Enhanced (−)-α-Bisabolol Productivity by Efficient Conversion of Mevalonate in Escherichia coli
Abstract
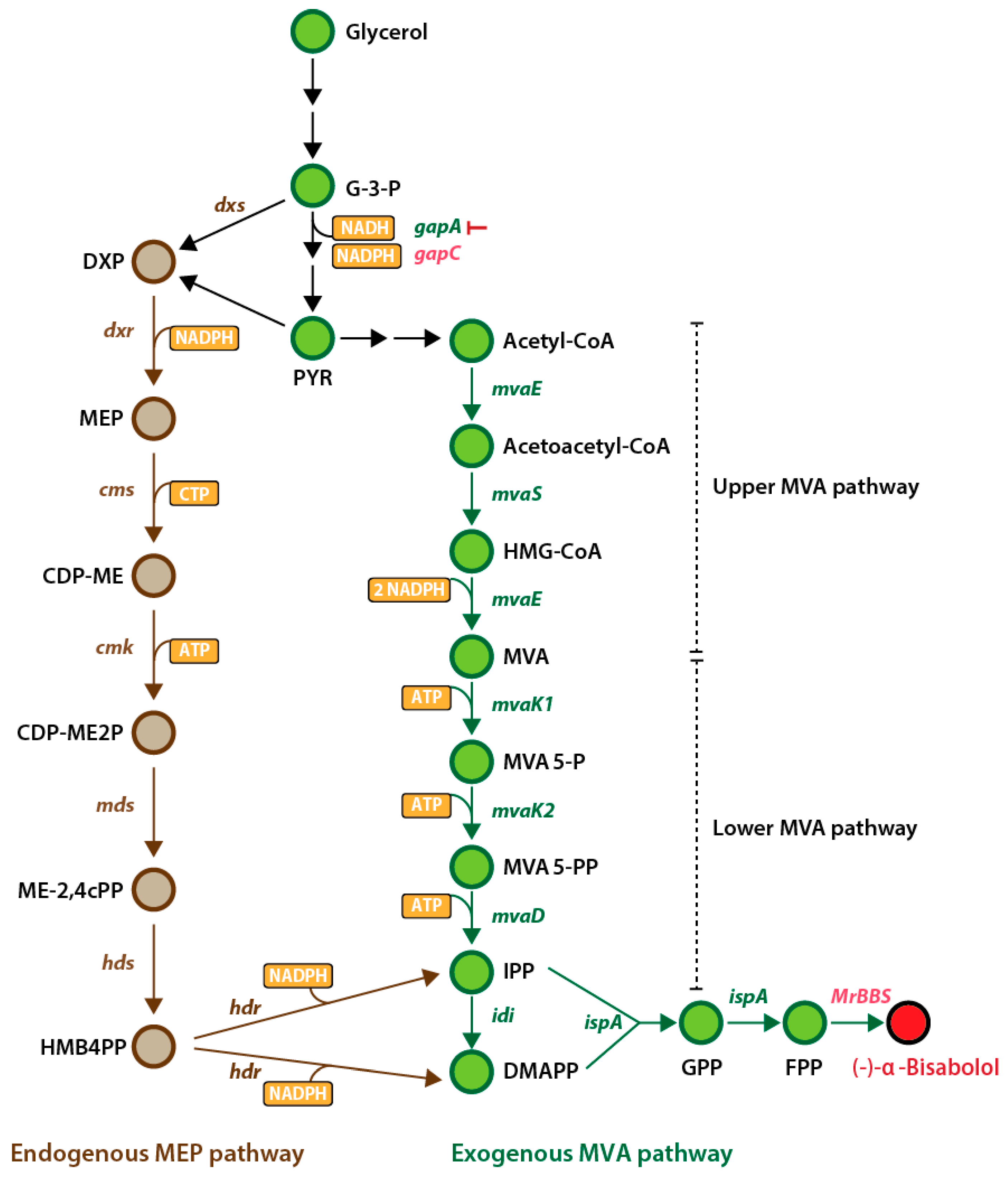
1. Introduction
2. Results
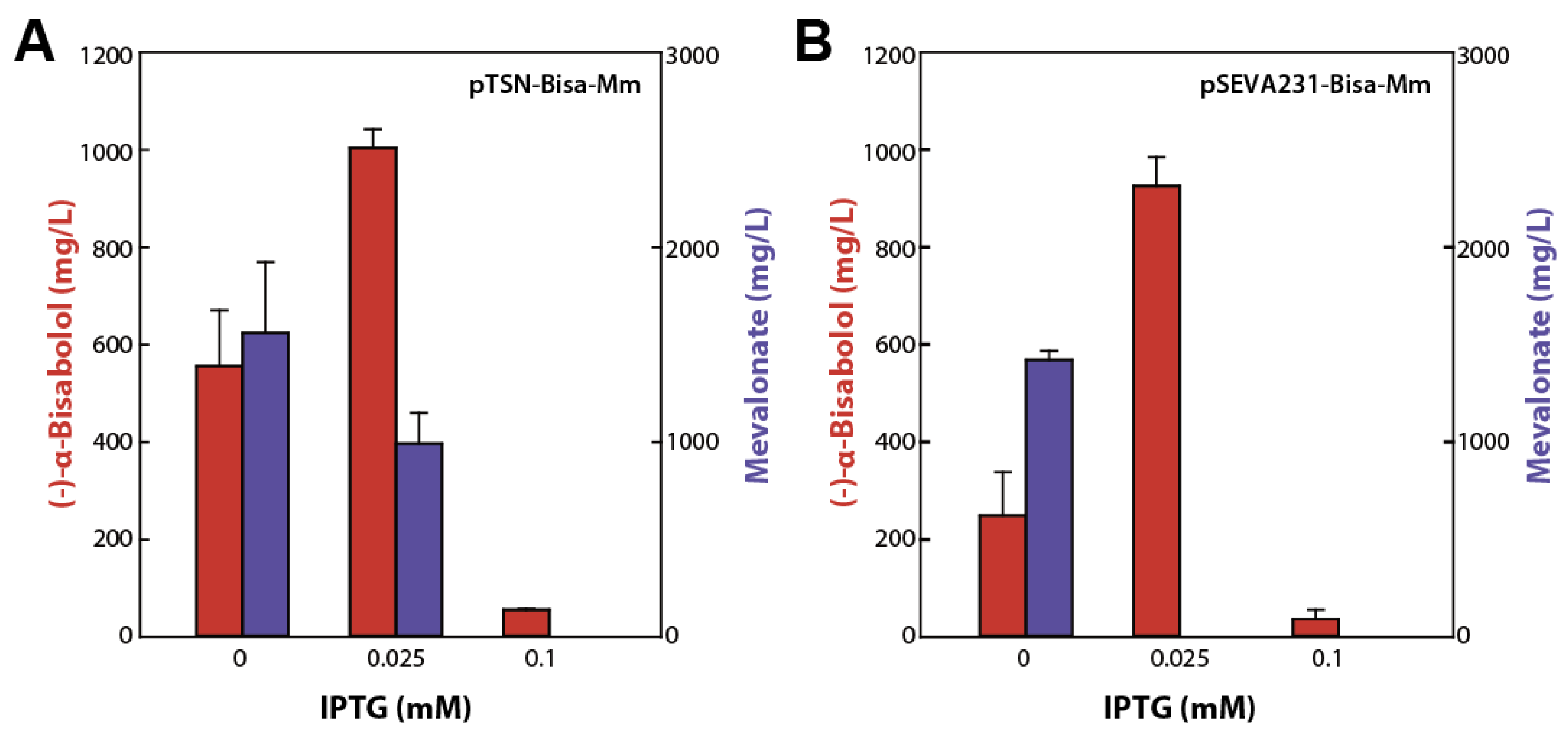
2.1. Feedback-Resistant MvaK1
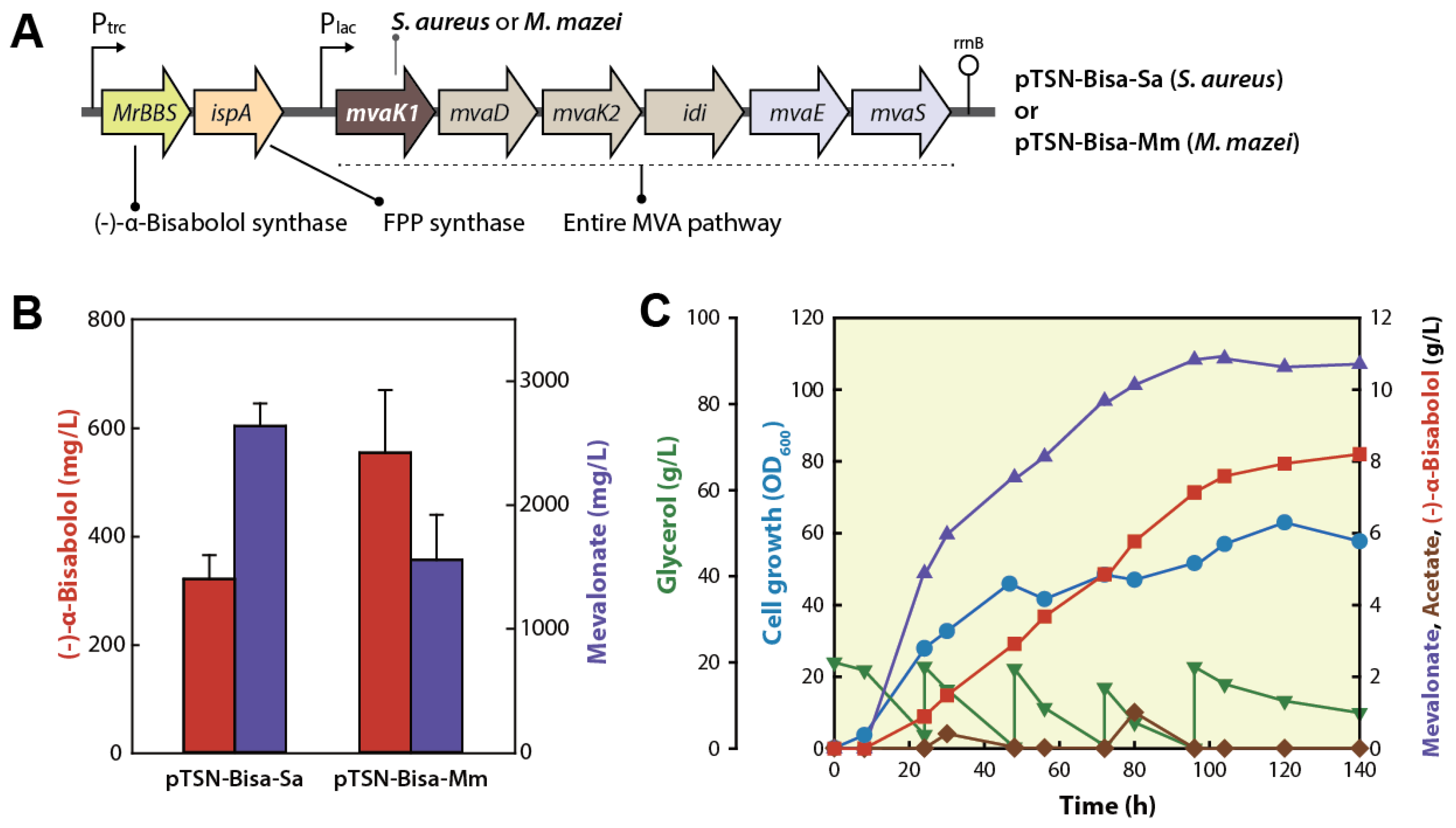
2.2. Overexpression of Entire MVA Pathway Genes
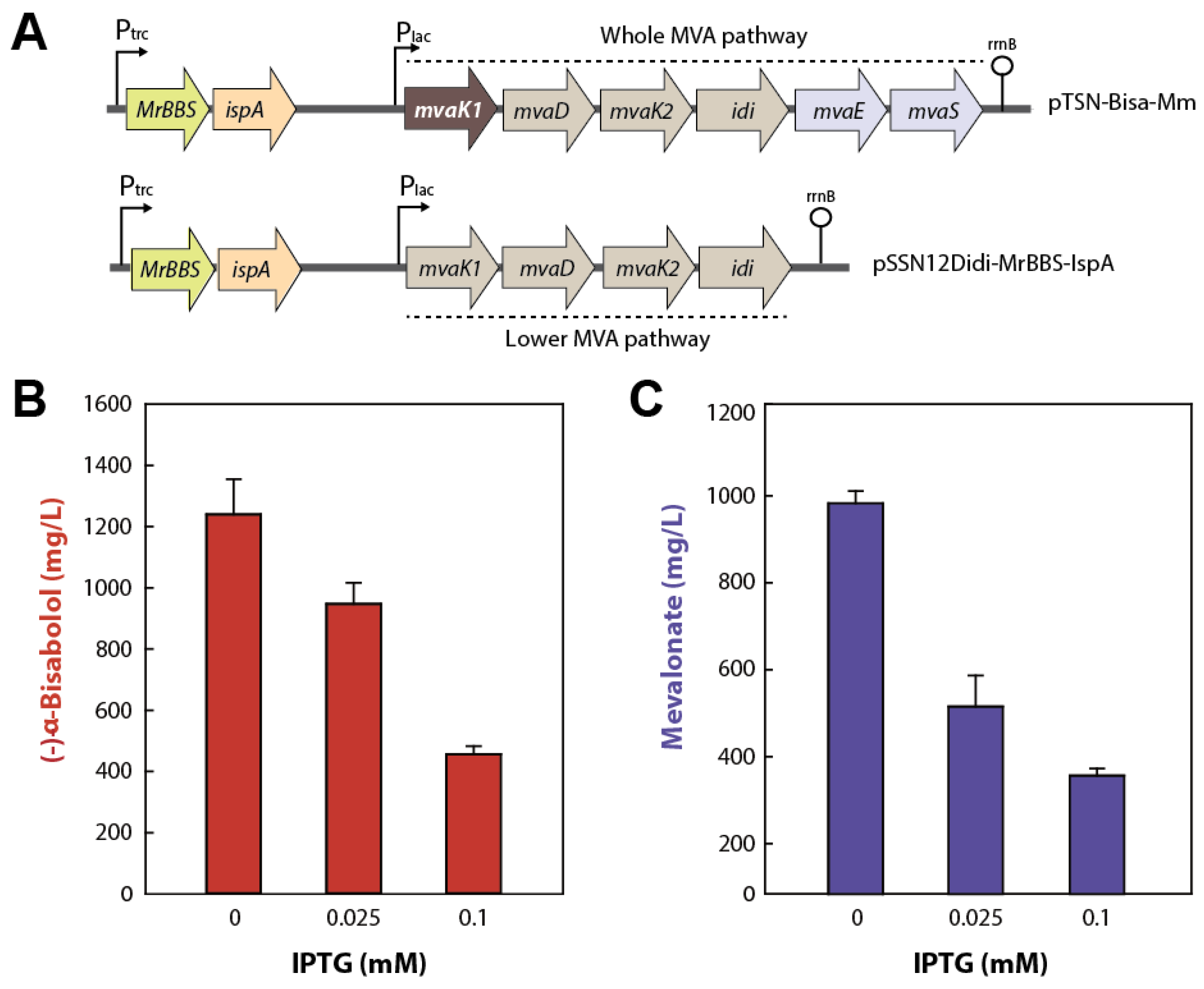
2.3. Reinforcement of the MVA Pathway
2.4. Sufficient Supply of reduced Nicotinamide Adenine Dinucleotide Phosphate (NADPH)
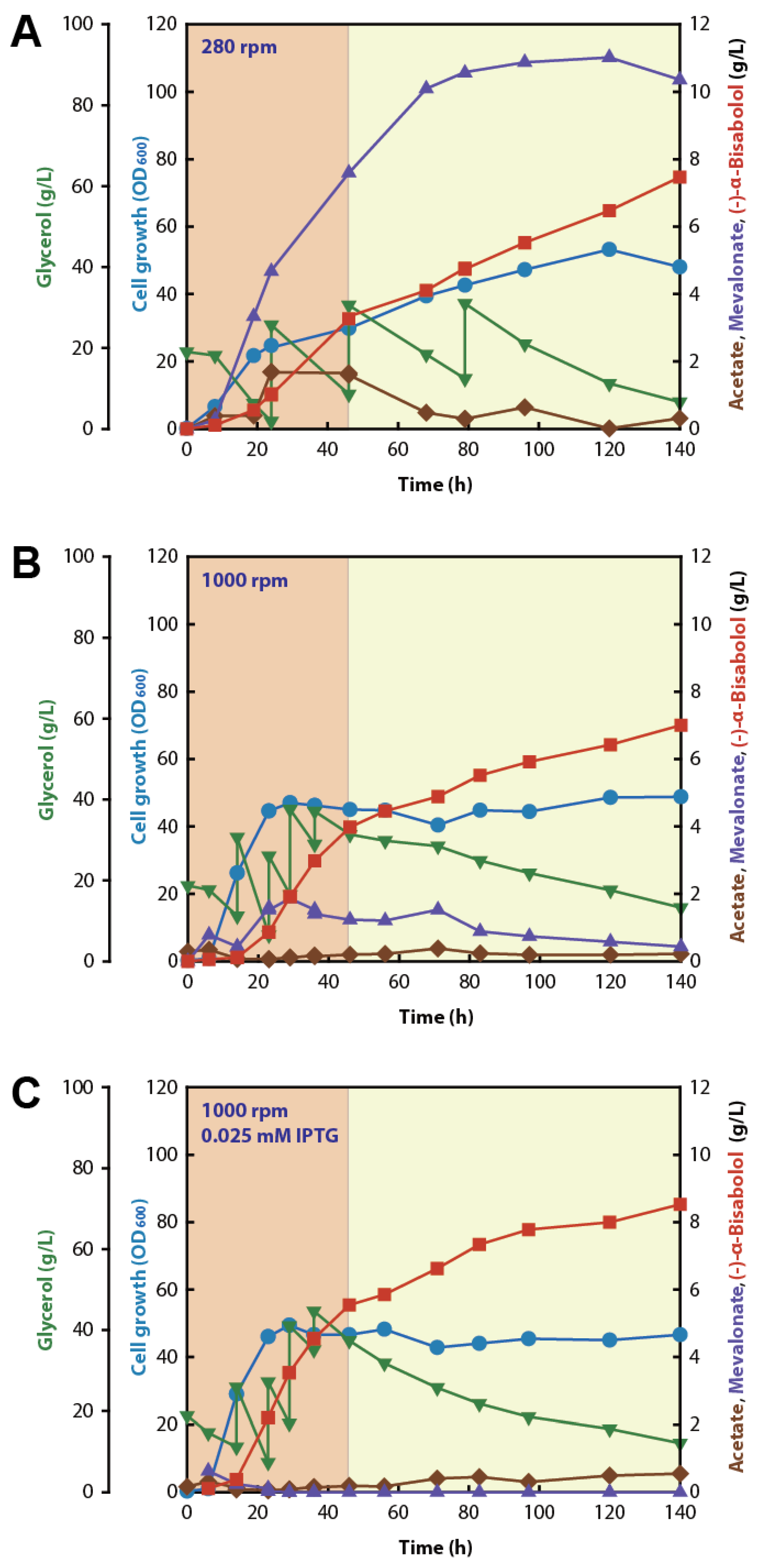
2.5. Effect of Aeration on (−)-α-Bisabolol Fermentation
3. Discussion
4. Materials and Methods
4.1. Strains and Culture Media
4.2. Plasmid Construction
4.3. Batch and Fed-Batch Fermentation
4.4. (−)-α-Bisabolol Quantification
4.5. Determination of Cell Growth and Metabolites
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Brehm-Stecher, B.F.; Johnson, E.A. Sensitization of Staphylococcus aureus and Escherichia coli to antibiotics by the sesquiterpenoids nerolidol, farnesol, bisabolol, and apritone. Antimicrob. Agents Chemother. 2003, 47, 3357–3360. [Google Scholar] [CrossRef] [PubMed]
- Forrer, M.; Kulik, E.M.; Filippi, A.; Waltimo, T. The antimicrobial activity of α-bisabolol and tea tree oil against Solobacterium moorei, a gram-positive bacterium associated with halitosis. Arch. Oral Biol. 2013, 58, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Kamatou, G.P.; Viljoen, A.M. A review of the application and pharmacological properties of α-bisabolol and α-bisabolol-rich oils. J. Am. Oil Chem. Soc. 2010, 87, 1–7. [Google Scholar] [CrossRef]
- Leite, G.d.O.; Leite, L.H.; Sampaio, R.d.S.; Araruna, M.K.A.; de Menezes, I.R.A.; da Costa, J.G.M.; Campos, A.R. (−)-α-Bisabolol attenuates visceral nociception and inflammation in mice. Fitoterapia 2011, 82, 208–211. [Google Scholar] [CrossRef] [PubMed]
- Russell, K.; Jacob, S.E. Bisabolol. Dermatitis 2010, 21, 57–58. [Google Scholar]
- Global One-Stop Reports Center. Available online: Http://www.Gosreports.Com/global-%ce%b1-bisabolol-market-worth-73-million-by-2020/ (accessed on 30 march 2019).
- Albertti, L.A.G.; Delatte, T.L.; de Farias, K.S.; Boaretto, A.G.; Verstappen, F.; van Houwelingen, A.; Cankar, K.; Carollo, C.A.; Bouwmeester, H.J.; Beekwilder, J. Identification of the bisabolol synthase in the endangered candeia tree (Eremanthus erythropappus (dc) mcleisch). Front. Plant Sci. 2018, 9, 1340. [Google Scholar] [CrossRef]
- de Souza, A.T.; Benazzi, T.L.; Grings, M.B.; Cabral, V.; da Silva, E.A.; Cardozo-Filho, L.; Antunes, O.A.C. Supercritical extraction process and phase equilibrium of candeia (Eremanthus erythropappus) oil using supercritical carbon dioxide. J. Supercrit. Fluids 2008, 47, 182–187. [Google Scholar] [CrossRef]
- Son, Y.J.; Kwon, M.; Ro, D.K.; Kim, S.U. Enantioselective microbial synthesis of the indigenous natural product (−)-α-bisabolol by a sesquiterpene synthase from chamomile (Matricaria recutita). Biochem. J. 2014, 463, 239–248. [Google Scholar] [CrossRef]
- Chatzivasileiou, A.O.; Stephanopoulos, G.; Ward, V.C.A. Metabolic engineering of Escherichia coli for the production of isoprenoids. FEMS Microbiol. Lett. 2018, 365, fny079. [Google Scholar]
- Ajikumar, P.K.; Xiao, W.H.; Tyo, K.E.; Wang, Y.; Simeon, F.; Leonard, E.; Mucha, O.; Phon, T.H.; Pfeifer, B.; Stephanopoulos, G. Isoprenoid pathway optimization for taxol precursor overproduction in Escherichia coli. Science 2010, 330, 70–74. [Google Scholar] [CrossRef]
- Han, G.H.; Kim, S.K.; Yoon, P.K.-S.; Kang, Y.; Kim, B.S.; Fu, Y.; Sung, B.H.; Jung, H.C.; Lee, D.H.; Kim, S.W. Fermentative production and direct extraction of (−)-α-bisabolol in metabolically engineered Escherichia coli. Microb. Cell Fact. 2016, 15, 185. [Google Scholar] [CrossRef]
- Voynova, N.E.; Rios, S.E.; Miziorko, H.M. Staphylococcus aureus mevalonate kinase: Isolation and characterization of an enzyme of the isoprenoid biosynthetic pathway. J. Bacteriol. 2004, 186, 61–67. [Google Scholar] [CrossRef]
- Andreassi, J.L.; Dabovic, K.; Leyh, T.S. Streptococcus pneumoniae isoprenoid biosynthesis is downregulated by diphosphomevalonate: An antimicrobial target. Biochemistry 2004, 43, 16461–16466. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.K.; Kim, S.H.; Subhadra, B.; Woo, S.G.; Rha, E.; Kim, S.W.; Kim, H.; Lee, D.H.; Lee, S.G. A genetically encoded biosensor for monitoring isoprene production in engineered Escherichia coli. ACS Synth. Biol. 2018, 7, 2379–2390. [Google Scholar] [CrossRef]
- Kim, S.K.; Han, G.H.; Seong, W.; Kim, H.; Kim, S.W.; Lee, D.H.; Lee, S.G. CRISPR interference-guided balancing of a biosynthetic mevalonate pathway increases terpenoid production. Metab. Eng. 2016, 38, 228–240. [Google Scholar] [CrossRef] [PubMed]
- Primak, Y.A.; Du, M.; Miller, M.C.; Wells, D.H.; Nielsen, A.T.; Weyler, W.; Beck, Z.Q. Characterization of a feedback-resistant mevalonate kinase from the archaeon Methanosarcina mazei. Appl. Environ. Microbiol. 2011, 77, 7772–7778. [Google Scholar] [CrossRef]
- Kang, A.; George, K.W.; Wang, G.; Baidoo, E.; Keasling, J.D.; Lee, T.S. Isopentenyl diphosphate (IPP)-bypass mevalonate pathways for isopentenol production. Metab. Eng. 2016, 34, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Pontrelli, S.; Chiu, T.Y.; Lan, E.I.; Chen, F.Y.; Chang, P.C.; Liao, J.C. Escherichia coli as a host for metabolic engineering. Metab. Eng. 2018, 50, 16–46. [Google Scholar] [CrossRef]
- Kazieva, E.; Yamamoto, Y.; Tajima, Y.; Yokoyama, K.; Katashkina, J.; Nishio, Y. Characterization of feedback-resistant mevalonate kinases from the methanogenic archaeons Methanosaeta concilii and Methanocella paludicola. Microbiology 2017, 163, 1283–1291. [Google Scholar] [CrossRef]
- Martin, V.J.; Pitera, D.J.; Withers, S.T.; Newman, J.D.; Keasling, J.D. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat. Biotechnol. 2003, 21, 796–802. [Google Scholar] [CrossRef] [PubMed]
- Kizer, L.; Pitera, D.J.; Pfleger, B.F.; Keasling, J.D. Application of functional genomics to pathway optimization for increased isoprenoid production. Appl. Environ. Microbiol. 2008, 74, 3229–3241. [Google Scholar] [CrossRef] [PubMed]
- Dahl, R.H.; Zhang, F.; Alonso-Gutierrez, J.; Baidoo, E.; Batth, T.S.; Redding-Johanson, A.M.; Petzold, C.J.; Mukhopadhyay, A.; Lee, T.S.; Adams, P.D. Engineering dynamic pathway regulation using stress-response promoters. Nat. Biotechnol. 2013, 31, 1039–1046. [Google Scholar] [CrossRef]
- Liu, C.L.; Dong, H.G.; Zhan, J.; Liu, X.; Yang, Y. Multi-modular engineering for renewable production of isoprene via mevalonate pathway in Escherichia coli. J. Appl. Microbiol. 2019, 126, 1128–1139. [Google Scholar] [CrossRef]
- Zhao, J.; Li, Q.; Sun, T.; Zhu, X.; Xu, H.; Tang, J.; Zhang, X.; Ma, Y. Engineering central metabolic modules of Escherichia coli for improving β-carotene production. Metab. Eng. 2013, 17, 42–50. [Google Scholar] [CrossRef]
- Choi, H.S.; Lee, S.Y.; Kim, T.Y.; Woo, H.M. In silico identification of gene amplification targets for improvement of lycopene production. Appl. Environ. Microbiol. 2010, 76, 3097–3105. [Google Scholar] [CrossRef] [PubMed]
- Alper, H.; Jin, Y.S.; Moxley, J.F.; Stephanopoulos, G. Identifying gene targets for the metabolic engineering of lycopene biosynthesis in Escherichia coli. Metab. Eng. 2005, 7, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Martinez, I.; Zhu, J.; Lin, H.; Bennett, G.N.; San, K.Y. Replacing Escherichia coli NAD-dependent glyceraldehyde 3-phosphate dehydrogenase (GAPDH) with a NADP-dependent enzyme from Clostridium acetobutylicum facilitates NADPH dependent pathways. Metab. Eng. 2008, 10, 352–359. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual; Sambrook, J., Russell, D.W., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001; Volume 3. [Google Scholar]
- Gibson, D.G.; Young, L.; Chuang, R.Y.; Venter, J.C.; Hutchison III, C.A.; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343. [Google Scholar] [CrossRef]

| Plasmids | Agitation (rpm) | IPTG (mM) | Final Titer (g/L) | Initial Yield * (g/g) | Initial Productivity * (g/L h) |
|---|---|---|---|---|---|
| pTSN-Bisa-Mm | 280 | 0 | 8.2 | 0.08 | 0.06 |
| pTSN-Bisa-Mm pSSN12Didi-MrBBS-IspA | 280 | 0 | 7.5 | 0.10 | 0.07 |
| 1000 | 0 | 7.0 | 0.07 | 0.09 | |
| 1000 | 0.025 | 8.5 | 0.11 | 0.12 | |
| pTSN-Bisa-Mm-GapC pSSN12Didi-MrBBS-IspA | 1000 | 0.025 | 5.3 | 0.07 | 0.07 |
| pTSN-Bisa-Mm-GapC pSSN12Didi-MrBBS-IspA pdCas9-sgRNA-GapA | 1000 | 0.025 | 3.6 | 0.05 | 0.05 |
| Name | Description | References |
|---|---|---|
| Strains | ||
| DH5α | F−, Φ80lacZ·ΔM15·ƒ(lacZYA−argF)U169 deoR recA1 endA1 hsdR17(rk−, mk+) phoA supE44 thi-1 gyrA96 relA1 | Enzynomics |
| Plasmids | ||
| pTrc99A | Ptrc promoter, AmpR, lacIq, pBR322 ori | GE Healthcare |
| pSTV28 | Plac promoter, CmR, p15A ori | Takara |
| pSECRi | PrhaBAD::cas9(D10A, H840A) and constitutive sgRNA expression cassette in pSEVA221 | [16] |
| pSNA-MrBBS-IspA | pTrc99A containing mvaE and mvaS of Enterococcus faecalis, mvaK1 and mvaK2 and mvaD of S. pneumoniae, idi, and ispA of E. coli, MrBBS of M. recutita | [12] |
| pTM-BBS | pTrc99A derivatives containing codon optimized Matricaria recutita MrBBS, mvaK1 of S. aureus, mvaD and mvaK2 of S. pneumoniae, idi of E. coli, mvaE and mvaS of E. faecali | [16] |
| pSSN12Didi | pSTV28 containing mvaK1, mvaK2 and mvaD from Streptococcus pneumoniae, idi of E. coli | [12] |
| pTSN-Bisa-Sa | pTrc99A containing mvaE and mvaS of Enterococcus faecalis, mvaK1 of S. aureus, mvaK2, and mvaD of S. pneumoniae, idi, and ispA of E. coli, MrBBS of M. recutita | This study |
| pTSN-Bisa-Mm | pTrc99A containing mvaE and mvaS of Enterococcus faecalis, mvaK1 of M. masei, mvaK2, and mvaD of S. pneumoniae, idi, and ispA of E. coli, MrBBS of M. recutita | This study |
| pSEVA231-Bisa-Mm | pTSN-Bisa-Mm with pBBR1 ori instead of pBR322 ori | This study |
| pSSN12Didi-MrBBS-IspA | pSSN12Didi containing ispA of E. coli and MrBBS of M. recutita | This study |
| pTSN-Bisa-Mm-GapC | pTSN-Bisa-Mm containing gapC of C. acetoburylicum | This study |
| pdCas9-sgRNA-GapA | pSECRi containing gRNA targeting gapA gene | This study |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.-J.; Kim, S.K.; Seong, W.; Woo, S.-G.; Lee, H.; Yeom, S.-J.; Kim, H.; Lee, D.-H.; Lee, S.-G. Enhanced (−)-α-Bisabolol Productivity by Efficient Conversion of Mevalonate in Escherichia coli. Catalysts 2019, 9, 432. https://doi.org/10.3390/catal9050432
Kim S-J, Kim SK, Seong W, Woo S-G, Lee H, Yeom S-J, Kim H, Lee D-H, Lee S-G. Enhanced (−)-α-Bisabolol Productivity by Efficient Conversion of Mevalonate in Escherichia coli. Catalysts. 2019; 9(5):432. https://doi.org/10.3390/catal9050432
Chicago/Turabian StyleKim, Soo-Jung, Seong Keun Kim, Wonjae Seong, Seung-Gyun Woo, Hyewon Lee, Soo-Jin Yeom, Haseong Kim, Dae-Hee Lee, and Seung-Goo Lee. 2019. "Enhanced (−)-α-Bisabolol Productivity by Efficient Conversion of Mevalonate in Escherichia coli" Catalysts 9, no. 5: 432. https://doi.org/10.3390/catal9050432
APA StyleKim, S.-J., Kim, S. K., Seong, W., Woo, S.-G., Lee, H., Yeom, S.-J., Kim, H., Lee, D.-H., & Lee, S.-G. (2019). Enhanced (−)-α-Bisabolol Productivity by Efficient Conversion of Mevalonate in Escherichia coli. Catalysts, 9(5), 432. https://doi.org/10.3390/catal9050432







