Deciphering the Role of V88L Substitution in NDM-24 Metallo-β-Lactamase
Abstract
1. Introduction
2. Results and Discussion
2.1. Phylogenetic Analysis
2.2. Functional Study
2.3. Characteristics of Enzyme Activity
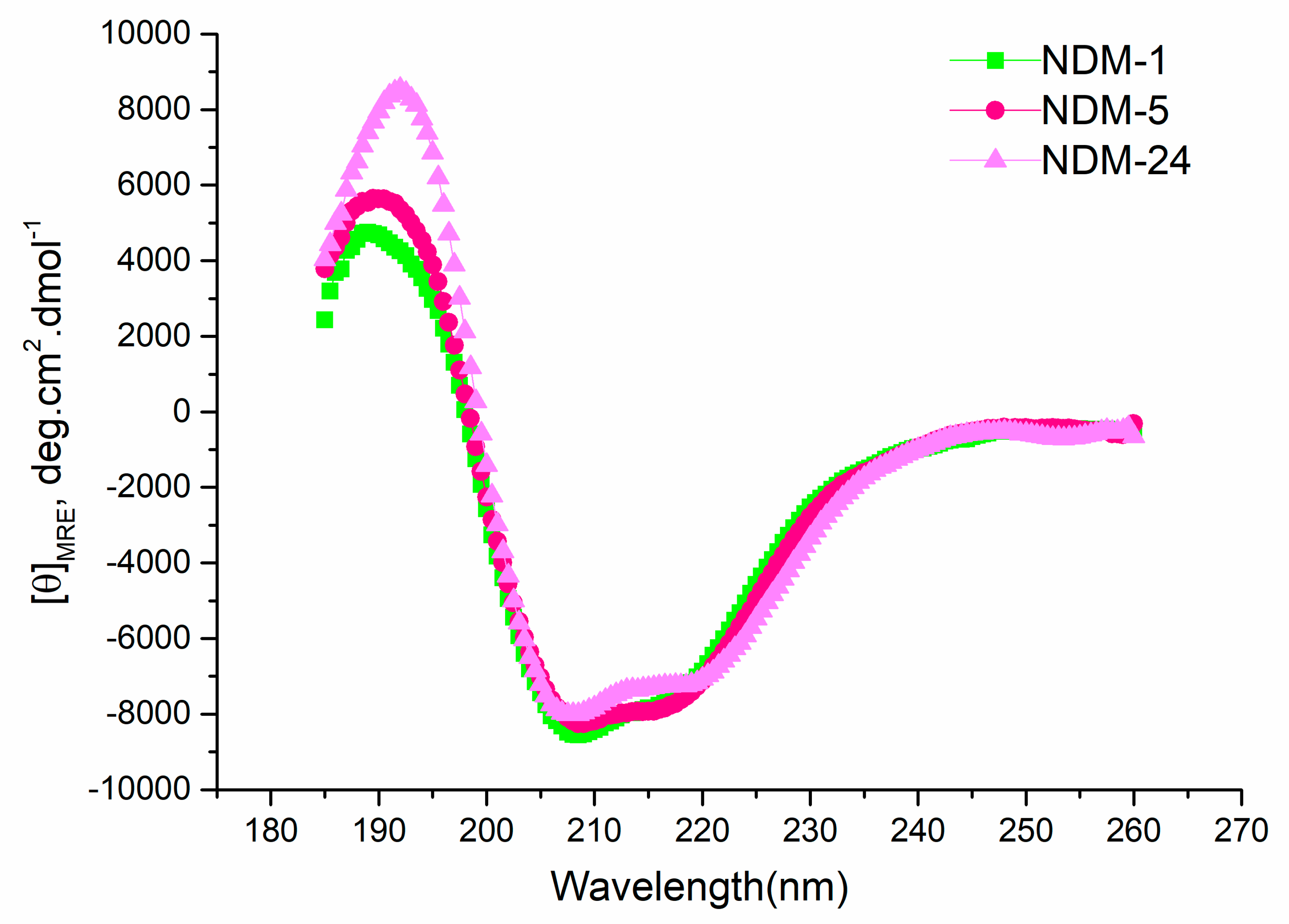
2.4. Thermal Stability
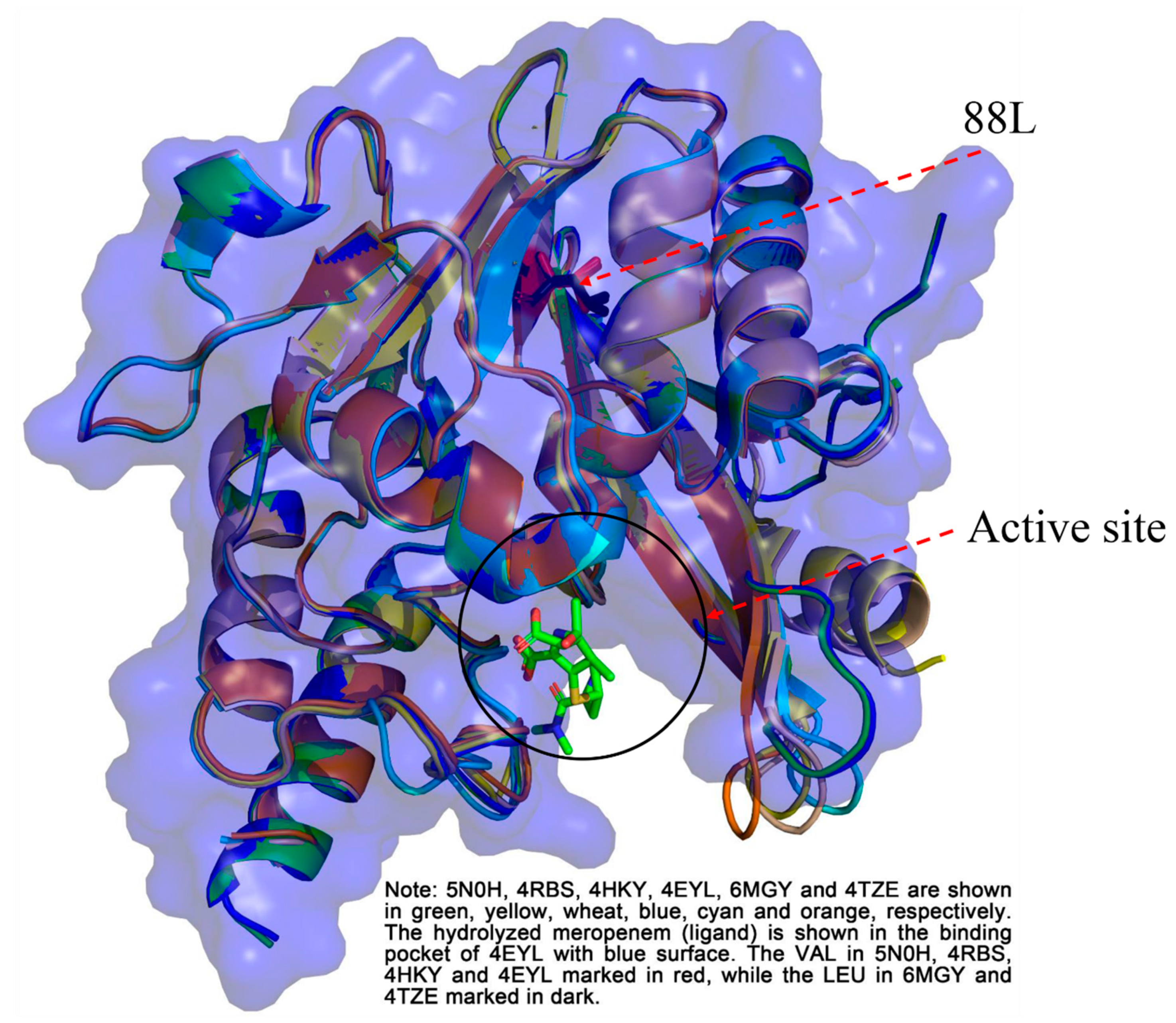
2.5. Structure Analysis
3. Material and Methods
3.1. Site-Directed Mutagenesis, Cloning and Expression of NDM Variants
3.2. Antimicrobial Susceptibility Tests
3.3. Production and Purification of NDM-1, NDM-5, and NDM-24
3.4. Determination of Kinetic Parameters
3.5. Circular Dichroism and Structure Analysis
3.6. Thermal Stability Testing
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Palzkill, T. Metallo-beta-lactamase structure and function. Ann. N. Y. Acad. Sci. 2013, 1277, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Garau, G.; García-Sáez, I.; Bebrone, C.; Anne, C.; Mercuri, P.; Galleni, M.; Frère, J.M.; Dideberg, O. Update of the standard numbering scheme for class B beta-lactamases. Antimicrob. Agents Chemother. 2004, 48, 2347–2349. [Google Scholar] [CrossRef] [PubMed]
- Yong, D.; Toleman, M.A.; Giske, C.G.; Cho, H.S.; Sundman, K.; Lee, K.; Walsh, T.R. Characterization of a New Metallo-β-Lactamase Gene, blaNDM-1, and a Novel Erythromycin Esterase Gene Carried on a Unique Genetic Structure in Klebsiella pneumoniae Sequence Type 14 from India. Antimicrob. Agents Chemother. 2009, 53, 5046–5054. [Google Scholar] [CrossRef] [PubMed]
- Bonnin, R.A.; Poirel, L.; Carattoli, A.; Nordmann, P. Characterization of an IncFII Plasmid Encoding NDM-1 from Escherichia coli ST131. PLoS ONE 2012, 7, 34752. [Google Scholar] [CrossRef] [PubMed]
- Dolejska, M.; Villa, L.; Poirel, L.; Nordmann, P.; Carattoli, A. Complete sequencing of an IncHI1 plasmid encoding the carbapenemase NDM-1, the ArmA 16S RNA methylase and a resistance-nodulation-cell division/multidrug efflux pump. J. Antimicrob. Chemother. 2013, 68, 34–39. [Google Scholar] [CrossRef] [PubMed]
- King, D.; Strynadka, N. Crystal structure of New Delhi metallo-betalactamase reveals molecular basis for antibiotic resistance. Protein Sci. 2011, 20, 1484–1491. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, L.J.; Bahr, G.; Nakashige, T.G.; Nolan, E.M.; Bonomo, R.A.; Vila, A.J. Membrane anchoring stabilizes and favors secretion of New Delhi metallo-beta-lactamase. Nat. Chem. Biol. 2016, 12, 516–522. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, L.J.; Bahr, G.; Vila, A.J. Lipidated beta-lactamases: From bench to bedside. Future Microbiol. 2016, 11, 1495–1498. [Google Scholar] [CrossRef]
- Khan, A.U.; Maryam, L.; Zarrilli, R. Structure, genetics and worldwide spread of New Delhi metallo-beta-lactamase (NDM): A threat to public health. BMC Microbiol. 2017, 17, 101. [Google Scholar] [CrossRef]
- Cheng, Z.; Thomas, P.W.; Ju, L.; Bergstrom, A.; Mason, K.; Clayton, D.; Miller, C.; Bethel, C.R.; Vanpelt, J.; Tierney, D.L.; et al. Evolution of New Delhi metallo-β-lactamase (NDM) in the clinic: Effects of NDM mutations on stability, zinc affinity, and mono-zinc activity. J. Biol. Chem. 2018, 293, 12606–12618. [Google Scholar] [CrossRef]
- Zhang, H.O.; Hau, Q. Crystal structure of NDM-1 reveals a common β-lactam hydrolysis mechanism. FASEB J. 2011, 25, 2574–2582. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Cunningham, M.A.; Mire, J.; Tesar, C.; Sacchettini, J.; Joachimiak, A. NDM-1, the ultimate promiscuous enzyme: Substrate recognition and catalytic mechanism. FASEB J. 2013, 27, 1917–1927. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Feng, Y.; McNally, A.; Zong, Z. bla NDM-21, a new variant of blaNDM in an Escherichia coli clinical isolate carrying blaCTX-M-55 and rmtB. J. Antimicrob. Chemother. 2018, 73, 2336–2339. [Google Scholar] [CrossRef] [PubMed]
- Bahr, G.; Vitor-Horen, L.; Bethel, C.R.; Bonomo, R.A.; González, L.J.; Vila, A.J. Clinical evolution of New Delhi Metallo-β-lactamase (NDM) optimizes resistance under Zn(II) deprivation. Antimicrob. Agents Chemother. 2018, 62, 1817–1849. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Li, J.; Wang, X.; Liu, D.; Ke, Y.; Wang, Y.; Shen, J. Novel Variant of New Delhi Metallo-β-lactamase, NDM-20, in Escherichia coli. Front. Microbiol. 2018, 9, 248. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wang, Y.; Walsh, T.R.; Liu, D.; Shen, Z.; Zhang, R.; Yin, W.; Yao, H.; Li, J.; Shen, J. Plasmid-mediated novel blaNDM-17 gene encoding a carbapenemase with enhanced activity in a ST48 Escherichia coli strain. Antimicrob. Agents Chemother. 2017, 61, 2216–2233. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, P.; Boulanger, A.E.; Poirel, L. NDM-4 Metallo-β-Lactamase with Increased Carbapenemase Activity from Escherichia coli. Antimicrob. Agents Chemother. 2012, 56, 2184–2186. [Google Scholar] [CrossRef]
- Zou, D.; Huang, Y.; Zhao, X.; Liu, W.; Dong, D.; Li, H.; Wang, X.; Huang, S.; Wei, X.; Yan, X.; et al. A Novel New Delhi Metallo-β-Lactamase Variant, NDM-14, Isolated in a Chinese Hospital Possesses Increased Enzymatic Activity against Carbapenems. Antimicrob. Agents Chemother. 2015, 59, 2450–2453. [Google Scholar] [CrossRef]
- Tada, T.; Miyoshi-Akiyama, T.; Dahal, R.K.; Sah, M.K.; Ohara, H.; Kirikae, T.; Pokhrel, B.M. NDM-8 Metallo-β-Lactamase in a Multidrug-Resistant Escherichia coli Strain Isolated in Nepal. Antimicrob. Agents Chemother. 2013, 57, 2394–2396. [Google Scholar] [CrossRef]
- Tada, T.; Shrestha, B.; Miyoshi-Akiyama, T.; Shimada, K.; Ohara, H.; Kirikae, T.; Pokhrel, B.M. NDM-12, a Novel New Delhi Metallo-β-Lactamase Variant from a Carbapenem-Resistant Escherichia coli Clinical Isolate in Nepal. Antimicrob. Agents Chemother. 2014, 58, 6302–6305. [Google Scholar] [CrossRef]
- Ines, S.; Emma, K.; Rudolf, R.; Rumyana, M.; Anne Marie, Q.; Adolf, B. VIM-15 and VIM-16, two new VIM-2-like metallo-beta-lactamases in Pseudomonas aeruginosa isolates from Bulgaria and Germany. Antimicrob. Agents Chemother. 2008, 52, 2977. [Google Scholar]
- Patricia, M.; Tomatis, P.E.; Mussi, M.A.; Fernando, P.; Viale, A.M.; Limansky, A.S.; Vila, A.J. Biochemical characterization of metallo-beta-lactamase VIM-11 from a Pseudomonas aeruginosa clinical strain. Antimicrob. Agents Chemother. 2008, 52, 2250. [Google Scholar]
- Jose-Manuel, R.M.; Patrice, N.; Nicolas, F.; Laurent, P. VIM-19, a metallo-beta-lactamase with increased carbapenemase activity from Escherichia coli and Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2010, 54, 471–476. [Google Scholar]
- Pierre, B.; Carine, B.; Te-Din, H.; Warda, B.; Yves, D.; Ariane, D.; Kurt, H.; Youri, G. Detection and characterization of VIM-31, a new variant of VIM-2 with Tyr224His and His252Arg mutations, in a clinical isolate of Enterobacter cloacae. Antimicrob. Agents Chemother. 2012, 56, 3283. [Google Scholar]
- Corbin, B.D.; Seeley, E.H.; Raab, A.; Feldmann, J.; Miller, M.R.; Torres, V.J.; Anderson, K.L.; Dattilo, B.M.; Dunman, P.M.; Gerads, R.; et al. Metal Chelation and Inhibition of Bacterial Growth in Tissue Abscesses. Science 2008, 319, 962–965. [Google Scholar] [CrossRef] [PubMed]
- Kumar, G.; Issa, B.; Kar, D.; Biswal, S.; Ghosh, A.S. E152A substitution drastically affects NDM-5 activity. FEMS Microbiol. Lett. 2017, 364. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Azam, M.W.; Khan, A.U. Non-active site mutation (Q123A) in New Delhi metallo-β-lactamase (NDM-1) enhanced its enzyme activity. Int. J. Biol. Macromol. 2018, 112, 1272–1277. [Google Scholar] [CrossRef] [PubMed]
- Carfi, A.; Pares, S.; Duée, E.; Galleni, M.; Duez, C.; Frère, J.M.; Dideberg, O. The 3-D structure of a zinc metallo-beta-lactamase from Bacillus cereus reveals a new type of protein fold. EMBO J. 1995, 14, 4914–4921. [Google Scholar] [CrossRef] [PubMed]
- Piccirilli, A.; Brisdelli, F.; Aschi, M.; Celenza, G.; Amicosante, G.; Perilli, M. Kinetic Profile and Molecular Dynamic Studies Show that Y229W Substitution in an NDM-1/L209F Variant Restores the Hydrolytic Activity of the Enzyme toward Penicillins, Cephalosporins, and Carbapenems. Antimicrob. Agents Chemother. 2019, 63, e02270-18. [Google Scholar] [CrossRef]
- Meini, M.-R.; Tomatis, P.E.; Weinreich, D.M.; Vila, A.J. Quantitative Description of a Protein Fitness Landscape Based on Molecular Features. Mol. Boil. Evol. 2015, 32, 1774–1787. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically: Approved Standard, 11th ed.; CLSI document M07; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 28th ed.; CLSI document M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- Crowder, M.W.; Walsh, T.R.; Banovic, L.; Pettit, M.; Spencer, J. Overexpression, purification, and characterization of the cloned metallo-beta-lactamase L1 from Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 1998, 42, 921. [Google Scholar] [CrossRef] [PubMed]
- De Meester, F.; Joris, B.; Reckinger, G. Automated analysis of enzyme inactivation phenomena. Application to β-lactamases and DD-peptidases. Biochem. Pharmacol. 1987, 36, 2393–2403. [Google Scholar] [CrossRef]
- Segel, I.H. Biochemical Calculations, 2nd ed.; John Wiley & Sons: New York, NY, USA, 1976; pp. 236–241. [Google Scholar]
- Liu, Z.; Zhang, R.; Li, W.; Yang, L.; Liu, D.; Wang, S.; Shen, J.; Wang, Y. Amino acid changes at the VIM-48 C-terminus result in increased carbapenem resistance, enzyme activity and protein stability. J. Antimicrob. Chemother. 2018, 74, 885–893. [Google Scholar] [CrossRef] [PubMed]
- Raussens, V.; Ruysschaert, J.-M.; Goormaghtigh, E. Protein concentration is not an absolute prerequisite for the determination of secondary structure from circular dichroism spectra: A new scaling method. Anal. Biochem. 2003, 319, 114–121. [Google Scholar] [CrossRef]
- Sreerama, N.; Venyaminov, S.Y.; Woody, R.W. Estimation of the number of α-helical and β-strand segments in proteins using circular dichroism spectroscopy. Protein Sci. 1999, 8, 370–380. [Google Scholar] [CrossRef] [PubMed]
- Sreerama, N.; Venyaminov, S.Y.; Woody, R.W. Analysis of Protein Circular Dichroism Spectra Based on the Tertiary Structure Classification. Anal. Biochem. 2001, 299, 271–274. [Google Scholar] [CrossRef] [PubMed]


| Antibiotic | MIC (μg/mL) | |||
|---|---|---|---|---|
| E. coli DH5α/pHSG398 | E. coli DH5α/pHSG398-NDM-24 | E. coli DH5α/pHSG398-NDM-1 | E. coli DH5α/pHSG398-NDM-5 | |
| Ampicillin | 2 | >256 | >256 | >256 |
| Penicillin G | 16 | >256 | >256 | >256 |
| Aztreonam | 0.031 | 0.031 | 0.031 | 0.031 |
| Cefepime | 0.031 | 2 | 1 | 2 |
| Cefotaxime | 0.062 | 32 | 64 | 64 |
| Cefoxitin | 2 | 128 | 128 | 128 |
| Ceftazidime | 0.125 | 256 | 256 | 256 |
| Cefazolin | 2 | 128 | 128 | 256 |
| Ertapenem | 0.015 | 1 | 0.25 | 2 |
| Imipenem | 0.062 | 2 | 2 | 2 |
| Meropenem | 0.031 | 1 | 1 | 2 |
| Kinetic Parameters | Enzyme | β-Lactams b | ||||||
|---|---|---|---|---|---|---|---|---|
| AMP | PEN | TAG | FEP | MEM | IPM | ETP | ||
| Km(μM) | NDM-24 | 638.79 ± 23.86 | 331.30 ± 29.43 | 173.85 ± 9.73 | 318.93 ± 10.86 | 266.24 ± 27.03 | 338.20 ± 24.23 | 125.23 ± 19.08 |
| NDM-1 | 1249.98 ± 210.94 | 224.57 ± 13.57 | 213.90 ± 11.01 | 173.55 ± 19.46 | 284.24 ± 7.87 | 234.83 ± 7.44 | 105.54 ± 3.09 | |
| NDM-5 | 825.00 ± 0.29 | 315.21 ± 46.68 | 76.45 ± 4.76 | 179.64 ± 12.19 | 275.16 ± 36.87 | 292.97 ± 13.76 | 82.18 ± 3.86 | |
| kcat (s−1) | NDM-24 | 259.94 ± 23.52 | 179.10 ± 8.17 | 43.13 ± 1.06 | 22.98 ± 0.34 | 151.75 ± 6.69 | 173.16 ± 8.83 | 110.31 ± 7.62 |
| NDM-1 | 254.34 ± 28.96 | 79.28 ± 1.96 | 26.73 ± 0.71 | 8.42 ± 0.63 | 75.18 ± 3.44 | 79.81 ± 5.15 | 62.89 ± 1.15 | |
| NDM-5 | 346.13 ± 31.30 | 214.13 ± 12.11 | 26.96 ± 0.75 | 13.05 ± 0.24 | 142.48 ± 17.91 | 149.63 ± 2.02 | 83.18 ± 1.67 | |
| kcat/Km (μM−1 s−1) | NDM-24 | 0.41 | 0.54 | 0.25 | 0.072 | 0.57 | 0.51 | 0.88 |
| NDM-1 | 0.20 | 0.35 | 0.13 | 0.046 | 0.26 | 0.34 | 0.60 | |
| NDM-5 | 0.40 | 0.68 | 0.35 | 0.073 | 0.52 | 0.51 | 1.01 | |
| kcat/Km (μM−1 s−1) ratio for: | NDM-24/NDM-1 | 2.00 | 1.53 | 1.98 | 1.49 | 2.16 | 1.51 | 1.46 |
| NDM-5/NDM-24 | 1.03 | 1.26 | 1.42 | 1.01 | 0.91 | 1.00 | 1.15 | |
| NDM-5/NDM-1 | 2.07 | 1.92 | 2.82 | 1.50 | 1.96 | 1.50 | 1.68 | |
| Program Algorithms a | Structural Elements b | SMP50(9) c | SP37(3) c | SP29(1) c | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| NDM-1 | NDM-5 | NDM-24 | NDM-1 | NDM-5 | NDM-24 | NDM-1 | NDM-5 | NDM-24 | ||
| SELCON3 | H(r) | 0.070 | 0.078 | 0.092 | 0.062 | 0.074 | 0.092 | 0.059 | 0.079 | 0.087 |
| H(d) | 0.085 | 0.088 | 0.089 | 0.081 | 0.088 | 0.089 | 0.078 | 0.087 | 0.086 | |
| S(r) | 0.215 | 0.199 | 0.195 | 0.228 | 0.214 | 0.195 | 0.231 | 0.191 | 0.196 | |
| S(d) | 0.115 | 0.109 | 0.108 | 0.117 | 0.113 | 0.108 | 0.118 | 0.107 | 0.108 | |
| Trn | 0.214 | 0.211 | 0.194 | 0.218 | 0.214 | 0.194 | 0.226 | 0.214 | 0.215 | |
| Unrd | 0.284 | 0.287 | 0.261 | 0.282 | 0.279 | 0.261 | 0.287 | 0.292 | 0.285 | |
| H(r)+H(d) | 0.155 | 0.166 | 0.181 | 0.143 | 0.162 | 0.181 | 0.137 | 0.166 | 0.173 | |
| S(r)+S(d) | 0.33 | 0.308 | 0.303 | 0.345 | 0.327 | 0.303 | 0.349 | 0.298 | 0.304 | |
| CONTINLL | H(r) | 0.054 | 0.075 | 0.091 | 0.046 | 0.079 | 0.097 | 0.071 | 0.078 | 0.093 |
| H(d) | 0.079 | 0.092 | 0.101 | 0.089 | 0.095 | 0.103 | 0.092 | 0.096 | 0.100 | |
| S(r) | 0.217 | 0.208 | 0.187 | 0.202 | 0.205 | 0.182 | 0.197 | 0.197 | 0.183 | |
| S(d) | 0.114 | 0.113 | 0.108 | 0.112 | 0.111 | 0.107 | 0.113 | 0.111 | 0.107 | |
| Trn | 0.233 | 0.220 | 0.220 | 0.248 | 0.216 | 0.216 | 0.231 | 0.222 | 0.225 | |
| Unrd | 0.303 | 0.292 | 0.293 | 0.304 | 0.293 | 0.294 | 0.297 | 0.297 | 0.292 | |
| H(r)+H(d) | 0.133 | 0.167 | 0.192 | 0.135 | 0.174 | 0.200 | 0.163 | 0.174 | 0.193 | |
| S(r)+S(d) | 0.331 | 0.321 | 0.295 | 0.314 | 0.316 | 0.289 | 0.310 | 0.308 | 0.290 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Piccirilli, A.; Liu, D.; Li, W.; Wang, Y.; Shen, J. Deciphering the Role of V88L Substitution in NDM-24 Metallo-β-Lactamase. Catalysts 2019, 9, 744. https://doi.org/10.3390/catal9090744
Liu Z, Piccirilli A, Liu D, Li W, Wang Y, Shen J. Deciphering the Role of V88L Substitution in NDM-24 Metallo-β-Lactamase. Catalysts. 2019; 9(9):744. https://doi.org/10.3390/catal9090744
Chicago/Turabian StyleLiu, Zhihai, Alessandra Piccirilli, Dejun Liu, Wan Li, Yang Wang, and Jianzhong Shen. 2019. "Deciphering the Role of V88L Substitution in NDM-24 Metallo-β-Lactamase" Catalysts 9, no. 9: 744. https://doi.org/10.3390/catal9090744
APA StyleLiu, Z., Piccirilli, A., Liu, D., Li, W., Wang, Y., & Shen, J. (2019). Deciphering the Role of V88L Substitution in NDM-24 Metallo-β-Lactamase. Catalysts, 9(9), 744. https://doi.org/10.3390/catal9090744





