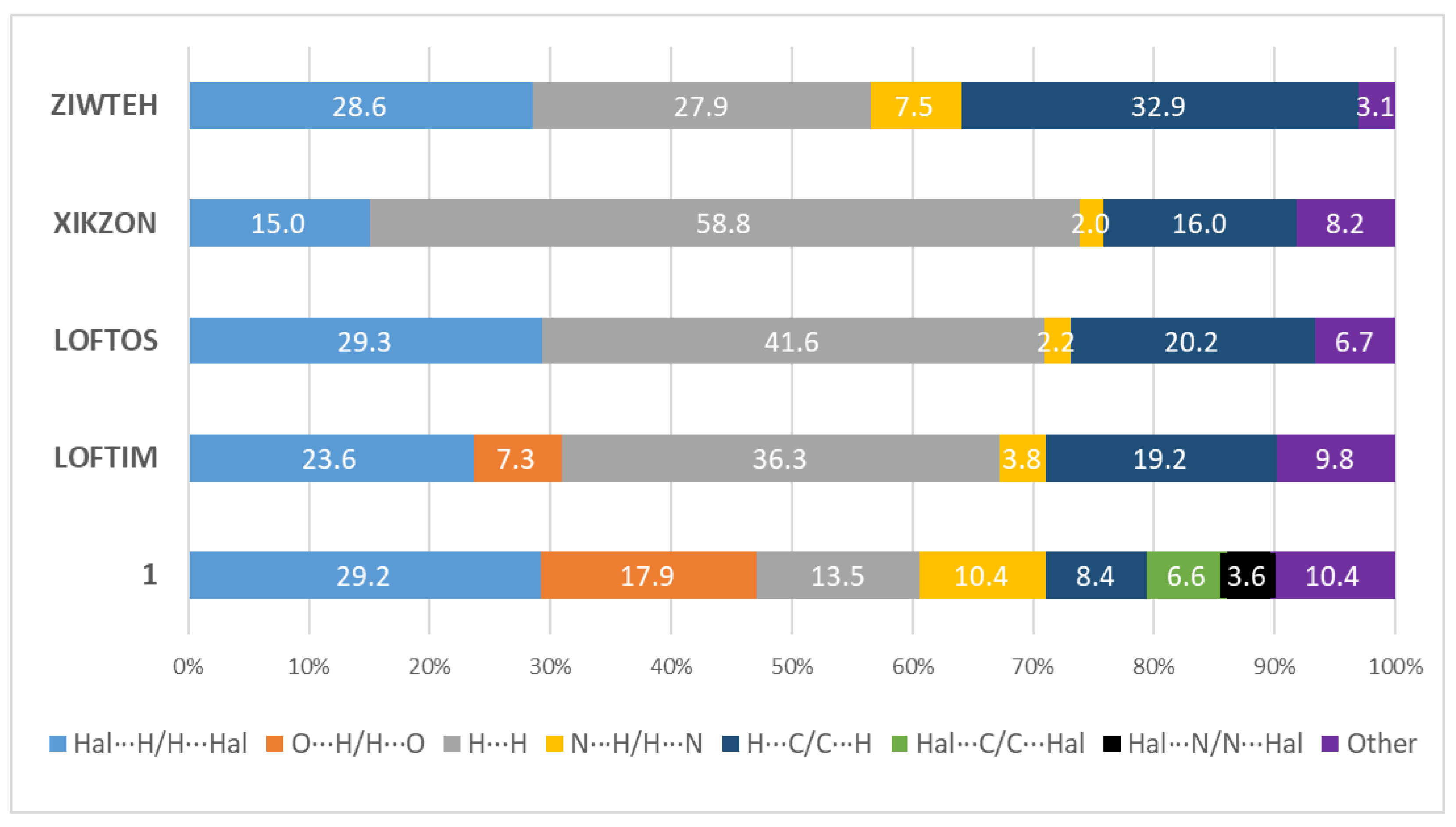
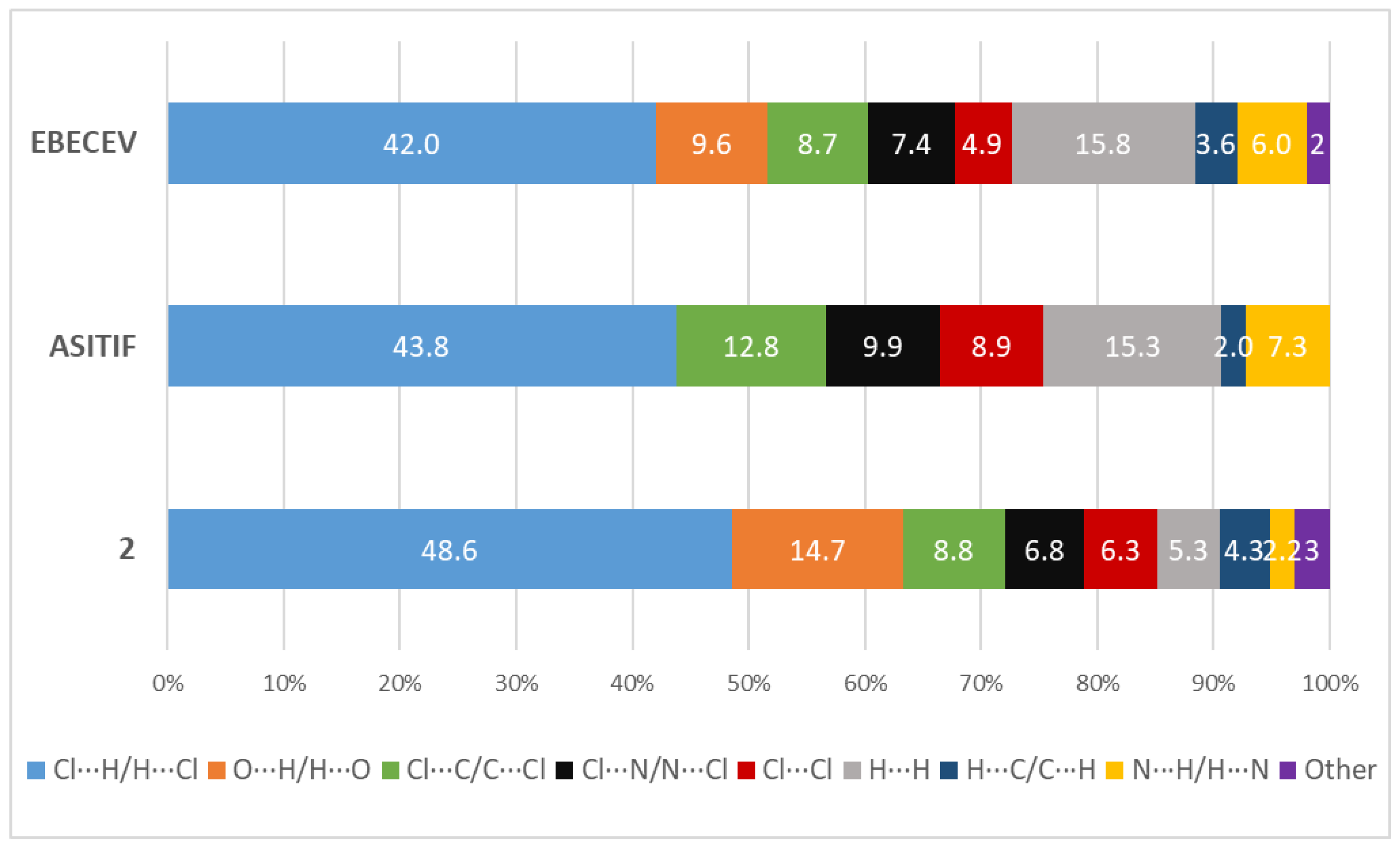
3.2. Hirshfeld Surface Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Richter, J.; Seidel, R.; Kirsch, R.; Mertig, M.; Pompe, W.; Plaschke, J.; Schackert, H.K. Nanoscale Palladium Metallization of DNA. Adv. Mater. 2000, 12, 507–510. [Google Scholar] [CrossRef]
- Van Nguyen, K.; Minteer, S.D. DNA-functionalized Pt nanoparticles as catalysts for chemically powered micromotors: Toward signal-on motion-based DNA biosensor. Chem. Commun. 2015, 51, 4782–4784. [Google Scholar] [CrossRef]
- Nguyen, K.; Monteverde, M.; Filoramo, A.; Goux-Capes, L.; Lyonnais, S.; Jegou, P.; Viel, P.; Goffman, M.; Bourgoin, J.-P. Synthesis of Thin and Highly Conductive DNA-Based Palladium Nanowires. Adv. Mater. 2008, 20, 1099–1104. [Google Scholar] [CrossRef]
- Ford, W.E.; Harnack, O.; Yasuda, A.; Wessels, J.M. Platinated DNA as Precursors to Templated Chains of Metal Nanoparticles. Adv. Mater. 2001, 13, 1793–1797. [Google Scholar] [CrossRef]
- Pekarik, V.; Peskova, M.; Duben, J.; Remes, M.; Heger, Z. Direct fluorogenic detection of palladium and platinum organometallic complexes with proteins and nucleic acids in polyacrylamide gels. Sci. Rep. 2020, 10, 12344. [Google Scholar] [CrossRef]
- Miller, M.A.; Askevold, B.; Mikula, H.; Kohler, R.H.; Pirovich, D.; Weissleder, R. Nano-palladium is a cellular catalyst for in vivo chemistry. Nat. Commun. 2017, 8, 15906. [Google Scholar] [CrossRef] [Green Version]
- Fong, C.W. Platinum anti-cancer drugs: Free radical mechanism of Pt-DNA adduct formation and anti-neoplastic effect. Free Radic. Biol. Med. 2016, 95, 216–229. [Google Scholar] [CrossRef]
- Clarke, M.J.; Zhu, F.; Frasca, D.R. Non-Platinum Chemotherapeutic Metallopharmaceuticals. Chem. Rev. 1999, 99, 2511–2533. [Google Scholar] [CrossRef]
- Hadizadeh, S.; Najafzadeh, N.; Mazani, M.; Amani, M.; Mansouri-Torshizi, H.; Niapour, A. Cytotoxic Effects of Newly Synthesized Palladium(II) Complexes of Diethyldithiocarbamate on Gastrointestinal Cancer Cell Lines. Biochem. Res. Int. 2014, 2014, 813457. [Google Scholar] [CrossRef]
- Hill, G.A.; Forde, G.; Gorb, L.; Leszczynski, J. cis-Diamminedichloropalladium and its interaction with guanine and guanine-cytosine base pair. Int. J. Quantum Chem. 2002, 90, 1121–1128. [Google Scholar] [CrossRef]
- Butour, J.L.; Wimmer, S.; Wimmer, F.; Castan, P. Palladium(II) compounds with potential antitumour properties and their platinum analogues: A comparative study of the reaction of some orotic acid derivatives with DNA in vitro. Chem. Biol. Interact. 1997, 104, 165–178. [Google Scholar] [CrossRef]
- Tikhomirov, A.G.; Ivanova, N.A.; Erofeeva, O.S.; Gorbacheva, L.B.; Efimenko, I.A. Interaction of Palladium(II) Acido Complexes with DNA. Russ. J. Coord. Chem. 2003 297 2003, 29, 489–493. [Google Scholar] [CrossRef]
- Tewari, B.B. Critical Reviews: Spectroscopic Studies On Palladium (Ii)-Complexes With Xanthine And Its Derivatives At Normal And High External Pressure Part-I: Spectroscopic Studies On Palladium (Ii)-Complexes With Xanthine And Its Derivatives At Normal Pressure. Boliv. J. Chem. 2013, 30, 122–130. [Google Scholar]
- Sigel, H. (Ed.) Metal Ions in Biological Systems Volume 42: Metal Complexes in Tumor Diagnosis and as Anticancer Agents, 1st ed.; CRC Press: Boca Raton, FL, USA, 2004; ISBN 9780824754945. [Google Scholar]
- Stone, P.J.; Kelman, A.D.; Sinex, F.M. Specific binding of antitumour drug cis-Pt(NH3)2Cl2 to DNA rich in guanine and cytosine. Nature 1974, 251, 736–737. [Google Scholar] [CrossRef]
- Cohen, G.L.; Ledner, J.A.; Bauer, W.R.; Ushay, H.M.; Caravana, C.; Lippard, S.J. Sequence dependent binding of cis-dichlorodiammineplatinum(II) to DNA. J. Am. Chem. Soc. 1980, 102, 2487–2488. [Google Scholar] [CrossRef]
- Tullius, T.D.; Lippard, S.J. cis-Diamminedichloroplatinum(II) binds in a unique manner to oligo(dG).oligo(dC) sequences in DNA—a new assay using exonuclease III. J. Am. Chem. Soc. 1981, 103, 4620–4622. [Google Scholar] [CrossRef]
- Galea, A.M.; Murray, V. The Anti-tumour Agent, Cisplatin, and its Clinically Ineffective Isomer, Transplatin, Produce Unique Gene Expression Profiles in Human Cells. Cancer Inform. 2008, 6, CIN.S802. [Google Scholar] [CrossRef]
- Kistenmacher, T.J.; Orbell, J.D.; Marzilli, L.G. Conformational Properties of Purine and Pyrimidine Complexes of cis-Platinum. In Platinum, Gold, and Other Metal Chemotherapeutic Agents; ACS Symposium Series; American Chemical Society: Washington, DC, USA, 1983; Volume 209, pp. 191–207. ISBN 9780841207585. [Google Scholar]
- Zeng, W.; Zhang, Y.; Zheng, W.; Luo, Q.; Han, J.; Liu, J.; Zhao, Y.; Jia, F.; Wu, K.; Wang, F. Discovery of Cisplatin Binding to Thymine and Cytosine on a Single-Stranded Oligodeoxynucleotide by High Resolution FT-ICR Mass Spectrometry. Molecules 2019, 24, 1852. [Google Scholar] [CrossRef] [Green Version]
- Baik, M.H.; Friesner, R.A.; Lippard, S.J. Theoretical Study of Cisplatin Binding to Purine Bases: Why Does Cisplatin Prefer Guanine over Adenine? J. Am. Chem. Soc. 2003, 125, 14082–14092. [Google Scholar] [CrossRef]
- Pezzano, H.; Podo, F. Structure of binary complexes of mono- and polynucleotides with metal ions of the first transition group. Chem. Rev. 1980, 80, 365–401. [Google Scholar] [CrossRef]
- Siddiqui, A.; Ceppi, P. A non-proliferative role of pyrimidine metabolism in cancer. Mol. Metab. 2020, 35. [Google Scholar] [CrossRef]
- Cochran, K.; Forde, G.; Hill, G.A.; Gorb, L.; Leszczynski, J. cis-Diamminodichloronickel and Its Interaction with Guanine and Guanine–Cytosine Base Pair. Struct. Chem. 2002, 13, 133–140. [Google Scholar] [CrossRef]
- Mikulski, C.M.; Mattucci, L.; Smith, Y.; Tran, T.B.; Karayannis, N.M. Guanine complexes with first row transition metal perchlorates. Inorg. Chim. Acta 1983, 80, 127–133. [Google Scholar] [CrossRef]
- Savchenkov, A.; Demina, L.; Safonov, A.; Grigoriev, M.; Solovov, R.; Abkhalimov, E. Syntheses and crystal structures of new aurate salts of adenine or guanine nucleobases. Acta Crystallogr. Sect. C Struct. Chem. 2020, 76, 139–147. [Google Scholar] [CrossRef] [Green Version]
- Efimenko, I.A.; Kurbakova, A.P.; Matović, Z.D.; Ponticelli, G. Synthesis and structure of palladium(II) mixed complexes with DNA purine or pyrimidine bases and imidazole derivatives. Part I. Transit. Met. Chem. 1994, 19, 539–541. [Google Scholar] [CrossRef]
- Trost, B.M.; Madsen, R.; Guile, S.D.; Brown, B. Palladium-Catalyzed Enantioselective Synthesis of Carbanucleosides. J. Am. Chem. Soc. 2000, 122, 5947–5956. [Google Scholar] [CrossRef]
- Ng, J.C.; Tan, C.Y.; Ong, B.H.; Matsuda, A.; Basirun, W.J.; Tan, W.K.; Singh, R.; Yap, B.K. Novel palladium-guanine-reduced graphene oxide nanocomposite as efficient electrocatalyst for methanol oxidation reaction. Mater. Res. Bull. 2019, 112, 213–220. [Google Scholar] [CrossRef]
- Uchida, K.; Toyama, A.; Tamura, Y.; Sugimura, M.; Mitsumori, F.; Furukawa, Y.; Takeuchi, H.; Harada, I. Interactions of guanine derivatives with ethylenediamine and diethylenetriamine complexes of palladium(II) in solution: Pd binding sites of the guanine ring and formation of a cyclic adduct, [{Pd(en)(guanine ring)}4]. Inorg. Chem. 1989, 28, 2067–2073. [Google Scholar] [CrossRef]
- Hadjiliadis, N. Pt(II) and Pd(II) interactions with nucleosides-binding sites-new compounds. Inorg. Chim. Acta 2016, 452, 279–284. [Google Scholar] [CrossRef]
- Gaballa, A.; Schmidt, H.; Hempel, G.; Reichert, D.; Wagner, C.; Rusanov, E.; Steinborn, D. Protonated nucleobase ligands: Synthesis, structure and characterization of 9-methyladeninium hexachloroplatinate and pentachloro(9-methyladeninium)platinum(IV). J. Inorg. Biochem. 2004, 98, 439–446. [Google Scholar] [CrossRef]
- Terzis, A.; Mentzafos, D. Trichloro(9-methylguaninium)platinum(II) hydrate and 9-methylguaninium hexachloroplatinate(IV) dihydrate: Synthesis and structure. Inorg. Chem. 1983, 22, 1140–1143. [Google Scholar] [CrossRef]
- Basallote, M.G.; Vilaplana, R. Palladium and platinum guanine complexes. Transit. Met. Chem. 1986, 11, 232–235. [Google Scholar] [CrossRef]
- Bruker AXS Inc. SAINT-Plus, Version 7.68; Bruker AXS Inc.: Madison, WI, USA, 2012. [Google Scholar]
- Sheldrick, G.M. SADABS; Bruker AXS: Madison, WI, USA, 2008. [Google Scholar]
- Sheldrick, G.M. SHELXT—Integrated space-group and crystal-structure determination. Acta Crystallogr. Sect. A Found. Adv. 2015, 71, 3–8. [Google Scholar] [CrossRef] [Green Version]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. Sect. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.K.; Puschmann, H. OLEX2: A complete structure solution, refinement and analysis program. J. Appl. Crystallogr. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Groom, C.R.; Bruno, I.J.; Lightfoot, M.P.; Ward, S.C. The Cambridge Structural Database. Acta Crystallogr. Sect. B Struct. Sci. Cryst. Eng. Mater. 2016, 72, 171–179. [Google Scholar] [CrossRef]
- De la Cruz, R.; Espinet, P.; Gallego, A.M.; Martín-Alvarez, J.M.; Martínez-Ilarduya, J.M. Structural and dynamic studies in solution of anionic dinuclear azolato-bridged palladium(II) complexes. J. Organomet. Chem. 2002, 663, 108–117. [Google Scholar] [CrossRef]
- Navarro-Ranninger, M.C.; Martínez-Carrera, S.; García-Blanco, S. Structure of trans-dichlorobis(1-methylimidazole)palladium(II), [Pd(C4H6N2)2Cl2]. Acta Crystallogr. Sect. C Cryst. Struct. Commun. 1983, 39, 186–188. [Google Scholar] [CrossRef]
- Liebing, P.; Edelmann, F.T. Trifluoromethylated 3-(Pyrazol-1-yl)propanamide (PPA) Ligands. Helv. Chim. Acta 2020, 103, e2000148. [Google Scholar] [CrossRef]
- Qin, Z.; Jennings, M.C.; Puddephatt, R.J. Self-Assembly of Polymer and Sheet Structures from Palladium(II) Complexes by Hydrogen Bonding between Carboxamide Substituents. Inorg. Chem. 2001, 40, 6220–6228. [Google Scholar] [CrossRef]
- Gupta, D.; Nowak, R.; Lippert, B. Pt(ii) complexes of unsubstituted guanine and 7-methylguanine. Dalt. Trans. 2010, 39, 73–84. [Google Scholar] [CrossRef]
- Spackman, P.R.; Turner, M.J.; McKinnon, J.J.; Wolff, S.K.; Grimwood, D.J.; Jayatilaka, D.; Spackman, M.A. CrystalExplorer: A program for Hirshfeld surface analysis, visualization and quantitative analysis of molecular crystals. J. Appl. Crystallogr. 2021, 54, 1006–1011. [Google Scholar] [CrossRef]
- Escobar, C.A.; Artigas, V.; Bacho, M.; Trujillo, A. π-halogen interaction on the crystalline packing of 1,3,5-tris(4-bromophenyl)-1,3,5-triazine-2,4,6-trione·[solvate]. J. Mol. Struct. 2022, 1247, 131307. [Google Scholar] [CrossRef]
- Lucas, X.; Bauzá, A.; Frontera, A.; Quiñonero, D. A thorough anion–π interaction study in biomolecules: On the importance of cooperativity effects. Chem. Sci. 2016, 7, 1038–1050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Youn, I.S.; Kim, D.Y.; Cho, W.J.; Madridejos, J.M.L.; Lee, H.M.; Kołaski, M.; Lee, J.; Baig, C.; Shin, S.K.; Filatov, M.; et al. Halogen−π Interactions between Benzene and X 2 /CX 4 (X = Cl, Br): Assessment of Various Density Functionals with Respect to CCSD(T). J. Phys. Chem. A 2016, 120, 9305–9314. [Google Scholar] [CrossRef]
- Zhuo, H.; Li, Q.; Li, W.; Cheng, J. Is π halogen bonding or lone pair⋯π interaction formed between borazine and some halogenated compounds? Phys. Chem. Chem. Phys. 2014, 16, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Kellett, C.W.; Kennepohl, P.; Berlinguette, C.P. π covalency in the halogen bond. Nat. Commun. 2020, 11, 3310. [Google Scholar] [CrossRef] [PubMed]
- Albright, E.; Cann, J.; Decken, A.; Eisler, S. Halogen⋯halogen interactions in diiodo-xylenes. CrystEngComm 2017, 19, 1024–1027. [Google Scholar] [CrossRef]
- Sadaf, H.; Imtiaz-ud-Din; Fettouhi, M.; Fazal, A.; Ahmad, S.; Farooqi, B.A.; Nadeem, S.; Ihsan-ul-Haq; Ahmad, W. Synthesis, crystal structures and biological activities of palladium(II) complexes of benzimidazole and 2-methylbenzimidazole. Polyhedron 2019, 170, 537–543. [Google Scholar] [CrossRef]
- Huang, Z.-J.; Du, L.; Xie, M.-J.; Chen, J. Dichlorido[bis(2-hexylsulfanyl)-1 H -benzimidazole-κ N 3 ]palladium(II). Acta Crystallogr. Sect. E Struct. Reports Online 2007, 63, m2474. [Google Scholar] [CrossRef]
- Sadaf, H.; Imtiaz-ud-Din; Zahra, S.S.; Ihsan-ul-Haq; Nadeem, S.; Tahir, M.N.; Ahmad, S.; Andleeb, S. Synthesis, X-ray structures and biological properties of palladium(II) complexes of 1,2-dimethylimidazole and benzimidazole. Polyhedron 2019, 160, 101–107. [Google Scholar] [CrossRef]
- Łakomska, I.; Fandzloch, M.; Wojtczak, A.; Szłyk, E. Platinum(IV) coordination compounds containing 5-methyl-1,2,4-triazolo[1,5-a]pyrimidin-7(4H)-one as nonleaving ligand. Molecular and cytotoxicity in vitro characterization. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2011, 79, 497–501. [Google Scholar] [CrossRef] [PubMed]
- Chongboriboon, N.; Samakun, K.; Dungkaew, W.; Kielar, F.; Sukwattanasinitt, M.; Chainok, K. Halogen-Bonding-Driven Self-Assembly of Solvates of Tetrabromoterephthalic Acid. Crystals 2021, 11, 198. [Google Scholar] [CrossRef]
- Politzer, P.; Murray, J.S.; Clark, T. Halogen bonding: An electrostatically-driven highly directional noncovalent interaction. Phys. Chem. Chem. Phys. 2010, 12, 7748. [Google Scholar] [CrossRef]






| Identification Code | 1 | 2 |
|---|---|---|
| Empirical formula | C10H14Cl4N10O3Pd | C5H10Cl5N5O3Pt |
| Formula weight | 570.51 | 560.52 |
| Temperature/K | 100(2) | 296(2) |
| Crystal system | Triclinic | Monoclinic |
| Space group | P-1 | P21/n |
| a/Å | 7.7566(4) | 7.9869(2) |
| b/Å | 10.7651(5) | 15.8000(5) |
| c/Å | 11.4193(5) | 13.0000(3) |
| α/° | 97.127(3) | 90 |
| β/° | 99.974(3) | 107.173(1) |
| γ/° | 90.128(3) | 90 |
| Volume/Å3 | 931.58(8) | 1567.37(7) |
| Z | 2 | 4 |
| ρcalcg/cm3 | 2.034 | 2.375 |
| μ/mm−1 | 1.608 | 9.812 |
| F(000) | 564.0 | 1048.0 |
| Crystal size/mm3 | 0.9 × 0.6 × 0.11 | 0.4 × 0.1 × 0.08 |
| Radiation | MoKα (λ = 0.71073) | MoKα (λ = 0.71073) |
| 2 Θ range for data collection/ | 8.23 to 60 | 8.348 to 60 |
| Index ranges | −10 ≤ h ≤ 10, −15 ≤ k ≤ 15, −15 ≤ l ≤ 16 | −10 ≤ h ≤ 11, −21 ≤ k ≤ 22, −17 ≤ l ≤ 18 |
| Reflections collected | 18,831 | 14,606 |
| Independent reflections | 5402 [Rint = 0.0711, Rsigma = 0.0736] | 4556 [Rint = 0.0263, Rsigma = 0.0286] |
| Data/restraints/parameters | 5402/0/257 | 4556/3/178 |
| Goodness-of-fit on F2 | 1.027 | 1.039 |
| Final R indexes [I ≥ 2σ (I)] | R1 = 0.0477, wR2 = 0.1168 | R1 = 0.0206, wR2 = 0.0408 |
| Final R indexes [all data] | R1 = 0.0778, wR2 = 0.1318 | R1 = 0.0293, wR2 = 0.0432 |
| Largest diff. peak/hole/e Å−3 | 1.30/−1.12 | 0.86/−0.97 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Novikov, A.P.; Volkov, M.A.; Safonov, A.V.; Grigoriev, M.S.; Abkhalimov, E.V. Synthesis and Characterization of New Guanine Complexes of Pt(IV) and Pd(II) by X-ray Diffraction and Hirshfeld Surface Analysis. Crystals 2021, 11, 1417. https://doi.org/10.3390/cryst11111417
Novikov AP, Volkov MA, Safonov AV, Grigoriev MS, Abkhalimov EV. Synthesis and Characterization of New Guanine Complexes of Pt(IV) and Pd(II) by X-ray Diffraction and Hirshfeld Surface Analysis. Crystals. 2021; 11(11):1417. https://doi.org/10.3390/cryst11111417
Chicago/Turabian StyleNovikov, Anton Petrovich, Mikhail Alexandrovich Volkov, Alexey Vladimirovich Safonov, Mikhail Semenovich Grigoriev, and Evgeny Vladilenovich Abkhalimov. 2021. "Synthesis and Characterization of New Guanine Complexes of Pt(IV) and Pd(II) by X-ray Diffraction and Hirshfeld Surface Analysis" Crystals 11, no. 11: 1417. https://doi.org/10.3390/cryst11111417
APA StyleNovikov, A. P., Volkov, M. A., Safonov, A. V., Grigoriev, M. S., & Abkhalimov, E. V. (2021). Synthesis and Characterization of New Guanine Complexes of Pt(IV) and Pd(II) by X-ray Diffraction and Hirshfeld Surface Analysis. Crystals, 11(11), 1417. https://doi.org/10.3390/cryst11111417








