Suitability of Recombinant Lipase Immobilised on Functionalised Magnetic Nanoparticles for Fish Oil Hydrolysis
Abstract
:1. Introduction
2. Results and Discussion
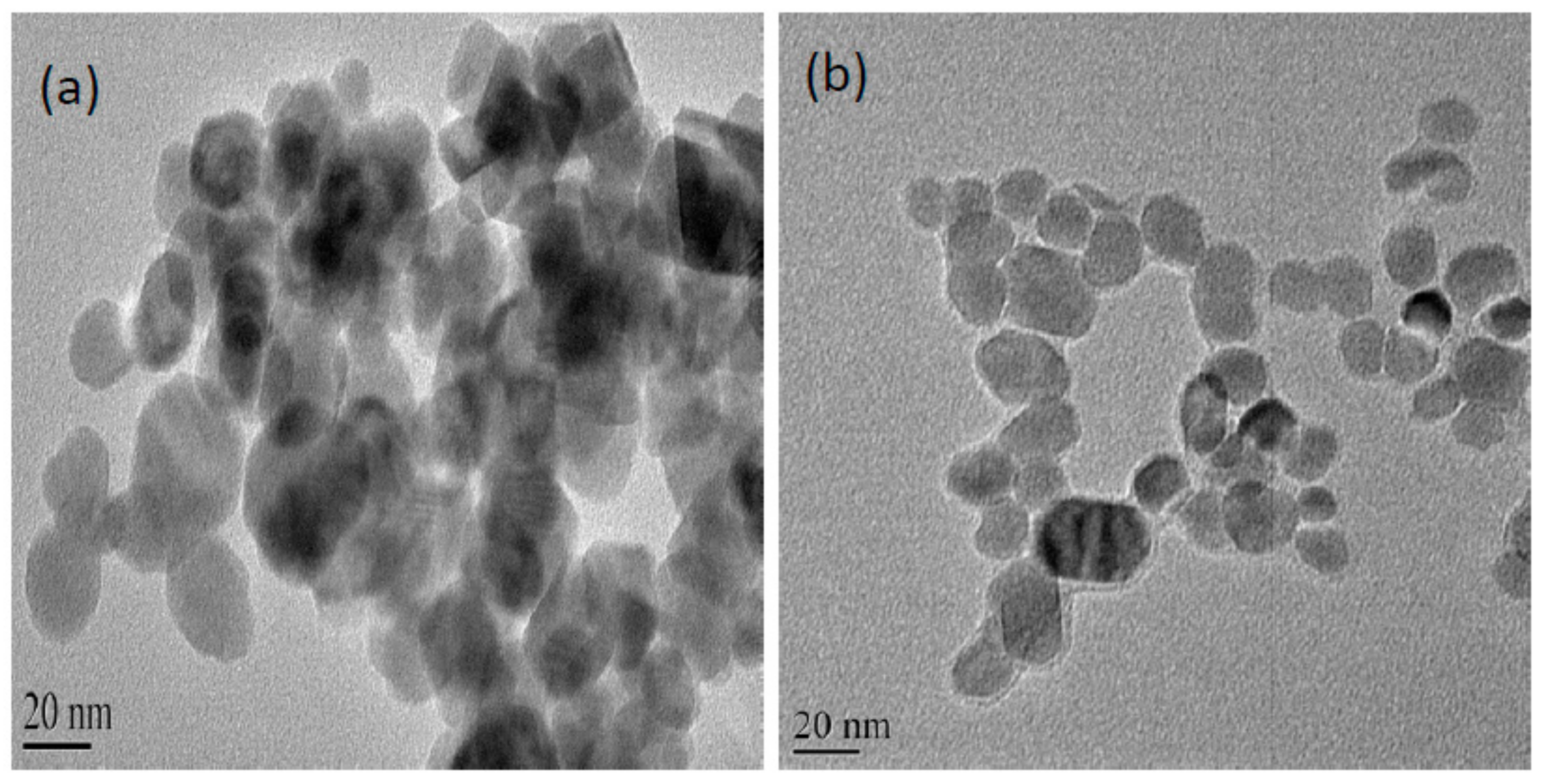
2.1. Characterisation of Zinc Doped Magnetic Nanoparticle
2.2. Enzyme Immobilisation Kinetics
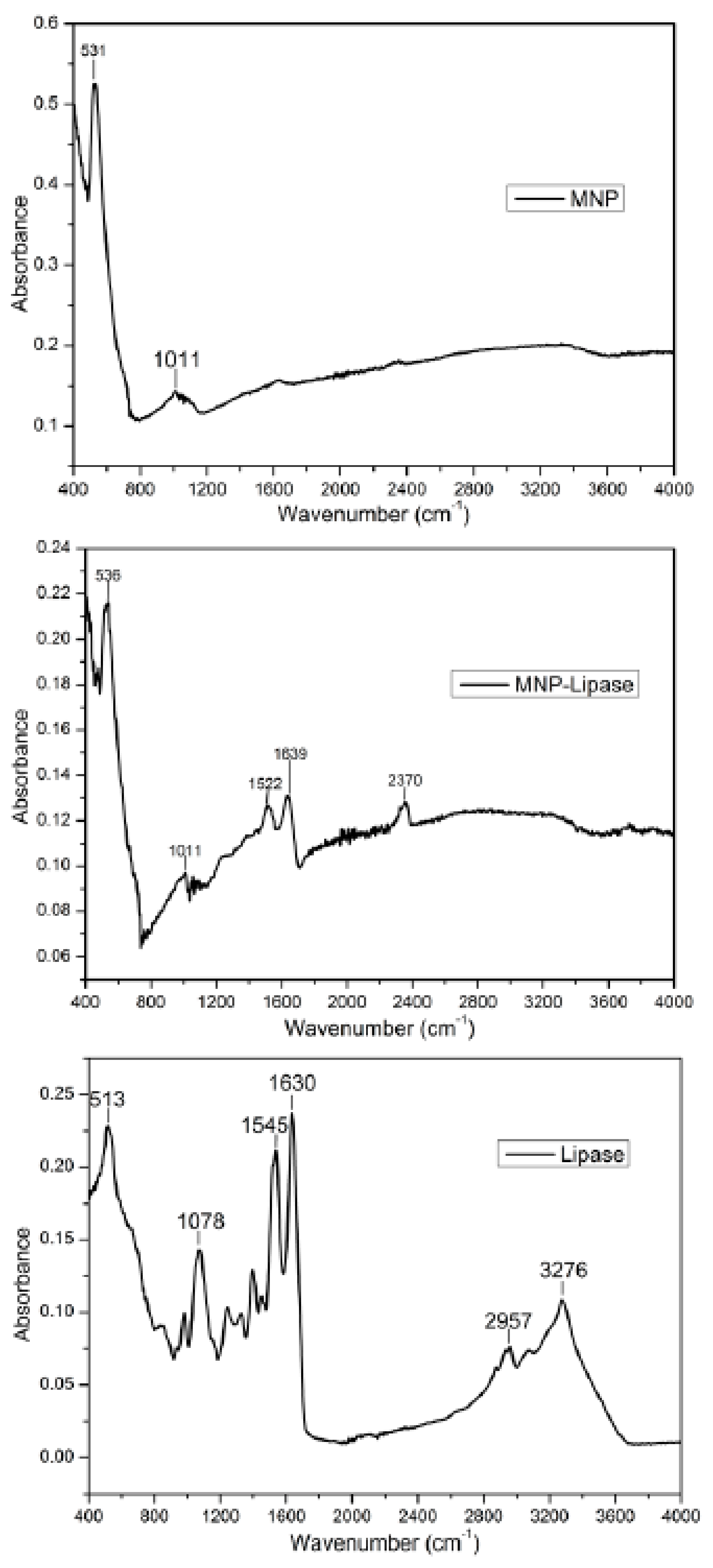
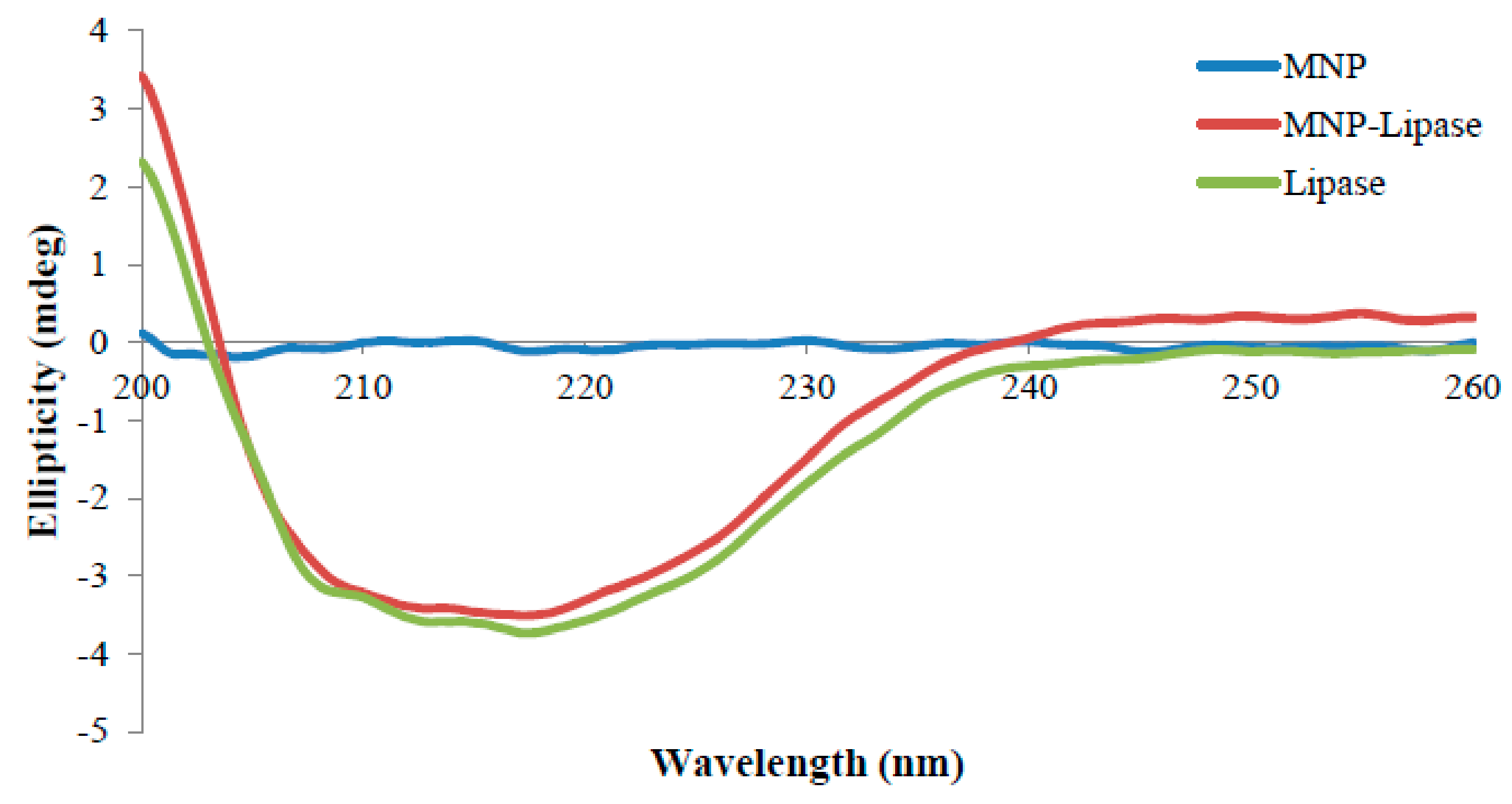
2.3. Structural Characterisation of Immobilised Enzyme
2.4. Biochemical Characterisation of the Immobilised and Free Lipase
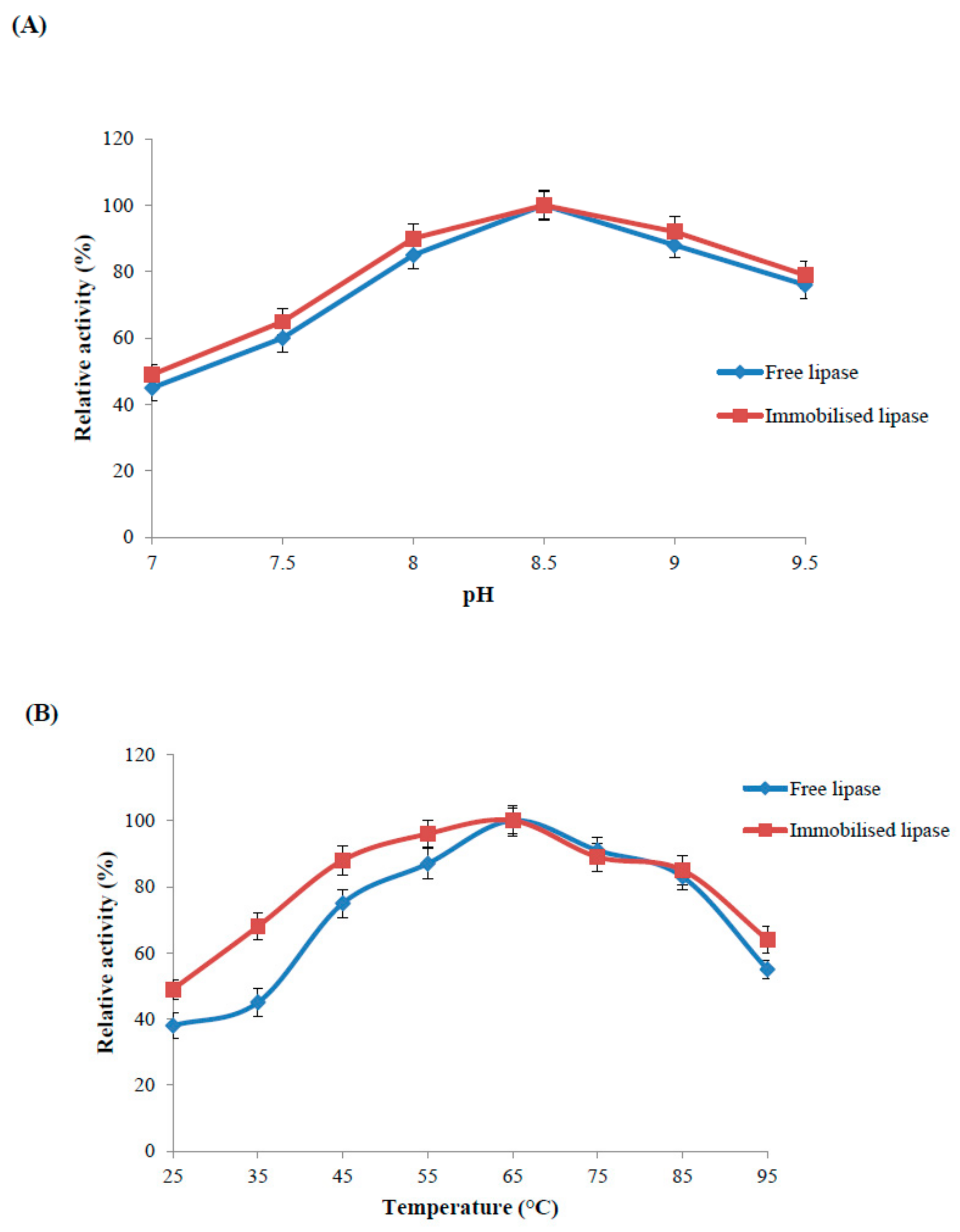
2.4.1. Optimal pH of the Immobilised and Free Lipase
2.4.2. Optimal Temperature and Thermal Stability of the Immobilised and Free Enzyme
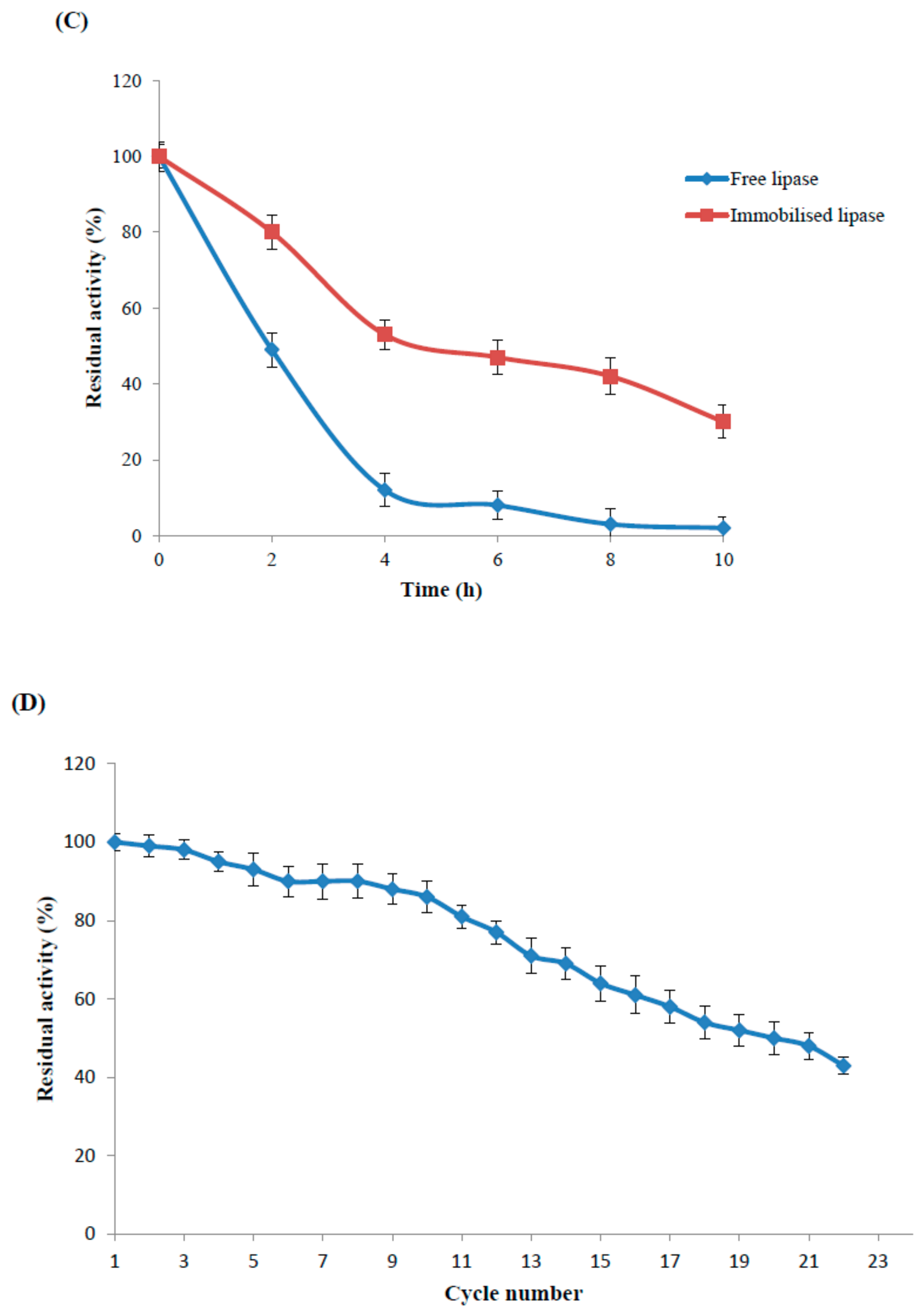
2.4.3. Reusability Study of the Maganetic Nanoparticle Immobilised Enzyme
2.4.4. Kinetics Study of Immobilised and Free Lipase Using Substrate (pNPP)
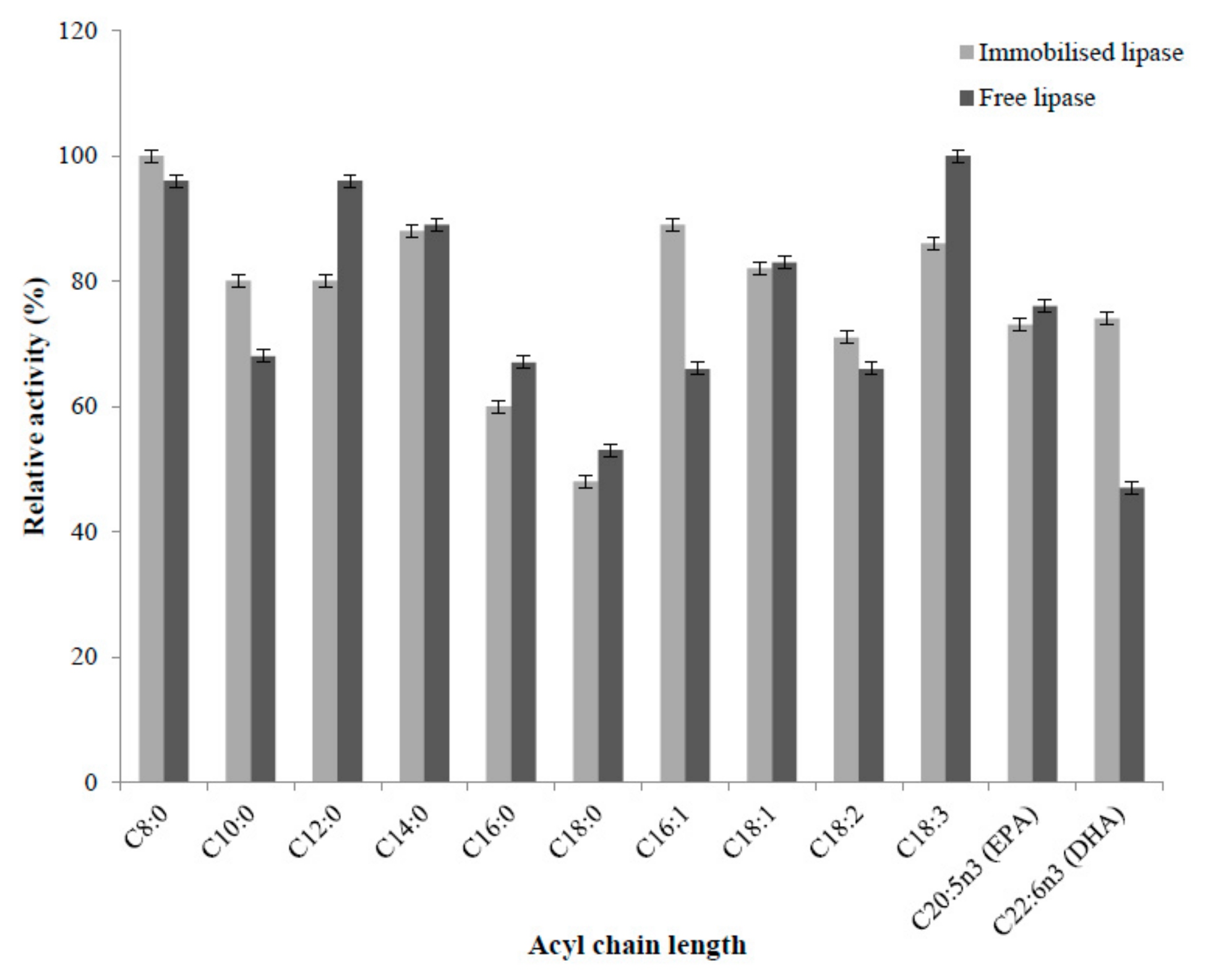
2.4.5. Substrate Selectivity of Immobilised and Free Enzyme
2.5. Fish Oil Hydrolysis of Immobilised and Free Enzyme
3. Materials and Methods
3.1. Materials
3.2. Synthesis and Characterisation of Zinc Doped Magnetic Iron Oxide Nanoparticles
3.3. Immobilisation of Recombinant Lipase on Functionalised Magnetic Nanoparticles
3.4. Biophysical Characterisation of Zinc Doped Nanoparticle Immobilised Enzyme
3.5. Determination of Immobilised Enzyme Activity and Protein Concentration
3.6. Biochemical Characterisation of the Zinc Doped Magnetic Nanoparticle Immobilised and Free Lipase
3.6.1. Effect of pH on the Enzyme Activity of Immobilised and Free Lipase
3.6.2. Optimum Temperature and Thermal Stability
3.6.3. Reusability of the Zinc Doped Magnetic Nanoparticle Immobilised Lipase
3.6.4. Determination of Enzyme Kinetics
3.6.5. Substrate Specificity for Long Chain Polyunsaturated Fatty Acids (PUFAs)
3.7. Fish Oil Hydrolysis Using Immobilised and Free Lipase
3.7.1. Analysis of Lipid Classes by Thin Layer Chromatography
3.7.2. Analysis of Lipid Classes by Capillary Chromatography (Iatroscan) and Gas Chromatography
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Porta, R.; Pandey, A.; Rosell, C.M. Enzymes as additives or processing aids in food biotechnology. Enzym. Res. 2010, 2010, 436859. [Google Scholar] [CrossRef]
- Kim, H.; Choi, N.; Oh, S.W.; Kim, Y.; Kim, B.H.; Kim, I.H. Synthesis of α-linolenic acid-rich triacylglycerol using a newly prepared immobilized lipase. Food Chem. 2017, 237, 654–658. [Google Scholar] [CrossRef]
- Xie, W.; Zang, X. Covalent immobilization of lipase onto aminopropyl-functionalized hydroxyapatite-encapsulated-γ-Fe2O3 nanoparticles: A magnetic biocatalyst for interesterification of soybean oil. Food Chem. 2017, 227, 397–403. [Google Scholar] [CrossRef]
- Xie, W.; Zang, X. Lipase immobilized on ionic liquid-functionalized magnetic silica composites as a magnetic biocatalyst for production of trans-free plastic fats. Food Chem. 2018, 257, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Ferreira-Dias, S.; Sandoval, G.; Plou, F.; Valero, F. The potential use of lipases in the production of fatty acid derivatives for the food and nutraceutical industries. Electron. J. Biotechnol. 2013, 16, 3. [Google Scholar]
- Kanwar, S.S.; Verma, M.L. Lipases. In Encyclopedia of Industrial Biotechnology: Bioprocess, Bioseparation, and Cell Technology; Wiley: Hoboken, NJ, USA, 2010; pp. 1–16. [Google Scholar]
- Fernandez-Lafuente, R. Lipase from Thermomyces lanuginosus: Uses and prospects as an industrial biocatalyst. J. Mol. Catal. B Enzym. 2010, 62, 197–213. [Google Scholar] [CrossRef]
- Kralovec, J.A.; Zhang, S.; Zhang, W.; Barrow, C.J. A review of the progress in enzymatic concentration and microencapsulation of omega-3 rich oil from fish and microbial source. Food Chem. 2012, 131, 639–644. [Google Scholar] [CrossRef]
- Xie, W.; Huang, J. Immobilization of Candida rugosa lipase onto graphene oxide Fe3O4 nanocomposite: Characterization and application for biodiesel production. Energy Convers. Manag. 2018, 159, 42–53. [Google Scholar] [CrossRef]
- Mateo, C.; Palomo, J.M.; Fernandez-Lorente, G.; Guisan, J.M.; Fernandez-Lafuente, R. Improvement of enzyme activity, stability and selectivity via immobilization techniques. Enzym. Microb. Technol. 2007, 40, 1451–1463. [Google Scholar] [CrossRef]
- Xie, W.; Wang, J. Enzymatic production of biodiesel from soybean oil by using immobilized lipase on Fe3O4/poly(styrene-methacrylic acid) magnetic microsphere as a biocatalyst. Energy Fuels 2014, 28, 2624–2631. [Google Scholar] [CrossRef]
- Verma, M.L.; Chaudhary, R.; Tsuzuki, T.; Barrow, C.J.; Puri, M. Immobilization of β-glucosidase on magnetic nanoparticle improves thermostability: Application in cellobiose hydrolysis. Bioresour. Technol. 2013, 135, 2–6. [Google Scholar] [CrossRef] [PubMed]
- Verma, M.L.; Naebe, M.; Barrow, C.J.; Puri, M. Enzyme immobilisation on amino-functionalised multi-walled carbon nanotubes: Structural and biocatalytic characterisation. PLoS ONE 2013, 8, e73642. [Google Scholar] [CrossRef]
- Xie, W.; Zang, X. Immobilized lipase on core-shell structured Fe3O4-MCM-41 nanocomposites as a magnetically recyclable biocatalyst for interesterification of soybean oil and lard. Food Chem. 2016, 194, 1283–1292. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Grate, J.W.; Wang, P. Nanobiocatalysis and its potential applications. Trends Biotechnol. 2008, 26, 639–646. [Google Scholar] [CrossRef] [PubMed]
- Ansari, S.A.; Husain, Q. Potential applications of enzymes immobilized on/in nanomaterials: A review. Biotechnol. Adv. 2012, 30, 512–523. [Google Scholar] [CrossRef]
- Verma, M.L.; Barrow, C.J.; Kennedy, J.F.; Puri, M. Immobilization of β-galactosidase from Kluyveromyces lactis on functionalized silicon dioxide nanoparticles: Characterization and lactose hydrolysis. Int. J. Biol. Macromol. 2012, 50, 432–437. [Google Scholar] [CrossRef]
- Singh, N.; Srivatava, G.; Talat, M.; Raghubanshi, H.; Srivastava, O.N.; Kayastha, A.M. Cicer α-galactosidase immobilization onto functionalized grapheme nanosheets using response surface methods and its applications. Food Chem. 2014, 142, 430–438. [Google Scholar] [CrossRef]
- Cao, M.; Li, Z.; Wang, J.; Ge, W.; Yue, T.; Li, R.; Colvin, V.L.; Yu, W.W. Food related applications of magnetic iron oxide nanoparticles: Enzyme immobilization, protein purification, and food analysis. Trends Food Sci. Technol. 2012, 27, 47–56. [Google Scholar] [CrossRef]
- Pavlidis, I.V.; Vorhaben, T.; Tsoufis, T.; Rudolf, P.; Bornscheuer, U.T.; Gournis, D.; Stamatis, H. Development of effective nanobiocatalytic systems through the immobilisation of hydrolases on functionalized carbon-based nanomaterials. Bioresour. Technol. 2012, 115, 164–171. [Google Scholar] [CrossRef]
- Hu, B.; Pan, J.; Yu, H.; Liu, J.; Xu, J. Immobilisation of Serratia marcescens lipase onto amino-functionalised magnetic nanoparticles for repeated use in enzymatic synthesis of Diltiazem intermediate. Process Biochem. 2009, 44, 1019–1024. [Google Scholar] [CrossRef]
- Koh, A.L.; Sinclair, R. TEM observations of bio-conjugated streptavidin-gold nanoparticles. MRS Proc. 2007, 1019. [Google Scholar] [CrossRef]
- Manning, M.C. Use of infrared spectroscopy to monitor protein structure and stability. Expert Rev. Proteom. 2005, 2, 731–743. [Google Scholar] [CrossRef] [PubMed]
- Bruce, I.J.; Sen, T. Surface modification of magnetic nanoparticles with alkoxysilanes and their application in magnetic bioseparations. Langmuir 2005, 21, 7029–7035. [Google Scholar] [CrossRef]
- Ganesan, A.; Moore, B.D.; Kelly, S.M.; Price, N.C.; Rolinski, O.J.; Birch, D.J.S.; Dunkin, I.R.; Halling, P.J. Optical spectroscopic methods for probing the conformational stability of immobilised enzymes. ChemPhysChem 2009, 10, 1492–1499. [Google Scholar] [CrossRef]
- Talbert, J.N.; Goddard, J.M. Characterization of lactase-conjugated magnetic nanoparticles. Process Biochem. 2013, 48, 656–662. [Google Scholar] [CrossRef]
- Jiang, Y.; Guo, C.; Xia, H.; Mahmood, I.; Liu, C.; Liu, H. Magnetic nanoparticles supported ionic liquids for lipase immobilization: Enzyme activity in catalysing esterification. J. Mol. Catal. B Enzym. 2009, 58, 103–109. [Google Scholar] [CrossRef]
- Xia, G.H.; Liu, W.; Jaing, X.P.; Wang, X.Y.; Zhang, Y.W.; Guo, J. Surface modification of Fe3O4@SiO2 magnetic nanoparticles for immobilization of lipase. J. Nanosci. Nanotechnol. 2017, 17, 370–376. [Google Scholar] [CrossRef]
- Hanefeld, U.; Gardossi, L.; Magner, E. Understanding enzyme immobilisation. Chem. Soc. Rev. 2009, 38, 453–468. [Google Scholar] [CrossRef]
- Matuoog, N.; Li, K.; Yan, Y. Thermomyces lanuginosus lipase immobilized on magnetic nanoparticles and its application in the hydrolysis of fish oil. J. Food Biochem. 2018, 42, e12549. [Google Scholar] [CrossRef]
- Sorensen, M.H.; Ng, J.B.S.; Bergstrom, L.; Alberius, P.C.A. Improved enzymative activity of Thermomyces lanuginosus lipase immobilized in a hydrophilic particulate mesoporous carrier. J. Colloid Interface Sci. 2010, 343, 359–365. [Google Scholar] [CrossRef]
- Karabulut, I.; Durmaz, G.; Hayaloglu, A. Fatty acid selectivity of lipases during acidolysis reaction between triolein and saturated fatty acids varying from caproic and behenic acids. J. Agric. Food Chem. 2009, 57, 7584–7590. [Google Scholar] [CrossRef]
- Matuoog, N.; Li, K.; Yan, Y. Immobilization of Thermomyces lanuginosus lipase on multi-walled carbon nanotubes and its application in the hydrolysis of fish oil. Mat. Res. Express 2017, 4, 25402. [Google Scholar] [CrossRef]
- Urrutia, P.; Arrieta, R.; Alvarez, L.; Cardenas, C.; Mesa, M.; Wilson, L. Immobilization of lipases in hydrophobic chitosan for selective hydrolysis of fish oil: The impact of support functionalization on lipase activity, selectivity and stability. Int. J. Biol Macromol. 2018, 108, 674–686. [Google Scholar] [CrossRef]
- Junior, W.G.M.; Fernandez-Lorente, G.; Guisan, J.M.; Ribeiro, E.J.; Resende, M.M.D. Production of omega-3 polyunsaturated fatty acids through hydrolysis of fish oil by Candida rugosa lipase immobilized and stabilized on different supports. Biocatal. Biotransform. 2017, 35, 63–73. [Google Scholar] [CrossRef]
- Aknabi, A.O.; Adcock, J.A.; Barrow, C.J. Selective concentration of EPA and DHA using Thermomyces lanuginosus lipase is due to fatty acid selectivity and not regioselectivity. Food Chem. 2013, 138, 615–620. [Google Scholar] [CrossRef]
- Ahmad, S.; Kamal, M.Z.; Sankaranarayanan, R.; Rao, N.M. Thermostable Bacillus subtilis lipases: In vitro evolution and structural insight. J. Mol. Biol. 2008, 381, 324–340. [Google Scholar] [CrossRef] [PubMed]
- Nalder, T.D.; Marshall, S.; Pfeffer, F.M.; Barrow, C.J. Characterisation of lipase fatty acid selectivity using novel omega-3 pNP-acyl esters. J. Funct. Foods 2014, 6, 259–269. [Google Scholar] [CrossRef]
- Chaudhary, R.; Roy, K.; Kanwar, R.K.; Walder, K.; Kanwar, J.R. Engineered atherosclerosis-specific zinc ferrite nanocomplexes-based MRI contrast agents. J. Biotechnol. 2016, 14, 6. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Winkler, U.K.; Stuckmann, M. Glucogen hyaluronate and some other polysaccharides greatly enhance the formation of exolipase by Serratia marcescens. J. Bacteriol. 1979, 138, 663–670. [Google Scholar] [PubMed]
- Christie, W.W.; Han, X. Lipid Analysis: Isolation, Separation, Identification and Lipidomic Analysis, 4th ed.; Oily Press, PJ Barnes & Associates: Bridgwater, UK, 2010. [Google Scholar]
- Craske, J.D.; Babbon, C.D. Gas liquid chromatography analysis of the fatty acid composition of fats and oils: A total system for high accuracy. J. Am. Oil Chem. Soc. 1987, 64, 1413–1417. [Google Scholar] [CrossRef]
| Structure | H(r) | H(d) | S(r) | S(d) | Trn | Unrd |
|---|---|---|---|---|---|---|
| Native enzyme | 0.272 | 0.143 | 0.038 | 0.057 | 0.197 | 0.293 |
| Immobilised enzyme | 0.228 | 0.157 | 0.027 | 0.075 | 0.197 | 0.315 |
| Kinetic Parameters | Free Lipase | Immobilised Lipase |
|---|---|---|
| KM (mM) | 12.5 | 14.7 |
| Vmax (U/mg) | 3.3 | 2.1 |
| Fatty Acid | Native Anchovy Oil Composition (%) | Free Lipase | Immobilised Lipase |
|---|---|---|---|
| Myristic acid (C14:0) | 8.34 | 5.49 | 7.09 |
| Palmitic acid (C16:0) | 16.69 | 11.82 | 14.97 |
| Palmitoleic acid (C16:1n7) | 9.78 | 9.16 | 9.32 |
| Oleic acid (C18:1n9) | 7.12 | 5.56 | 7.11 |
| EPA (C20:5n3) | 16.97 | 21.63 | 20.15 |
| DHA (C22:6n3) | 13.01 | 17.01 | 12.38 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Verma, M.L.; Rao, N.M.; Tsuzuki, T.; Barrow, C.J.; Puri, M. Suitability of Recombinant Lipase Immobilised on Functionalised Magnetic Nanoparticles for Fish Oil Hydrolysis. Catalysts 2019, 9, 420. https://doi.org/10.3390/catal9050420
Verma ML, Rao NM, Tsuzuki T, Barrow CJ, Puri M. Suitability of Recombinant Lipase Immobilised on Functionalised Magnetic Nanoparticles for Fish Oil Hydrolysis. Catalysts. 2019; 9(5):420. https://doi.org/10.3390/catal9050420
Chicago/Turabian StyleVerma, Madan L, Nalam M Rao, Takuya Tsuzuki, Colin J Barrow, and Munish Puri. 2019. "Suitability of Recombinant Lipase Immobilised on Functionalised Magnetic Nanoparticles for Fish Oil Hydrolysis" Catalysts 9, no. 5: 420. https://doi.org/10.3390/catal9050420
APA StyleVerma, M. L., Rao, N. M., Tsuzuki, T., Barrow, C. J., & Puri, M. (2019). Suitability of Recombinant Lipase Immobilised on Functionalised Magnetic Nanoparticles for Fish Oil Hydrolysis. Catalysts, 9(5), 420. https://doi.org/10.3390/catal9050420







