Synthesis, X-ray Structure, Hirshfeld Analysis of Biologically Active Mn(II) Pincer Complexes Based on s-Triazine Ligands
Abstract
:1. Introduction
2. Materials and Methods
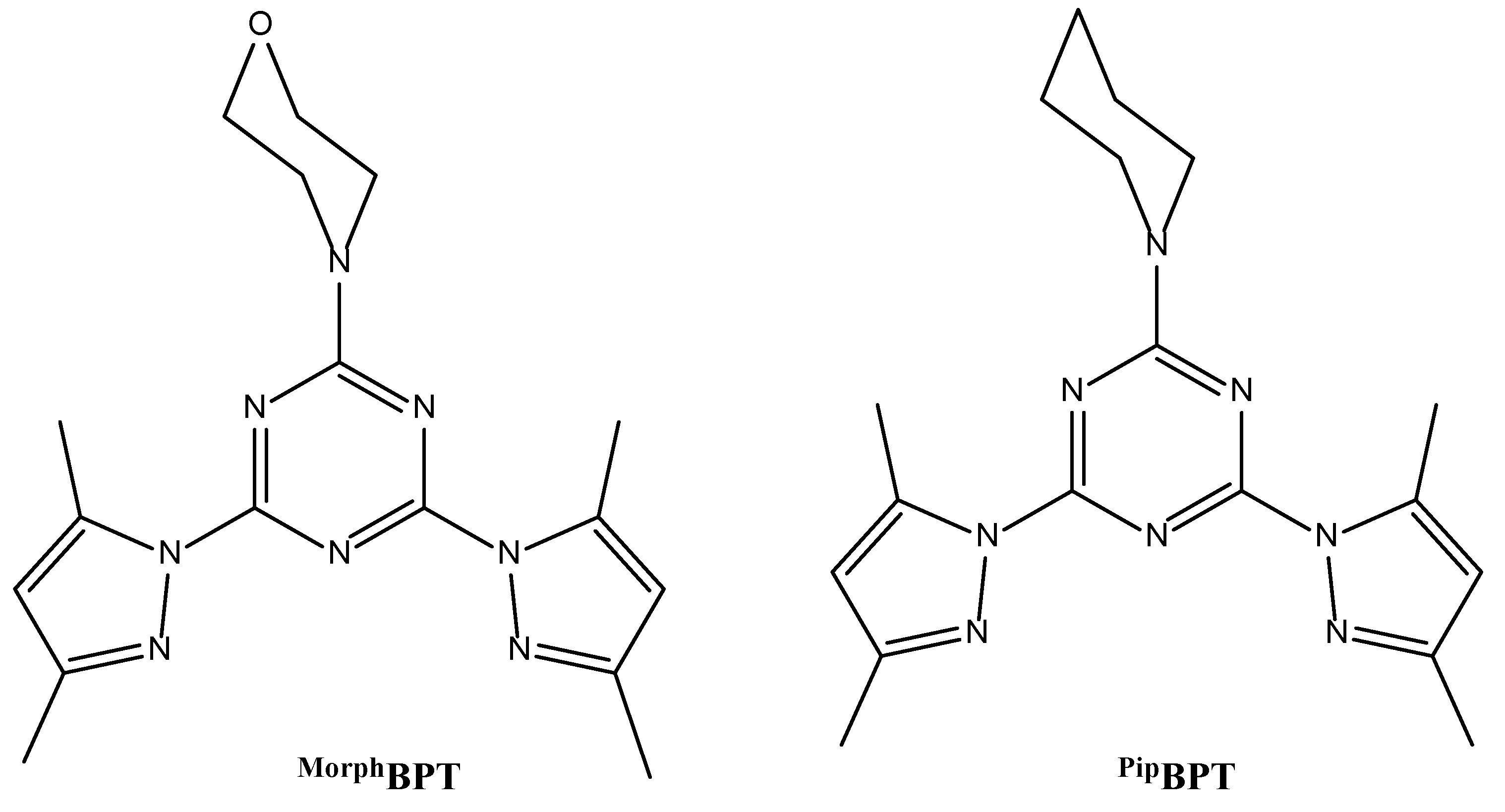
2.1. Preparation of the Organic Ligands
2.2. Syntheses of [Mn(MorphBPT)(H2O)2NO3]NO3; (1) and [Mn(PipBPT)(H2O)2NO3]NO3; (2)
2.3. Crystal Structure Determination
2.4. Antimicrobial Studies
2.5. Density Functional Theory (DFT) Calculations
3. Results and Discussion
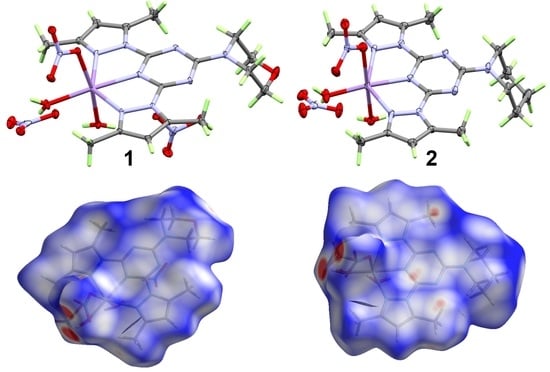
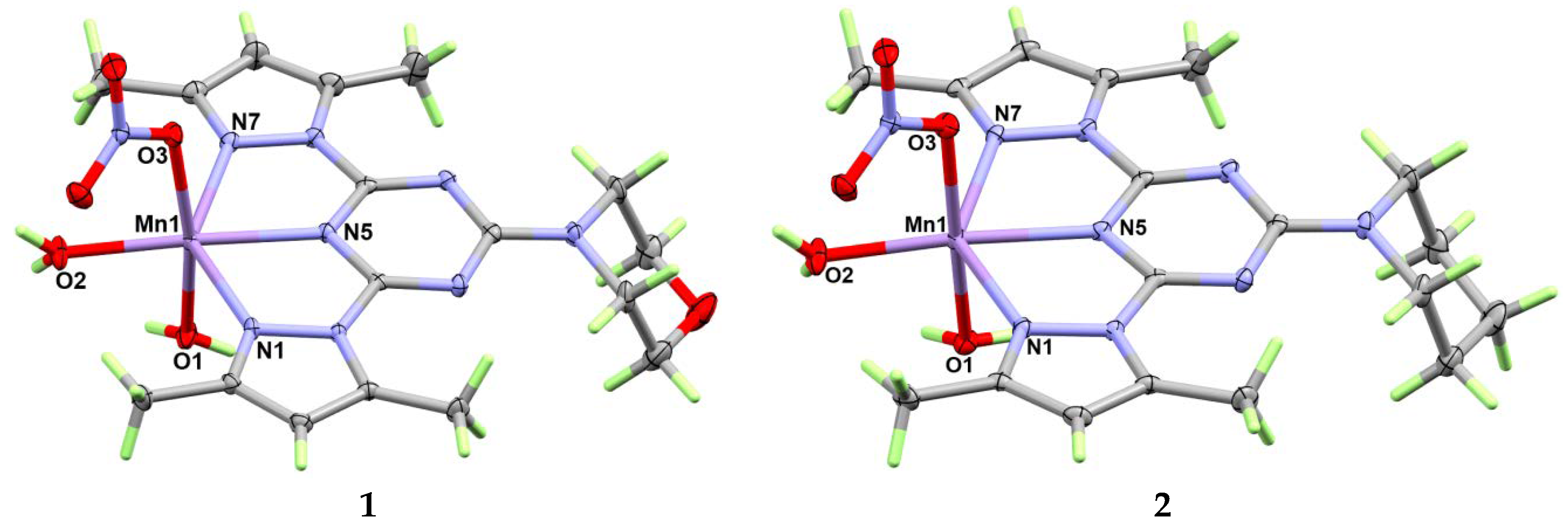
3.1. X-ray Crystal Structure Description
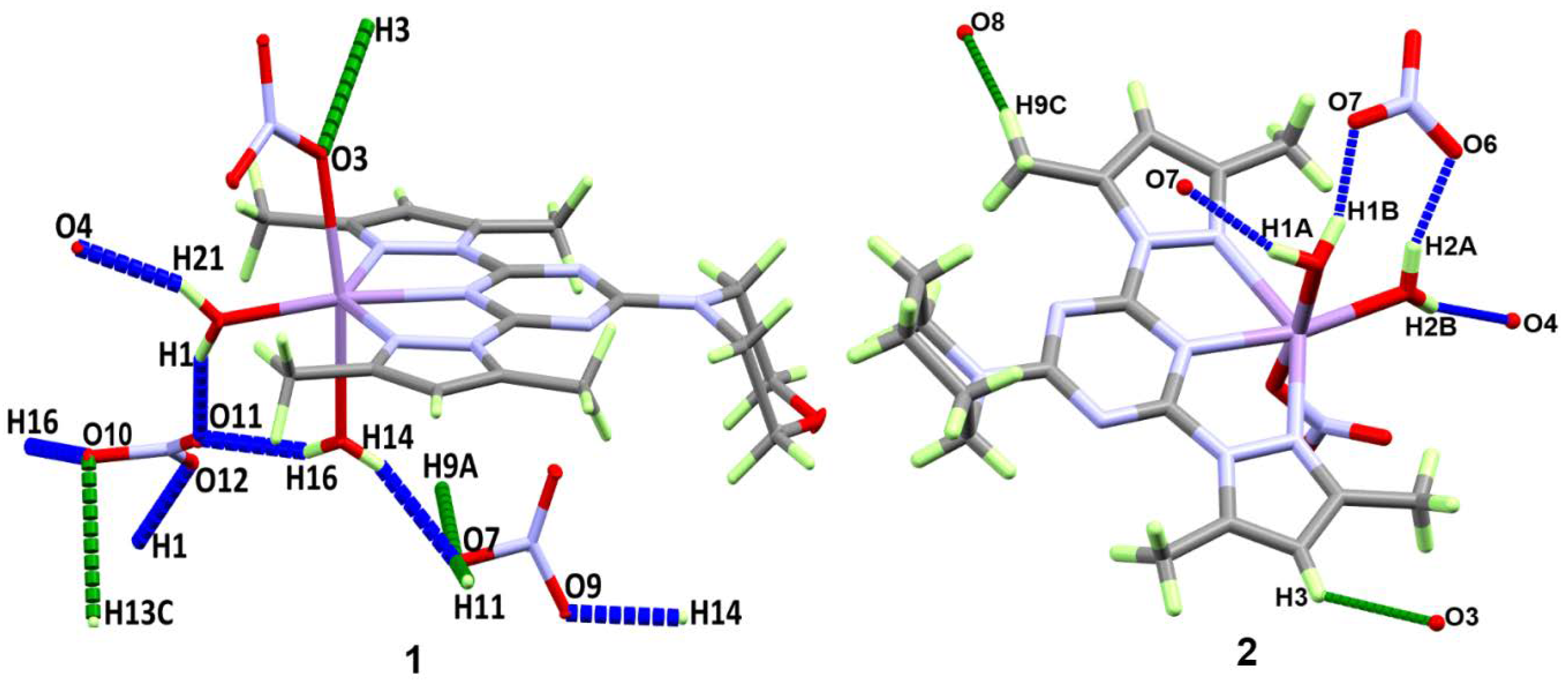
3.2. Analysis of Molecular Packing
3.3. AIM Topology Analysis
3.4. Antimicrobial Activity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ronconi, L.; Sadler, P.J. Using coordination chemistry to design new medicines. Coord. Chem. Rev. 2007, 251, 1633–1648. [Google Scholar] [CrossRef]
- Storr, T.; Thompson, K.H.; Orvi, C. Design of targeting ligands in medicinal inorganic chemistry. Chem. Soc. Rev. 2006, 35, 534–544. [Google Scholar] [CrossRef]
- Biswas, B.; Kole, N.; Patra, M.; Shampa Dotta, S.; Ganguly, M. Synthesis, structural characterization and biological activity of a trinuclear zinc(II) complex: DNA interaction study and antimicrobial activity. J. Chem. Sci. 2013, 125, 1445–1453. [Google Scholar] [CrossRef] [Green Version]
- Patel, M.N.; Joshi, H.N.; Patel, C.R. Cytotoxic, DNA binding, DNA cleavage and antibacterial studies of ruthenium–fluoroquinolone complexes. J. Chem. Sci. 2014, 126, 739–749. [Google Scholar] [CrossRef] [Green Version]
- Madalan, A.M.; Kravtsov, V.C.; Pajic, D.; Zadro, K.; Simonov, Y.A.; Stanica, N.; Ouahab, L.; Lipkowski, J.; Andruh, M. Chemistry at the apical position of square-pyramidal copper(II) complexes: Synthesis, crystal structures, and magnetic properties of mononuclear Cu(II), and heteronuclear Cu(II)–Hg(II) and Cu(II)–Co(II) complexes containing [Cu(AA)(BB)]+ moieties (AA = acetylacetonate, salicylaldehydate; BB = 1,10-phenanthroline, Me2bipy = 4,4′-dimethyl-2,2′-bipyridine). Inorg. Chim. Acta 2004, 357, 4151–4164. [Google Scholar]
- Paulovicova, A.; El-Ayaan, U.; Fukuda, Y. Synthesis, characterization and X-ray crystal structures of two five-coordinate ternary copper(II) complexes containing acetylacetonate with 1,10-phenanthroline and 2,9-dimethyl phenanthroline. Inorg. Chim. Acta 2001, 321, 56–62. [Google Scholar] [CrossRef]
- Triller, M.U.; Hsieh, W.Y.; Pecoraro, V.L.; Rompel, A.; Krebs, B. Preparation of Highly Efficient Manganese Catalase Mimics‖. Inorg. Chem. 2002, 4, 5544–5554. [Google Scholar] [CrossRef]
- Wu, A.J.; Penner-Hahn, J.E.; Pecoraro, V.L. Structural, Spectroscopic, and Reactivity Models for the Manganese Catalases. Chem. Rev. 2004, 104, 903–938. [Google Scholar] [CrossRef]
- Reddig, N.; Pursche, D.; Kloskowski, M.; Slinn, C.; Baldeau, S.M.; Rompel, A. Tuning the Catalase Activity of Dinuclear Manganese Complexes by Utilizing Different Substituted Tripodal Ligands. Eur. J. Inorg. Chem. 2004, 2004, 879–887. [Google Scholar] [CrossRef]
- Signorella, S.; Rompel, A.; Buldt-Karentzopoulos, K.; Krebs, B.; Pecoraro, V.L.; Tuchagues, J.P. Reevaluation of the Kinetics of Polynuclear Mimics for Manganese Catalases. Inorg. Chem. 2007, 46, 10864–10868. [Google Scholar] [CrossRef]
- Signorella, S.; Hureau, C. Bioinspired functional mimics of the manganese catalases. Coord. Chem. Rev. 2012, 256, 1229–1245. [Google Scholar] [CrossRef]
- Biava, H.; Palopoli, C.; Duhayon, C.; Tuchagues, J.P.; Signorella, S. Synthesis, Structure, and Catalase-Like Activity of Dimanganese(III) Complexes of 1,5-Bis[(2-hydroxy-5-X-benzyl)(2-pyridylmethyl)amino]pentan-3-ol (X = H, Br, OCH3). Inorg. Chem. 2009, 48, 3205–3214. [Google Scholar] [CrossRef] [PubMed]
- Biju, A.R.; Rajasekharan, M.V. Structure, magnetic properties, catalase activity and DFT studies of [Mn2(μ-RCOO)2(μ-OR)2]2+ type dinuclear manganese(III,III) complexes. Inorg. Chim. Acta 2011, 372, 275–280. [Google Scholar] [CrossRef]
- Vazquez-Fernandez, M.A.; Bermejo, M.R.; Fernandez-Garcia, M.I.; Gonzalez-Riopedre, G.; Rodriguez-Doutun, M.J.; Maneiro, M.J. Influence of the geometry around the manganese ion on the peroxidase and catalase activities of Mn(III)–Schiff base complexes. Inorg. Biochem. 2011, 105, 1538–1547. [Google Scholar] [CrossRef] [PubMed]
- Kani, I.; Atlier, Ö.; Güven, K. Mn(II) complexes with bipyridine, phenanthroline and benzoic acid: Biological and catalase-like activity. J. Chem. Sci. 2016, 128, 523–536. [Google Scholar] [CrossRef] [Green Version]
- Devereux, M.; McCann, M.; Leonl, V.; Geraghty, M.; McKee, V.; Wikaira, J. Synthesis and Biological Activity of Manganese (II) Complexes of Phthalic and Isophthalic Acid: X-Ray Crystal Structures of [Mn(ph)(Phen)2(H2O)] 4H2O, [Mn(Phen)2(H2O)2]2(Isoph)2(Phen) 12H2O and {[Mn(Isoph)(bipy)]4·2.75biby}n(phH2 = Phthalic Acid; isoph = Isophthalic Acid; phen = 1,10-Phenanthroline; bipy = 2,2-Bipyridine). Metal Based Drugs 2000, 7, 275–288. [Google Scholar]
- Avila, D.S.; Puntel, R.L.; Aschner, M. Interrelations between Essential Metal Ions and Human Diseases; Springer: Berlin, Germany, 2013; pp. 199–227. [Google Scholar]
- Yoon, H.; Kim, D.S.; Lee, G.H.; Kim, K.W.; Kim, H.R.; Chae, H.J. Apoptosis Induced by Manganese on Neuronal SK-N-MC Cell Line: Endoplasmic Reticulum (ER) Stress and Mitochondria Dysfunction. Environ. Health Toxicol. 2011, 26, e2011017. [Google Scholar] [CrossRef]
- Yadamani, S.; Neamati, A.; Homayouni-Tabrizi, M.; Beyramabadi, S.A.; Yadamani, S.; Gharib, A.; Morsali, A.; Khashi, M. Treatment of the breast cancer by using low frequency electromagnetic fields and Mn(II) complex of a Schiff base derived from the pyridoxal. Breast 2018, 41, 107–112. [Google Scholar] [CrossRef]
- Tabatabayi, Z.S.; Tabrizi, M.H.; Neamati, A.; Beyramabadi, S.A. Mn(II) complex of a vitamin B6 Schiff base as an exclusive apoptosis inducer in human MCF7 and HepG2 cancer cells: Synthesis, characterization, and biological studies. J. Cell. Biochem. 2020, 121, 2677–2689. [Google Scholar] [CrossRef]
- Soliman, S.M.; Almarhoon, Z.; El-Faham, A. Synthesis, Molecular and Supramolecular Structures of New Cd(II) Pincer-Type Complexes with s-Triazine Core Ligand. Crystals 2019, 9, 226. [Google Scholar] [CrossRef] [Green Version]
- Soliman, S.M.; Elsilk, S.E.; El-Faham, A. Synthesis, structure and biological activity of zinc(II) pincer complexes with 2,4-bis(3,5-dimethyl-1H-pyrazol-1-yl)-6-methoxy-1,3,5-triazine. Inorg. Chim. Acta 2020, 508, 119627. [Google Scholar] [CrossRef]
- Soliman, S.M.; Elsilk, S.E.; El-Faham, A. Syntheses, structure, Hirshfeld analysis and antimicrobial activity of four new Co(II) complexes with s-triazine-based pincer ligand. Inorg. Chim. Acta 2020, 510, 119753. [Google Scholar] [CrossRef]
- Soliman, S.M.; Elsilk, S.E.; El-Faham, A. Novel one-dimensional polymeric Cu(II) complexes via Cu(II)-assisted hydrolysis of the 2,4-bis(3,5-dimethyl-1H-pyrazol-1-yl)-6-methoxy-1,3,5-triazine pincer ligand: Synthesis, structure, and antimicrobial activities. Appl. Organometall. Chem. 2020, 510, 119753. [Google Scholar]
- Chen, W.; Chu, J.F.; Wang, Y.Q. Synthesis, characterization and preliminary reactivity behaviors with transitional metals of a new polydentate N-donor ligand. J. Mol. Struct. 2014, 1068, 237–244. [Google Scholar] [CrossRef]
- Chu, J.F.; Zhang, M.Y.; Wang, Y.Q. Syntheses, Crystal Structures, and Properties of Three Copper(II) Complexes Constructed by 4-(4,6-Bis(1H-pyrazol-1-yl)-1,3,5-triazin-2-yl)morpholine. Z. Anorg. Allg. Chem. 2017, 643, 1101–1106. [Google Scholar] [CrossRef]
- Aleksanyan, D.V.; Churusova, S.G.; Brunova, V.V.; Rybalkina, E.; Susova, O.; Peregudov, A.S.; Klemenkova, Z.S.; Denisov, G.L.; Kozlov, V.A. Synthesis, characterization, and cytotoxic activity of N -metallated rhenium(I) pincer complexes with (thio)phosphoryl pendant arms. J. Organometall. Chem. 2020, 926, 121498. [Google Scholar] [CrossRef]
- Shakirova, J.R.; Hendi, Z.; Zhukovsky, D.D.; Sokolov, V.V.; Jamali, S.; Pavlovskiy, V.V.; Porsev, V.V.; Evarestov, R.A.; Tunik, S.P. NIR emitting platinum pincer complexes based on the N^N^C ligand containing {benz[4,5]imidazo[1,2-a]pyrazin} aromatic system; synthesis, characterization and photophysical study. Inorg. Chim. Acta 2020, 511, 119776. [Google Scholar] [CrossRef]
- Fayoumi, A.; Lyubov, D.M.; Tolpygin, A.O.; Shavyrin, A.S.; Cherkasov, A.V.; Ob’edkov, A.M.; Trifonov, A.A. Sc and Y Hetero-alkyl Complexes with NCsp3N Pincer Type Diphenylmethanido Ligand: Synthesis, Structure and Reactivity. Eur. J. Inorg. Chem. 2020, 2020, 3259–3267. [Google Scholar] [CrossRef]
- Gafurov, Z.N.; Bekmukhamedov, G.E.; Kagilev, A.A.; Kantyukov, A.O.; Sakhapov, I.F.; Mikhailov, I.K.; Khayarov, K.R.; Zaripov, R.B.; Islamov, D.R.; Usachev, K.S.; et al. Unsymmetrical pyrazole-based PCN pincer Ni(II) halides: Reactivity and catalytic activity in ethylene oligomerization. J. Organometall. Chem. 2020, 912, 121163. [Google Scholar] [CrossRef]
- Safronov, S.V.; Gutsul, E.I.; Golub, I.E.; Dolgushin, F.M.; Nelubina, Y.V.; Filippov, O.A.; Epstein, L.M.; Peregudov, A.S.; Belkova, N.V.; Shubin, S.E. Synthesis, Structural Properties and Reactivity of Ruthenocene-based Pincer Pd(II) Tetrahydroborate. Dalton Trans. 2019, 48, 12720–12729. [Google Scholar] [CrossRef]
- Sharma, A.; Ghabbour, H.; Khan, S.T.; de la Torre, B.G.; Albericio, F.; El-Faham, A. Novel pyrazolyl-s-triazine derivatives, molecular structure and antimicrobial activity. J. Mol. Struct. 2017, 1145, 244–253. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Program for Empirical Absorption Correction of Area Detector Data; University of Göttingen: Göttingen, Germany, 1996. [Google Scholar]
- Sheldrick, G.M. SHELXT-Integrated space-group and crystal-structure determination. Acta Cryst. A 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spek, A.L. Structure validation in chemical crystallography. Acta Cryst. D 2009, 65, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Turner, M.J.; McKinnon, J.J.; Wolff, S.K.; Grimwood, D.J.; Spackman, P.R.; Jayatilaka, D.; Spackman, M.A. Crystal Explorer 17; University of Western Australia: Perth, Australia, 2017. Available online: http://hirshfeldsurface.net (accessed on 12 June 2017).
- Spackman, M.A.; Jayatilaka, D. Hirshfeld surface analysis. Cryst. Eng. Comm. 2009, 11, 19–32. [Google Scholar] [CrossRef]
- Spackman, M.A.; McKinnon, J.J. Fingerprinting intermolecular interactions in molecular crystals. Cryst. Eng. Commun. 2002, 4, 378–392. [Google Scholar] [CrossRef]
- Bernstein, J.; Davis, R.E.; Shimoni, L.; Chang, N.-L. Patterns in hydrogen bonding: Functionality and graph set analysis in crystals. Angew. Chem. Int. Ed. Engl. 1995, 34, 1555–1573. [Google Scholar] [CrossRef]
- McKinnon, J.J.; Jayatilaka, D.; Spackman, M.A. Towards quantitative analysis of intermolecular interactions with Hirshfeld surfaces. Chem. Commun. 2007, 37, 3814–3816. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. GAUSSIAN 09.Revision A02; Gaussian Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Adamo, C.; Barone, V. Exchange functionals with improved long-range behavior and adiabatic connection methods without adjustable parameters: The mPWmPW and mPW1PWmPW1PW models. J. Chem. Phys. 1998, 108, 664–675. [Google Scholar] [CrossRef]
- Glendening, E.D.; Reed, A.E.; Carpenter, J.E.; Weinhold, F. NBO Version 3.1, CI; University of Wisconsin: Madison, WI, USA, 1998. [Google Scholar]
- Lu, T.; Chen, F. Multiwfn: A multifunctional wavefunction analyzer. J. Comput. Chem. 2012, 33, 580–592. [Google Scholar] [CrossRef]
- Bader, R.F.W. Atoms in Molecules: A Quantum Theory; Oxford University Press: Oxford, UK, 1990. [Google Scholar]
- Matta, C.F.; Hernandez-Trujillo, J.; Tang, T.-H.; Bader, R.F.W. Hydrogen-hydrogen bonding: A stabilizing interaction in molecules and crystals. Chem. Eur. J. 2003, 9, 1940–1951. [Google Scholar] [CrossRef]
- Grabowski, S.J.; Pfitzner, A.; Zabel, M.; Dubis, A.T.; Palusiak, M. Intramolecular H–H interactions for the Crystal Structures of [4-((E)-But-1-enyl)-2,6-dimethoxyphenyl]pyridine-3-carboxylate and [4-((E)-Pent-1-enyl)-2,6-dimethoxyphenyl]pyridine-3-carboxylate; DFT calculations on modeled styrene derivatives. J. Phys. Chem. B 2004, 108, 1831–1837. [Google Scholar] [CrossRef]
- Matta, C.F.; Castillo, N.; Boyd, R.J. Characterization of a closed-shell fluorine-fluorine bonding interaction in aromatic compounds on the basis of the electron density. J. Phys. Chem. A 2005, 109, 3669–3681. [Google Scholar] [CrossRef] [PubMed]
- Pendás, A.M.; Francisco, E.; Blanco, M.A.; Gatti, C. Bond paths as privileged exchange channels. Chem. Eur. J. 2007, 13, 9362–9371. [Google Scholar] [CrossRef] [PubMed]
- Bobrov, M.F.; Popova, G.V.; Tsirelson, V.G. A topological analysis of electron density and chemical bonding in cyclophosphazenes PnNnX2n (X = H, F, Cl; n = 2, 3, 4). Russ. J. Phys. Chem. 2006, 80, 584–590. [Google Scholar] [CrossRef]
- Gatti, C. Chemical bonding in crystals: New directions. Zeitschrift für Kristallographie 2005, 220, 399–457. [Google Scholar] [CrossRef]
- Gibbs, G.V.; Downs, R.T.; Cox, D.F.; Ross, N.L.; Boisen, M.B., Jr.; Rosso, K.M. Shared and closed-shell O-O interactions in silicates. J. Phys. Chem. A 2008, 112, 3693–3699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Espinosa, E.; Molins, E.; Lecomte, C. Hydrogen bond strengths revealed by topological analyses of experimentally observed electron densities. Chem. Phys. Lett. 1998, 285, 170–173. [Google Scholar] [CrossRef]
- Cremer, D.; Kraka, E. Chemical Bonds without Bonding Electron Density—Does the Difference Electron-Density Analysis Suffice for a Description of the Chemical Bond? Angew. Chem. Int. Ed. Engl. 1984, 23, 627–628. [Google Scholar] [CrossRef]
- De Souza, S.M.; Delle Monache, F.; Smânia, A. Antibacterial activity of coumarins. Zeitschrift fuer Naturforschung C 2005, 60, 693–700. [Google Scholar] [CrossRef]
- Arshad, M.; Bhat, A.R.; Hoi, K.K.; Choi, I.; Athar, F. Synthesis, characterization and antibacterial screening of some novel 1,2,4-triazine derivatives. Chin. Chem. Lett. 2017, 28, 1559–1565. [Google Scholar] [CrossRef]
- Torshizi, H.M.; Jahromi, S.Z. Synthesis, Characterization and in Vitro Antimicrobial Screening of the Xanthate Derivatives and their Iron(II) Complexes. Iran. J. Chem. Chem. Eng. 2017, 36, 43–54. [Google Scholar]
- Dhokale, N.T.; Karale, B.K.; Nagawadw, A.V. Synthesis, Characterization and Antibacterial Studies on Mn(II) and Fe(II) Complexes of N, O Donor Salicyloyl Pyrazole Oxime Schiff Bases. Orient. J. Chem. 2017, 33, 165–172. [Google Scholar] [CrossRef] [Green Version]

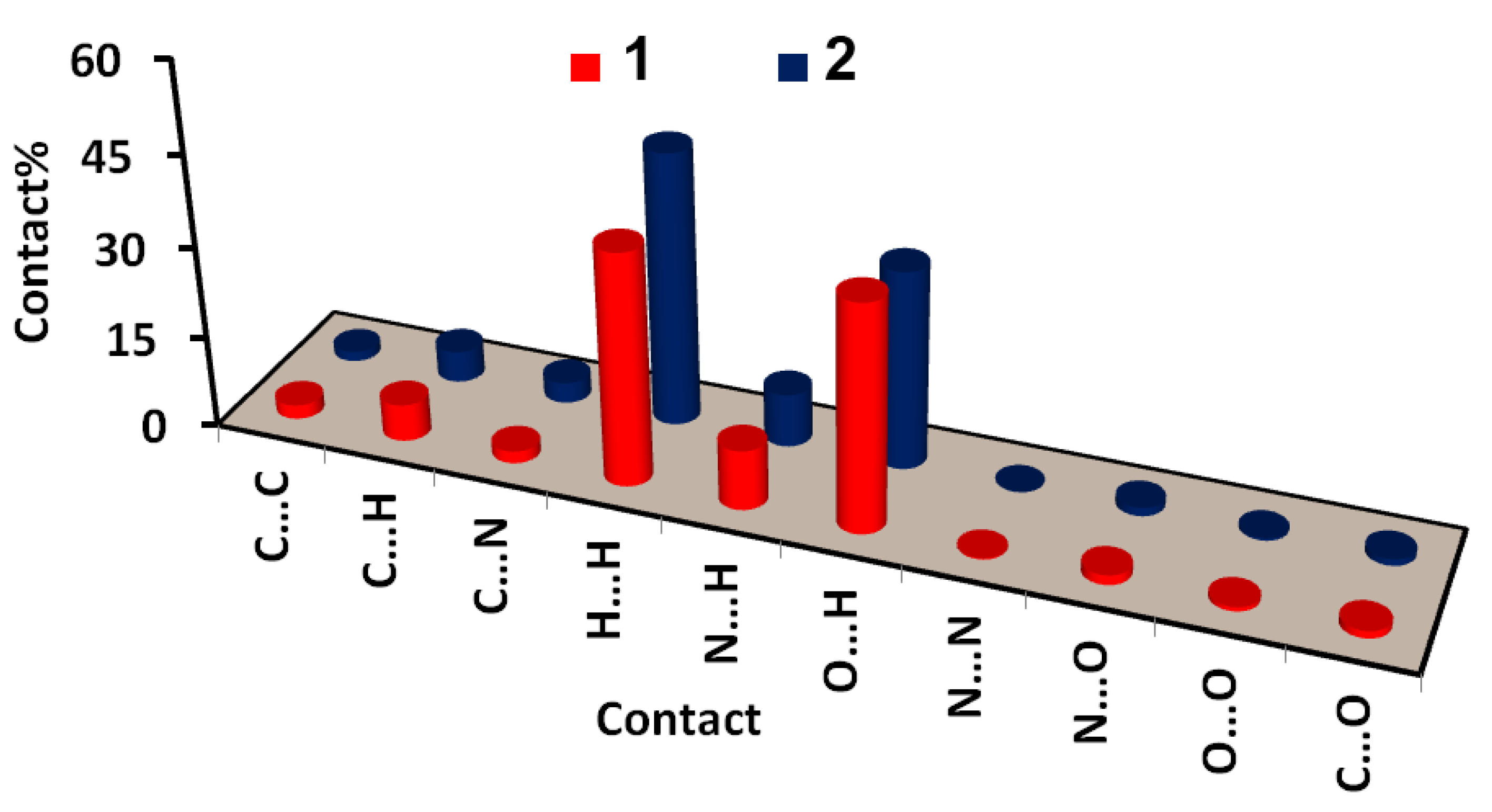
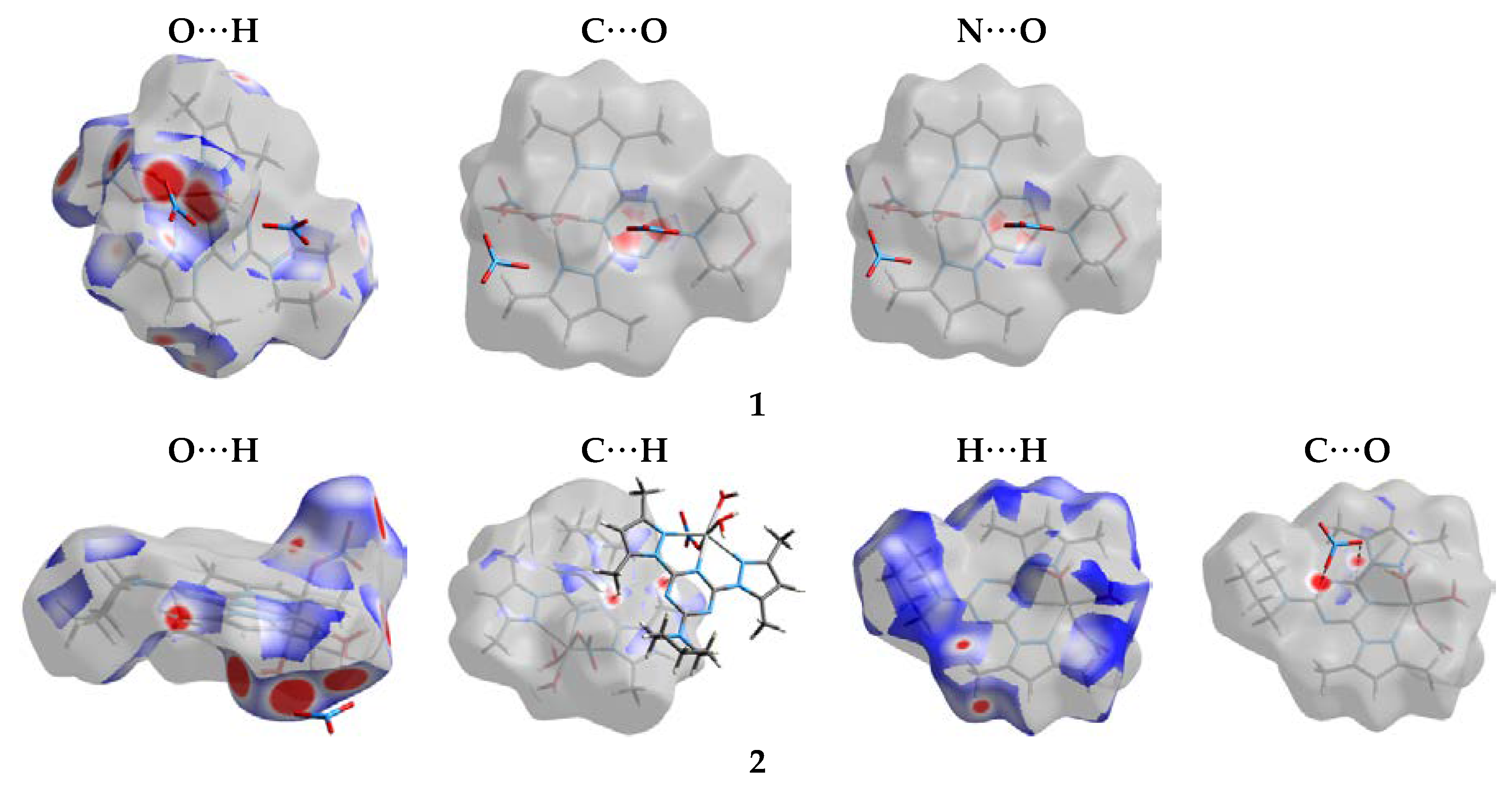
| Compound | 1 | 2 |
|---|---|---|
| Empirical formula | C17H26MnN10O9 | C18H28MnN10O8 |
| Formula weight (g/mol) | 569.42 | 567.44 |
| Temperature (K) | 124(2) | 117(2) |
| λ (Mo-Kα, Å) | 0.71073 | 0.71073 |
| Crystal system | Triclinic | Triclinic |
| Space group | P-1 | P-1 |
| Unit cell dimensions | a = 8.2794(5) Å | a = 7.830(5) Å |
| b = 12.1167(7) Å | b = 12.951(7) Å | |
| c = 12.1738(7) Å | c = 13.990(7) Å | |
| α = 88.3660(19)° | α = 116.533(10)° | |
| β = 89.605(2)° | β = 94.832(14)° | |
| γ = 79.184(2)° | γ = 101.694(16)° | |
| Volume (Å3) | 1199.08(12) | 1217.9(12) |
| Z | 2 | 2 |
| Density (calc. g/cm3) | 1.577 | 1.547 |
| Absorption coefficient (mm−1) | 0.621 | 0.608 |
| F(000) | 590 | 590 |
| Crystal size (mm3) | 0.29 × 0.21 × 0.11 | 0.31 × 0.23 × 0.19 |
| θ range for data collection | 2.36 to 26.34° | 2.71 to 25.31° |
| Index ranges | −10 ≤ h ≤ 10, −15 ≤ k ≤ 15, −15 ≤ l ≤ 15 | −9 ≤ h ≤ 9, −15 ≤ k ≤ 15, −16 ≤ l ≤ 16 |
| Reflections collected | 21,451 | 19,570 |
| Independent reflections | 4863 [R(int) = 0.0315] | 4437 [R(int) = 0.0554] |
| Completeness to θ | 99.30% | 99.80% |
| Refinement method | Full-matrix least-squares on F2 | |
| Data/restraints/parameters | 4863/0/385 | 4437/0/351 |
| Goodness-of-fit on F2 | 0.894 | 1.06 |
| Final R indices [I>2sigma(I)] | R1 = 0.0275, wR2 = 0.0671 | R1 = 0.0298, wR2 = 0.0732 |
| R indices (all data) | R1 = 0.0341, wR2 = 0.0721 | R1 = 0.0371, wR2 = 0.0775 |
| Extinction coefficient | 0.0100(8) | 0.0157(17) |
| Largest diff. peak and hole | 0.371 and −0.318 | 0.409 and −0.308 |
| CCDC | 2025609 | 2025610 |
| Atoms | 1 | 2 |
|---|---|---|
| Mn1–O2 | 2.1416(12) | 2.1288(19) |
| Mn1–O3 | 2.1683(11) | 2.1604(16) |
| Mn1–O1 | 2.2159(12) | 2.2306(17) |
| Mn1–N5 | 2.2172(12) | 2.1973(18) |
| Mn1–N1 | 2.3188(13) | 2.3097(19) |
| Mn1–N7 | 2.3267(13) | 2.3580(18) |
| O2–Mn1–O3 | 90.35(5) | 96.78(6) |
| O2–Mn1–O1 | 80.70(5) | 83.33(6) |
| O3–Mn1–O1 | 167.76(5) | 172.64(5) |
| O2–Mn1–N5 | 170.23(5) | 166.35(6) |
| O3–Mn1–N5 | 97.55(4) | 94.69(6) |
| O1–Mn1–N5 | 90.62(5) | 84.34(6) |
| O2–Mn1–N1 | 115.16(5) | 116.12(6) |
| O3–Mn1–N1 | 98.37(5) | 94.40(6) |
| O1–Mn1–N1 | 93.02(5) | 92.13(6) |
| N5–Mn1–N1 | 69.49(4) | 70.18(5) |
| O2–Mn1-N7 | 106.03(5) | 103.93(6) |
| O3–Mn1–N7 | 85.47(4) | 86.40(6) |
| O1–Mn1–N7 | 88.98(5) | 86.44(6) |
| N5–Mn1–N7 | 69.09(4) | 69.39(5) |
| N1–Mn1–N7 | 138.55(4) | 139.49(6) |
| Atoms | D–H (Å) | H···A (Å) | D···A (Å) | D–H···A (°) |
|---|---|---|---|---|
| 1 | ||||
| O2–H1···O11 | 0.82(2) | 1.89(2) | 2.662(3) | 157(2) |
| O2–H1···O12 | 0.82(2) | 1.95(2) | 2.766(3) | 175(2) |
| O1–H14···O9 | 0.83(2) | 1.97(2) | 2.728(3) | 153(2) |
| O1–H14···O7i | 0.83(2) | 1.96(2) | 2.758(3) | 161(2) |
| O1–H16···O10 | 0.85(2) | 2.07(2) | 2.884(3) | 161(2) |
| O1–H16···O11 | 0.85(2) | 2.07(2) | 2.831(4) | 150(2) |
| O2–H21···O5 | 0.85(2) | 2.54(2) | 2.924(2) | 109(2) |
| O2–H21···O4ii | 0.85(2) | 2.01(2) | 2.816(2) | 159(2) |
| C3–H3···O3iii | 0.95 | 2.52 | 3.319(2) | 142 |
| C9–H9A···O7iv | 0.98 | 2.49 | 3.455(3) | 170 |
| C11–H11···O7v | 0.95 | 2.39 | 3.328(3) | 171 |
| C13–H13C···O10 | 0.98 | 2.49 | 3.271(3) | 136 |
| Symmetry Code: (i) 1-x,1-y,2-z; (ii) -x,2-y,1-z; (iii) 1+x,y,z; (iv) -1+x,y,z; (v) -x,1-y,2-z | ||||
| 2 | ||||
| O1–H1A···O7i | 0.83(3) | 2.04(3) | 2.839(3) | 162(3) |
| O1–H1B···O7 | 0.86(2) | 1.89(2) | 2.723(3) | 163(2) |
| O2–H2A···O6 | 0.87(2) | 1.92(2) | 2.758(3) | 161(2) |
| O2–H2B···O4ii | 0.83(4) | 1.99(4) | 2.795(3) | 162(4) |
| C3–H3···O3iii | 0.95 | 2.53 | 3.240(3) | 132 |
| C9–H9C···O8iv | 0.98 | 2.42 | 3.329(3) | 154 |
| Symmetry Code: (i) 1-x,1-y,-z; (ii) 1-x,1-y,1-z; (iii) 1+x,y,z; (iv) -x,1-y,-z | ||||
| 1 | 2 | ||
|---|---|---|---|
| C7···O8i | 2.940(3) | C8···O7ii | 3.090(2) |
| N3···O8i | 3.077(3) | C7···O8ii | 2.982(2) |
| N4···O8i | 2.995(3) | ||
| C8···O8i | 3.044(3) | ||
| C6···O8i | 3.141(3) | ||
| C6···O9i | 3.052(3) | ||
| N5···O9i | 2.831(3) | ||
| C8···O9i | 2.889(3) | ||
| Symmetry Code: (i) x,y,z in 1 (ii) 1-x,1-y,-z in 2 | |||
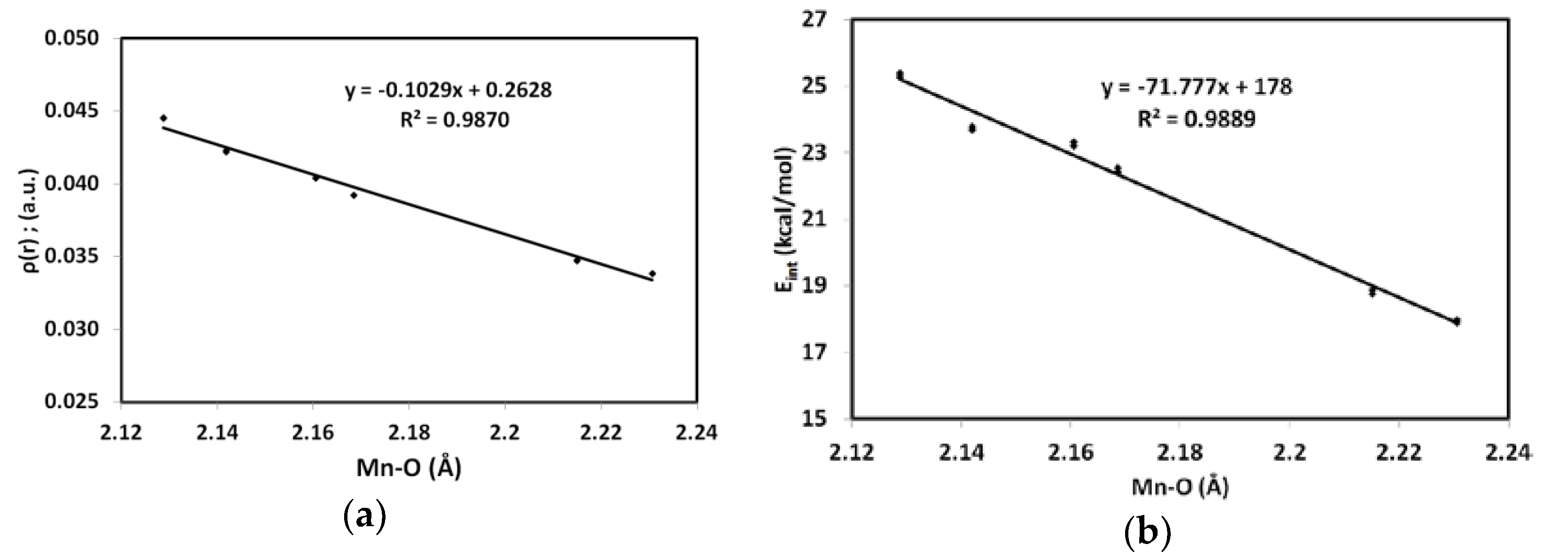
| Bond | Ρ(r) | G(r) | V(r) | Eint a | H(r) b | V(r)/G(r) c |
|---|---|---|---|---|---|---|
| 1 (MPW1PW91) | ||||||
| Mn1–N1 | 0.0315 | 0.0526 | −0.0511 | 16.0302 | 0.0015 | 0.9712 |
| Mn1–N7 | 0.0310 | 0.0508 | −0.0495 | 15.5195 | 0.0013 | 0.9737 |
| Mn1–N5 | 0.0446 | 0.0730 | −0.0749 | 23.5085 | −0.0019 | 1.0257 |
| Mn1–O1 | 0.0348 | 0.0617 | −0.0602 | 18.8813 | 0.0015 | 0.9754 |
| Mn1–O2 | 0.0423 | 0.0764 | −0.0759 | 23.8078 | 0.0006 | 0.9927 |
| Mn1–O3 | 0.0392 | 0.0731 | −0.0719 | 22.5571 | 0.0012 | 0.9835 |
| 1 (WB97XD) | ||||||
| Mn1–N1 | 0.0315 | 0.0524 | −0.0507 | 15.9153 | 0.0016 | 0.9689 |
| Mn1–N7 | 0.0309 | 0.0506 | −0.0491 | 15.4165 | 0.0014 | 0.9714 |
| Mn1–N5 | 0.0445 | 0.0729 | −0.0746 | 23.3928 | −0.0017 | 1.0234 |
| Mn1–O1 | 0.0347 | 0.0615 | −0.0599 | 18.7798 | 0.0016 | 0.9734 |
| Mn1–O2 | 0.0422 | 0.0763 | −0.0755 | 23.6955 | 0.0007 | 0.9904 |
| Mn1–O3 | 0.0392 | 0.0728 | −0.0715 | 22.4445 | 0.0013 | 0.9820 |
| 2 (MPW1PW91) | ||||||
| Mn1–N1 | 0.0323 | 0.0536 | −0.0522 | 16.3859 | 0.0014 | 0.9746 |
| Mn1–N7 | 0.0285 | 0.0464 | −0.0449 | 14.0784 | 0.0016 | 0.9662 |
| Mn1–N5 | 0.0465 | 0.0774 | −0.0796 | 24.9777 | −0.0022 | 1.0283 |
| Mn1–O1 | 0.0339 | 0.0592 | −0.0574 | 17.9982 | 0.0018 | 0.9698 |
| Mn1–O2 | 0.0446 | 0.0815 | −0.0810 | 25.4111 | 0.0005 | 0.9940 |
| Mn1–O3 | 0.0404 | 0.0754 | −0.0743 | 23.3263 | 0.0010 | 0.9862 |
| 2 (WB97XD) | ||||||
| Mn1–N1 | 0.0323 | 0.0533 | −0.0518 | 16.2604 | 0.0015 | 0.9724 |
| Mn1–N7 | 0.0284 | 0.0462 | −0.0445 | 13.9667 | 0.0017 | 0.9638 |
| Mn1–N5 | 0.0464 | 0.0772 | −0.0792 | 24.8540 | −0.0020 | 1.0260 |
| Mn1–O1 | 0.0338 | 0.0590 | −0.0570 | 17.8993 | 0.0019 | 0.9676 |
| Mn1–O2 | 0.0445 | 0.0812 | −0.0806 | 25.2873 | 0.0006 | 0.9921 |
| Mn1–O3 | 0.0404 | 0.0751 | −0.0740 | 23.2066 | 0.0012 | 0.9846 |
| Bond | MPW1PW91 | WB97XD | MPW1PW91 | WB97XD |
|---|---|---|---|---|
| 1 | 2 | |||
| Mn1–N1 | 0.143 | 0.144 | 0.142 | 0.143 |
| Mn1–N7 | 0.136 | 0.137 | 0.122 | 0.123 |
| Mn1–N5 | 0.198 | 0.199 | 0.206 | 0.207 |
| Mn1–O1 | 0.129 | 0.129 | 0.119 | 0.119 |
| Mn1–O2 | 0.163 | 0.163 | 0.170 | 0.171 |
| Mn1–O3 | 0.157 | 0.158 | 0.159 | 0.161 |
| Atom | MorphBPT | 1 | PipBPT | 2 |
|---|---|---|---|---|
| N5 | −0.4618(−0.4740) | −0.5921(−0.6022) | −0.4671(−0.4794) | −0.5892(−0.5987) |
| N1 | −0.2237(−0.2271) | −0.3163(−0.3191) | −0.2243(−0.2275) | −0.3042(−0.3069) |
| N7 | −0.2015(−0.2275) | −0.3084(−0.3106) | −0.2224(−0.2256) | −0.3085(−0.3114) |
| Compound | S. aureus | E. coli | C. albicans |
|---|---|---|---|
| MorphBPT | - | - | - |
| PipBPT | 11 | - | 12 |
| 1 | 21 | 17 | 15 |
| 2 | 27 | 20 | 17 |
| Fluconazole | - | - | 14 |
| Gentamycin | 28 | 21 | - |
| Microbes | 1 | 2 | ||
|---|---|---|---|---|
| MIC (µg/mL) | MBC (µg/mL) | MIC (µg/mL) | MBC (µg/mL) | |
| S. aureus | 8.3 | 16.5 | 6.5 | 13.5 |
| E. coli | 8.3 | 16.5 | 8.3 | 16.8 |
| C. albicans | 18.5 | 100 | 18.5 | 100 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soliman, S.M.; Al-Rasheed, H.H.; El-Faham, A. Synthesis, X-ray Structure, Hirshfeld Analysis of Biologically Active Mn(II) Pincer Complexes Based on s-Triazine Ligands. Crystals 2020, 10, 931. https://doi.org/10.3390/cryst10100931
Soliman SM, Al-Rasheed HH, El-Faham A. Synthesis, X-ray Structure, Hirshfeld Analysis of Biologically Active Mn(II) Pincer Complexes Based on s-Triazine Ligands. Crystals. 2020; 10(10):931. https://doi.org/10.3390/cryst10100931
Chicago/Turabian StyleSoliman, Saied M., Hessa H. Al-Rasheed, and Ayman El-Faham. 2020. "Synthesis, X-ray Structure, Hirshfeld Analysis of Biologically Active Mn(II) Pincer Complexes Based on s-Triazine Ligands" Crystals 10, no. 10: 931. https://doi.org/10.3390/cryst10100931