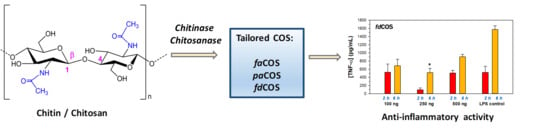
Tailored Enzymatic Synthesis of Chitooligosaccharides with Different Deacetylation Degrees and Their Anti-Inflammatory Activity
Abstract
:1. Introduction
2. Results and Discussion
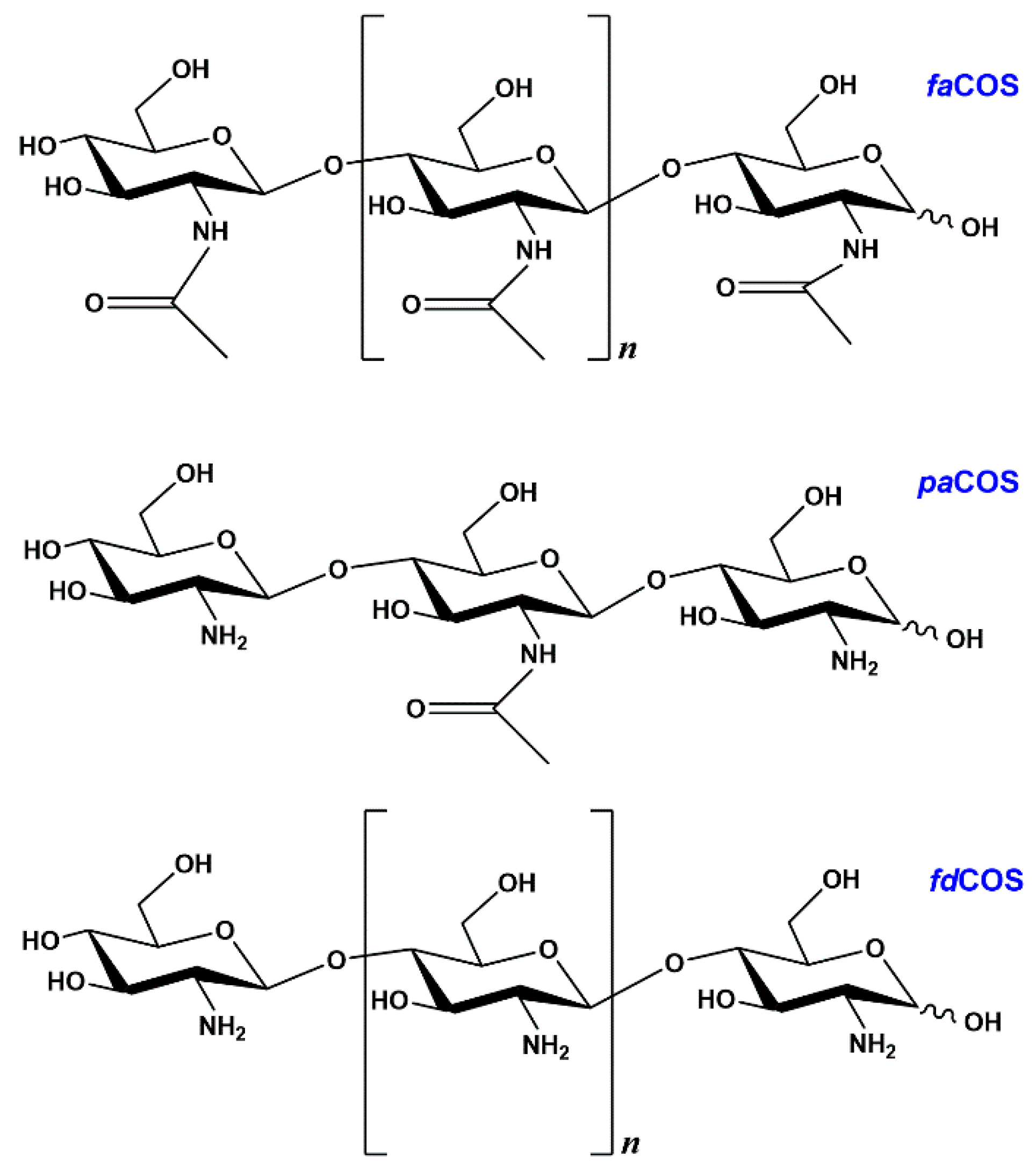
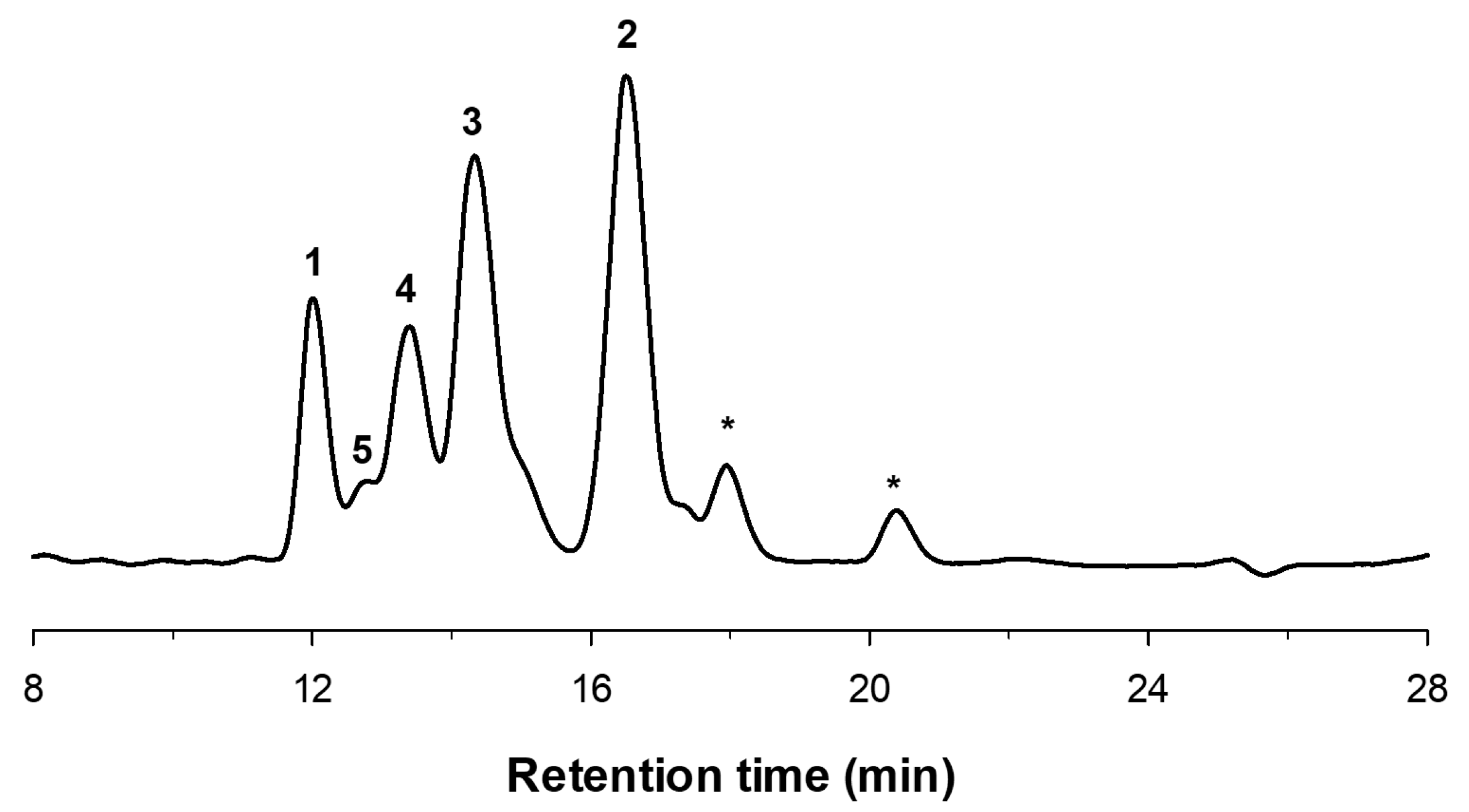
2.1. Enzymatic Production and Characterization of fdCOS
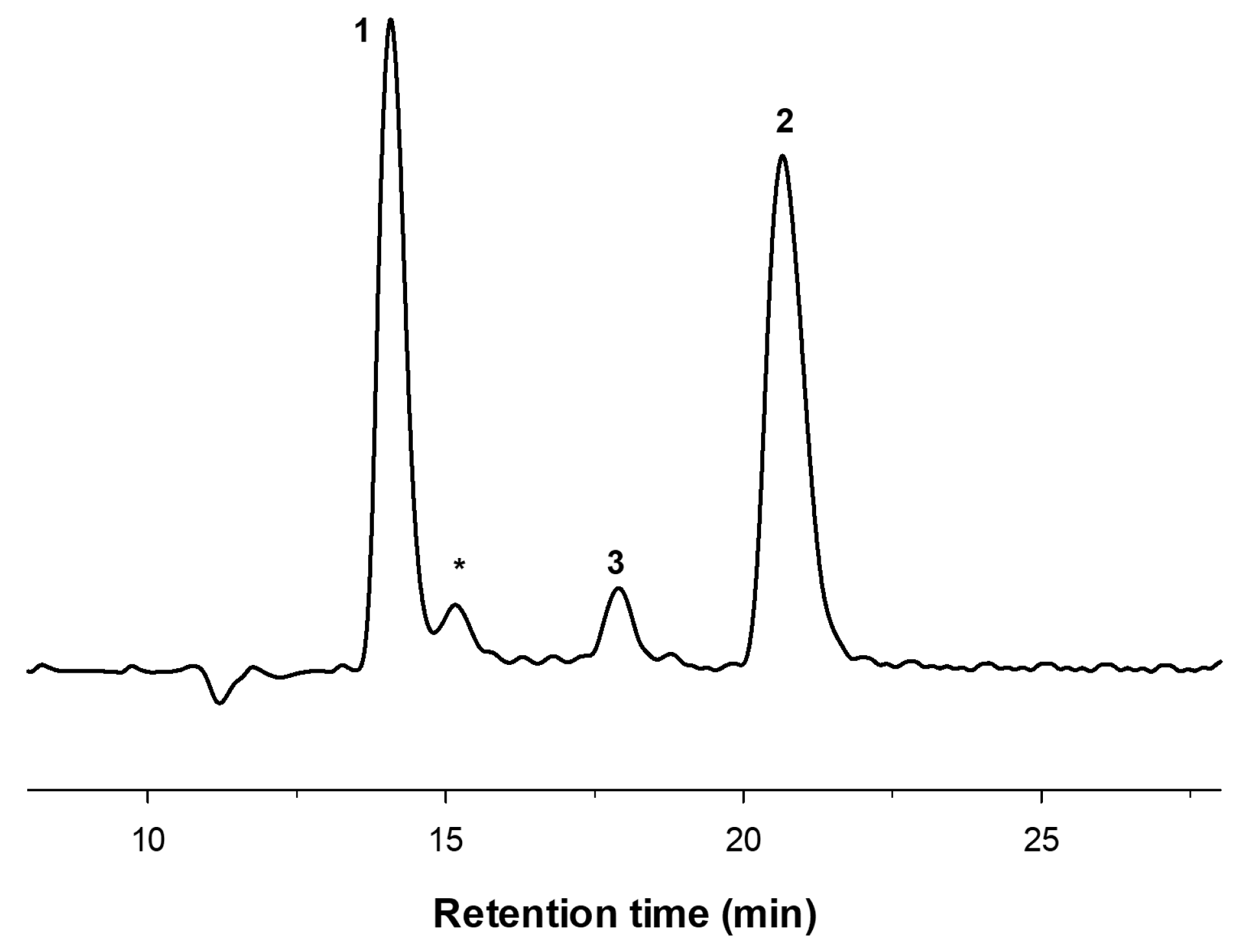
2.2. Enzymatic Production and Characterization of faCOS
2.3. Enzymatic Production and Characterization of paCOS
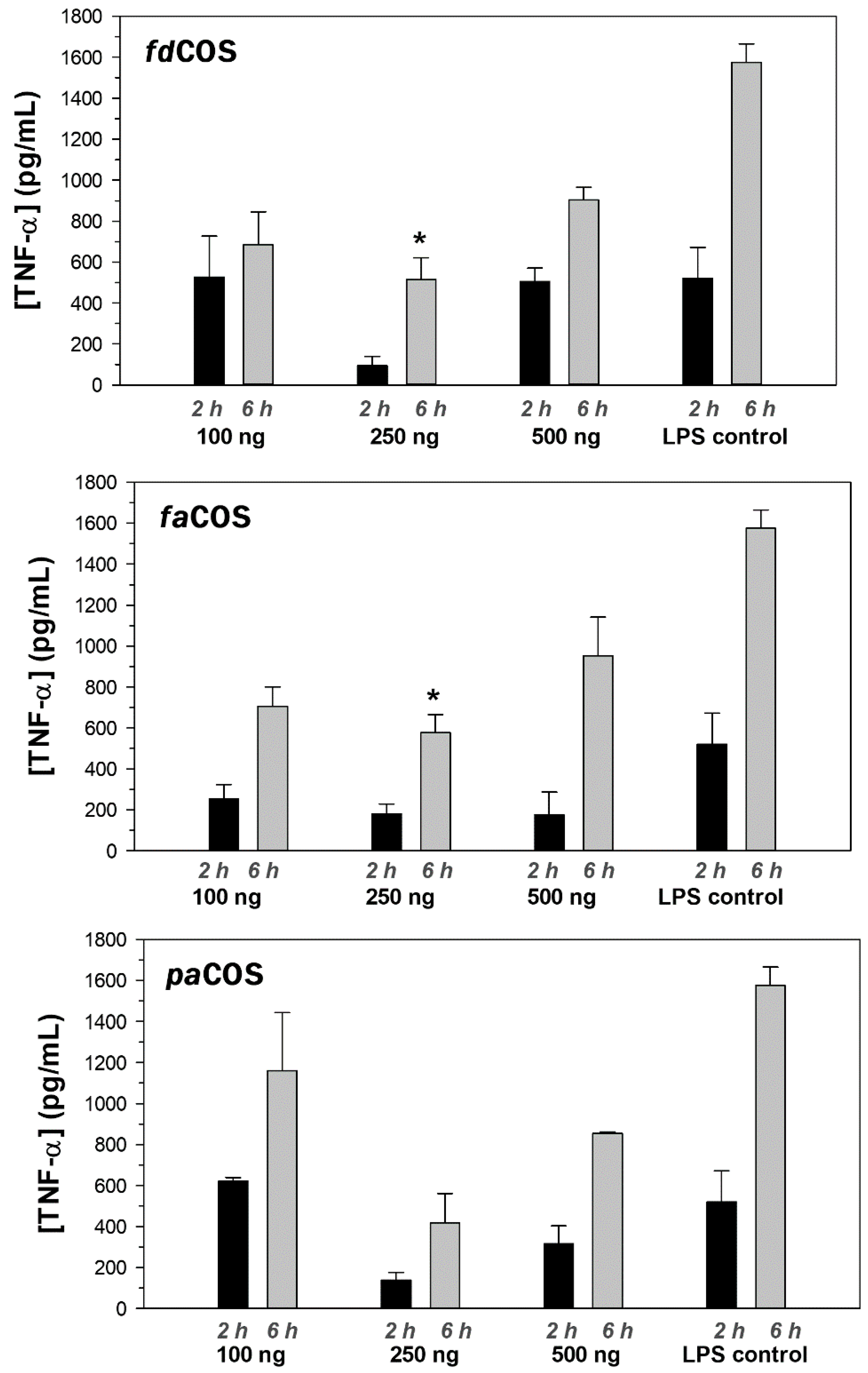
2.4. Anti-Inflammatory Activity of fdCOS, faCOS, and paCOS
3. Materials and Methods
3.1. Enzymes and Reagents
3.2. Preparation of Colloidal Chitin
3.3. COS Production and Purification
3.4. COS Characterization by HPAEC-PAD and MALDI-TOF
3.5. Anti-Inflammatory Activity of COS
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hamed, I.; Özogul, F.; Regenstein, J.M. Industrial applications of crustacean by-products (chitin, chitosan, and chitooligosaccharides): A review. Trends Food Sci. Technol. 2016, 48, 40–50. [Google Scholar] [CrossRef]
- Gortari, M.C.; Hours, R.A. Biotechnological processes for chitin recovery out of crustacean waste: A mini-review. Electron. J. Biotechnol. 2013, 16, 1–14. [Google Scholar]
- Kumar, M.; Brar, A.; Vivekanand, V.; Pareek, N. Bioconversion of chitin to bioactive chitooligosaccharides: Amelioration and coastal pollution reduction by microbial resources. Mar. Biotechnol. 2018, 20, 269–281. [Google Scholar] [CrossRef] [PubMed]
- Hamer, S.N.; Cord-Landwehr, S.; Biarnés, X.; Planas, A.; Waegeman, H.; Moerschbacher, B.M.; Kolkenbrock, S. Enzymatic production of defined chitosan oligomers with a specific pattern of acetylation using a combination of chitin oligosaccharide deacetylases. Sci. Rep. 2015, 5, 8716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liaqat, F.; Eltem, R. Chitooligosaccharides and their biological activities: A comprehensive review. Carbohydr. Polym. 2018, 184, 243–259. [Google Scholar] [CrossRef]
- Je, J.Y.; Kim, S.K. Chitooligosaccharides as potential nutraceuticals: production and bioactivities. Adv. Food Nutr. Res. 2012, 65, 321–336. [Google Scholar]
- Xia, W.; Liu, P.; Zhang, J.; Chen, J. Biological activities of chitosan and chitooligosaccharides. Food Hydrocolloids 2011, 25, 170–179. [Google Scholar] [CrossRef]
- Fernandes, J.C.; Spindola, H.; de Sousa, V.; Santos-Silva, A.; Pintado, M.E.; Malcata, F.X.; Carvalho, J.E. Anti-inflammatory activity of chitooligosaccharides in vivo. Mar. Drugs 2010, 8, 1763–1768. [Google Scholar] [CrossRef]
- Jiang, M.; Guo, Z.; Wang, C.; Yang, Y.; Liang, X.; Ding, F. Neural activity analysis of pure chito-oligomer components separated from a mixture of chitooligosaccharides. Neurosci. Lett. 2014, 581, 32–36. [Google Scholar] [CrossRef]
- Wu, S.-J.; Pan, S.-K.; Wang, H.-B.; Wu, J.-H. Preparation of chitooligosaccharides from cicada slough and their antibacterial activity. Int. J. Biol. Macromol. 2013, 62, 348–351. [Google Scholar] [CrossRef]
- Artan, M.; Karadeniz, F.; Karagozlu, M.Z.; Kim, M.M.; Kim, S.K. Anti-HIV-1 activity of low molecular weight sulfated chitooligosaccharides. Carbohydr. Res. 2010, 345, 656–662. [Google Scholar] [CrossRef]
- Huang, R.; Mendis, E.; Kim, S.K. Improvement of ACE inhibitory activity of chitooligosaccharides (COS) by carboxyl modification. Bioorg. Med. Chem. 2005, 13, 3649–3655. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Aam, B.B.; Wang, W.; Norberg, A.L.; Sørlie, M.; Eijsink, V.G.H.; Du, Y. Inhibition of angiogenesis by chitooligosaccharides with specific degrees of acetylation and polymerization. Carbohydr. Polym. 2012, 89, 511–518. [Google Scholar] [CrossRef] [Green Version]
- Kim, E.K.; Je, J.Y.; Lee, S.J.; Kim, Y.S.; Hwang, J.W.; Sung, S.H.; Moon, S.H.; Jeon, B.T.; Kim, S.K.; Jeon, Y.J.; et al. Chitooligosaccharides induce apoptosis in human myeloid leukemia HL-60 cells. Bioorg. Med. Chem. Lett. 2012, 22, 6136–6138. [Google Scholar] [CrossRef] [PubMed]
- Mengíbar, M.; Mateos-Aparicio, I.; Miralles, B.; Heras, A. Influence of the physico-chemical characteristics of chito-oligosaccharides (COS) on antioxidant activity. Carbohydr. Polym. 2013, 97, 776–782. [Google Scholar] [CrossRef] [Green Version]
- Yarullina, L.G.; Sorokan, A.V.; Burkhanova, G.F.; Cherepanova, E.A.; Maksimov, I.V. Influence of chitooligosaccharides with different acetylation degrees on the H2O2 content and the activity of pathogenesis-related proteins in potato plants infected with Phytophthora infestans. Appl. Biochem. Microbiol. 2018, 54, 528–534. [Google Scholar] [CrossRef]
- Liang, S.; Sun, Y.X.; Dai, X.L. A review of the preparation, analysis and biological functions of chitooligosaccharide. Int. J. Mol. Sci. 2018, 19, 2197. [Google Scholar] [CrossRef] [PubMed]
- Santos-Moriano, P.; Fernandez-Arrojo, L.; Mengibar, M.; Belmonte-Reche, E.; Peñalver, P.; Acosta, F.N.; Ballesteros, A.O.; Morales, J.C.; Kidibule, P.; Fernandez-Lobato, M.; et al. Enzymatic production of fully deacetylated chitooligosaccharides and their neuroprotective and anti-inflammatory properties. Biocatal. Biotransform. 2018, 36, 57–67. [Google Scholar] [CrossRef]
- de Araujo, N.K.; de Assis, C.F.; Dos Santos, E.S.; de Macedo, G.R.; de Farias, L.F.; Arimateia, H., Jr.; de Freitas Fernandes Pedrosa, M.; Pagnoncelli, M.G. Production of enzymes by Paenibacillus chitinolyticus and Paenibacillus ehimensis to obtain chitooligosaccharides. Appl. Biochem. Biotechnol. 2013, 170, 292–300. [Google Scholar] [CrossRef] [PubMed]
- Plou, F.J.; Gómez de Segura, A.; Ballesteros, A. Application of glycosidases and transglycosidases for the synthesis of oligosaccharides. In Industrial enzymes: Structure, Function and Application; Polaina, J., MacCabe, A.P., Eds.; Springer: New York, NY, USA, 2007; pp. 141–157. [Google Scholar]
- Fernandez-Arrojo, L.; Marin, D.; Gomez de Segura, A.; Linde, D.; Alcalde, M.; Gutierrez-Alonso, P.; Ghazi, I.; Plou, F.J.; Fernandez-Lobato, M.; Ballesteros, A. Transformation of maltose into prebiotic isomaltooligosaccharides by a novel alpha-glucosidase from Xantophyllomyces dendrorhous. Process Biochem. 2007, 42, 1530–1536. [Google Scholar] [CrossRef]
- Linde, D.; Rodriguez-Colinas, B.; Estevez, M.; Poveda, A.; Plou, F.J.; Fernandez-Lobato, M. Analysis of neofructooligosaccharides production mediated by the extracellular beta-fructofuranosidase from Xanthophyllomyces dendrorhous. Bioresour. Technol. 2012, 109, 123–130. [Google Scholar] [CrossRef]
- Liu, S.; Shao, S.; Li, L.; Cheng, Z.; Tian, L.; Gao, P.; Wang, L. Substrate-binding specificity of chitinase and chitosanase as revealed by active-site architecture analysis. Carbohydr. Res. 2015, 418, 50–56. [Google Scholar] [CrossRef]
- de Abreu, M.; Alvaro-Benito, M.; Sanz-Aparicio, J.; Plou, F.J.; Fernandez-Lobato, M.; Alcalde, M. Synthesis of 6-kestose using an efficient beta-fructofuranosidase engineered by directed evolution. Adv. Synth. Catal. 2013, 355, 1698–1702. [Google Scholar] [CrossRef]
- Song, J.Y.; Alnaeeli, M.; Park, J.K. Efficient digestion of chitosan using chitosanase immobilized on silica-gel for the production of multisize chitooligosaccharides. Process Biochem. 2014, 49, 2107–2113. [Google Scholar] [CrossRef]
- Alcalde, M.; Ferrer, M.; Plou, F.J. Environmental biocatalysis: From remediation with enzymes to novel green processes. Biocatal. Biotransform. 2007, 25, 113. [Google Scholar] [CrossRef]
- Olicón-Hernández, D.R.; Vázquez-Landaverde, P.A.; Cruz-Camarillo, R.; Rojas-Avelizapa, L.I. Comparison of chito-oligosaccharide production from three different colloidal chitosans using the endochitonsanolytic system of Bacillus thuringiensis. Prep. Biochem. Biotechnol. 2017, 47, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Santos-Moriano, P.; Woodley, J.M.; Plou, F.J. Continuous production of chitooligosaccharides by an immobilized enzyme in a dual-reactor system. J. Mol. Catal. B Enzym. 2016, 133, 211–217. [Google Scholar] [CrossRef]
- Li, H.; Fei, Z.; Gong, J.; Yang, T.; Xu, Z.; Shi, J. Screening and characterization of a highly active chitosanase based on metagenomic technology. J. Mol. Catal. B Enzym. 2015, 111, 29–35. [Google Scholar] [CrossRef]
- Thadathil, N.; Velappan, S.P. Recent developments in chitosanase research and its biotechnological applications: A review. Food Chem. 2014, 150, 392–399. [Google Scholar] [CrossRef]
- Sinha, S.; Chand, S.; Tripathi, P. Production, purification and characterization of a new chitosanase enzyme and improvement of chitosan pentamer and hexamer yield in an enzyme membrane reactor. Biocatal. Biotransform. 2014, 32, 208–213. [Google Scholar] [CrossRef]
- Santos-Moriano, P.; Kidibule, P.E.; Alleyne, E.; Ballesteros, A.O.; Heras, A.; Fernandez-Lobato, M.; Plou, F.J. Efficient conversion of chitosan into chitooligosaccharides by a chitosanolytic activity from Bacillus thuringiensis. Process Biochem. 2018, 73, 102–108. [Google Scholar] [CrossRef]
- Kittur, F.S.; Vishu Kumar, A.B.; Varadaraj, M.C.; Tharanathan, R.N. Chitooligosaccharides—Preparation with the aid of pectinase isozyme from Aspergillus niger and their antibacterial activity. Carbohydr. Res. 2005, 340, 1239–1245. [Google Scholar] [CrossRef]
- Xia, W.; Liu, P.; Liu, J. Advance in chitosan hydrolysis by non-specific cellulases. Bioresour. Technol. 2008, 99, 6751–6762. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.B.V.; Tharanathan, R.N. A comparative study on depolymerization of chitosan by proteolytic enzymes. Carbohydr. Polym. 2004, 58, 275–283. [Google Scholar]
- Oyeleye, A.; Normi, Y.M. Chitinase: Diversity, limitations, and trends in Engineering for suitable applications. Biosci. Rep. 2018, 38, 4. [Google Scholar] [CrossRef]
- Moon, C.; Seo, D.J.; Song, Y.S.; Hong, S.H.; Choi, S.H.; Jung, W.J. Antifungal activity and patterns of N-acetyl-chitooligosaccharide degradation via chitinase produced from Serratia marcescens PRNK-1. Microb. Pathog. 2017, 113, 218–224. [Google Scholar] [CrossRef] [PubMed]
- Sha, L.; Shao, E.; Guan, X.; Huang, Z. Purification and partial characterization of intact and truncated chitinase from Bacillus thuringiensis HZP7 expressed in Escherichia coli. Biotechnol. Lett. 2016, 38, 279–284. [Google Scholar] [CrossRef]
- Kidibule, P.E.; Santos-Moriano, P.; Jiménez-Ortega, E.; Ramírez-Escudero, M.; Limón, M.C.; Remacha, M.; Plou, F.J.; Sanz-Aparicio, J.; Fernández-Lobato, M. Use of chitin and chitosan to produce new chitooligosaccharides by chitinase Chit42: Enzymatic activity and structural basis of protein specificity. Microb. Cell Fact. 2018, 17, 47. [Google Scholar] [CrossRef] [PubMed]
- Wei, P.; Ma, P.; Xu, Q.S.; Bai, Q.H.; Gu, J.G.; Xi, H.; Du, Y.G.; Yu, C. Chitosan oligosaccharides suppress production of nitric oxide in lipopolysaccharide-induced N9 murine microglial cells in vitro. Glycoconjugate J. 2012, 29, 285–295. [Google Scholar] [CrossRef]
- Yoon, H.J.; Moon, M.E.; Park, H.S.; Im, S.Y.; Kim, Y.H. Chitosan oligosaccharide (COS) inhibits LPS-induced inflammatory effects in RAW 264.7 macrophage cells. Biochem. Biophys. Res. Commun. 2007, 358, 954–959. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, H.T.; Xu, Q.S.; Du, Y.G.; Xu, J. Chitosan oligosaccharides block LPS-induced O-GlcNAcylation of NF-kappa B and endothelial inflammatory response. Carbohydr. Polym. 2014, 99, 568–578. [Google Scholar] [CrossRef] [PubMed]
- Yousef, M.; Pichyangkura, R.; Soodvilai, S.; Chatsudthipong, V.; Muanprasat, C. Chitosan oligosaccharide as potential therapy of inflammatory bowel disease: Therapeutic efficacy and possible mechanisms of action. Pharmacol. Res. 2012, 66, 66–79. [Google Scholar] [CrossRef]
- Lee, H.J.; Park, J.M.; Han, Y.M.; Gil, H.K.; Kim, J.; Chang, J.Y.; Jeong, M.; Go, E.J.; Hahm, K.B. The role of chronic inflammation in the development of gastrointestinal cancers: reviewing cancer prevention with natural anti-inflammatory intervention. Expert Rev. Gastroenterol. Hepatol. 2016, 10, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.J.; Wang, L.; Feng, P.P.; Yin, L.H.; Wang, C.; Zhi, S.X.; Dong, J.Y.; Wang, J.Y.; Lin, Y.; Chen, D.P.; et al. Inhibition of epithelial TNF-alpha receptors by purified fruit bromelain ameliorates intestinal inflammation and barrier dysfunction in colitis. Front. Immunol. 2017, 8, 1468. [Google Scholar] [CrossRef] [PubMed]
- Pangestuti, R.; Bak, S.S.; Kim, S.K. Attenuation of pro-inflammatory mediators in LPS-stimulated BV2 microglia by chitooligosaccharides via the MAPK signaling pathway. Int. J. Biol. Macromol. 2011, 49, 599–606. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, Á.; Mengíbar, M.; Fernández, M.; Alemany, S.; Heras, A.; Acosta, N. Influence of preparation methods of chitooligosaccharides on their physicochemical properties and their anti-inflammatory effects in mice and in RAW 264.7 macrophages. Mar. Drugs 2018, 16, 430. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-H.; Senevirathne, M.; Ahn, C.-B.; Kim, S.-K.; Je, J.-Y. Factors affecting anti-inflammatory effect of chitooligosaccharides in lipopolysaccharides-induced RAW264.7 macrophage cells. Bioorg. Med. Chem. 2009, 19, 6655–6658. [Google Scholar]
- Jeuniaux, C. Chitinases. Methods Enzymol. 1966, 8, 644–650. [Google Scholar]
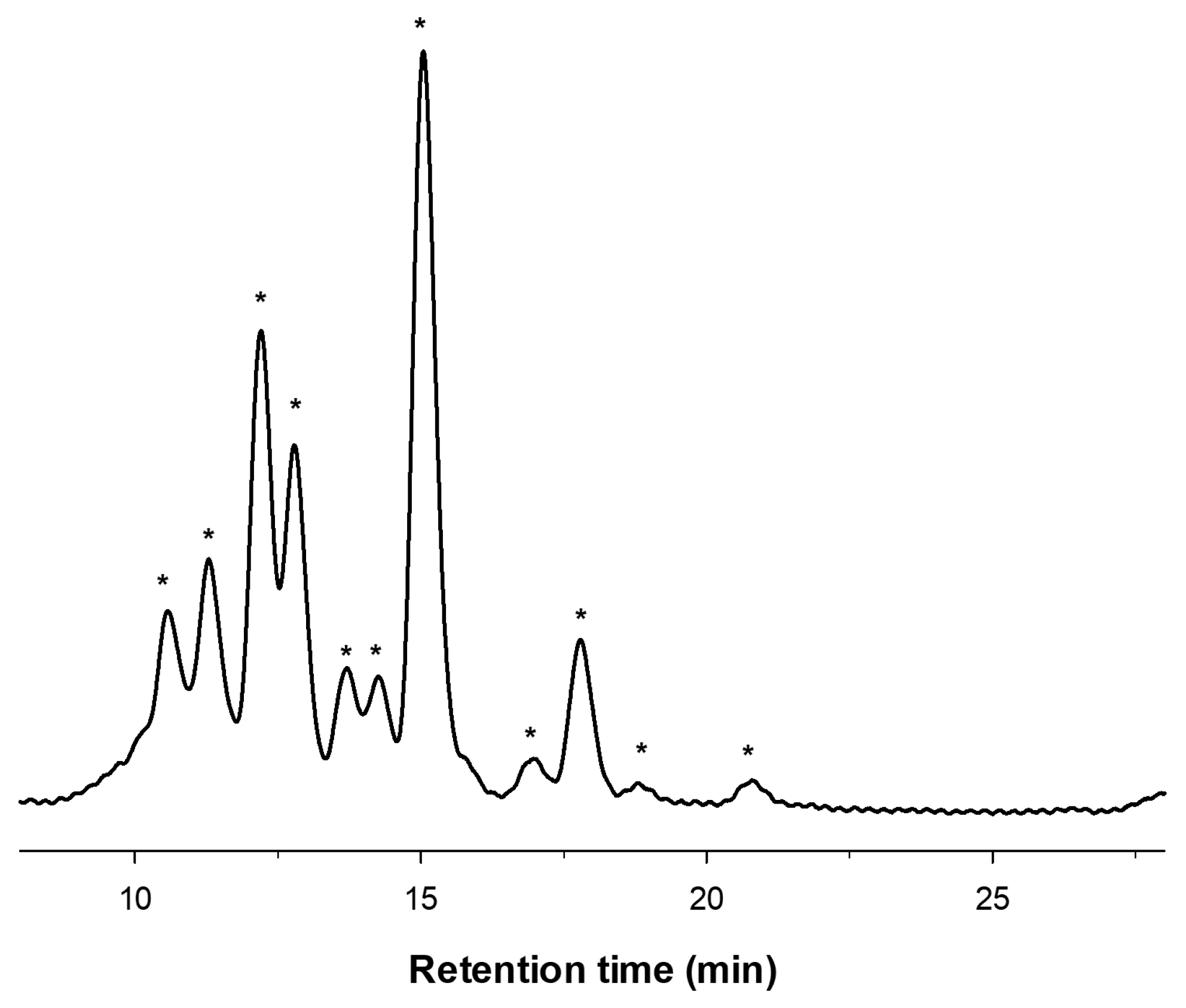
| m/z | Assignation |
|---|---|
| 180.0 | GlcN + H+ |
| 363.1 | (GlcN)2 + Na+ |
| 524.2/540.2 | (GlcN)3 + Na+/K+ |
| 566.2/582.2 | (GlcN)2-GlcNAc + Na+/K+ |
| 685.3/701.3 | (GlcN)4 + Na+/ K+ |
| 727.3/743.3 | (GlcN)3-GlcNAc + Na+/K+ |
| 846.3/862.2 | (GlcN)5 + Na+/ K+ |
| 888.3/904.3 | (GlcN)4-GlcNAc + Na+/K+ |
| 1023.3 | (GlcN)6 + K+ |
| 1049.4 | (GlcN)5-GlcNAc + Na+ |
| 1210.4 | (GlcN)6-GlcNAc + Na+ |
| m/z | Assignation |
|---|---|
| 405.2 | GlcN-GlcNAc + Na+ |
| 447.2/463.2 | (GlcNAc)2 + Na+/K+ |
| 608.3/624.2 | GlcN-(GlcNAc)2 + Na+ |
| 769.3/785.2 | (GlcN)2-(GlcNAc)2 + Na+/K+ |
| 811.3 | GlcN-(GlcNAc)3 + Na+ |
| 853.3 | (GlcNAc)4 + Na+ |
| m/z | Assignation |
|---|---|
| 405.2/421.2 | GlcN-GlcNAc + Na+/K+ |
| 447.2/463.2 | (GlcNAc)2 + Na+/K+ |
| 566.3/582.2 | (GlcN)2-GlcNAc + Na+/K+ |
| 608.3/624.3 | GlcN-(GlcNAc)2 + Na+/K+ |
| 727.3/743.3 | (GlcN)3-GlcNAc + Na+/K+ |
| 769.3/785.3 | (GlcN)2-(GlcNAc)2 + Na+/K+ |
| 811.3/827.3 | GlcN-(GlcNAc)3 + Na+/K+ |
| 888.4/904.3 | (GlcN)4-GlcNAc + Na+/K+ |
| 930.4/946.3 | (GlcN)3-(GlcNAc)2 + Na+/K+ |
| 1049.4/1065.4 | (GlcN)5-GlcNAc + Na+/K+ |
| 1091.4/1107.4 | (GlcN)4-(GlcNAc)2 + Na+/K+ |
| 1133.4/1149.4 | (GlcN)3-(GlcNAc)3 + Na+/K+ |
| 1210.4/1226.4 | (GlcN)6-GlcNAc + Na+/K+ |
| 1252.5/1268.4 | (GlcN)5-(GlcNAc)2 + Na+/K+ |
| 1294.5/1310.4 | (GlcN)4-(GlcNAc)3 + Na+/K+ |
| 1413.5/1429.5 | (GlcN)6-(GlcNAc)2 + Na+/K+ |
| 1532.6/1548.5 | (GlcN)8-GlcNAc + Na+/K+ |
| Enzyme | Substrate | Reaction Conditions | Main Products |
|---|---|---|---|
| Chit42 | Colloidal chitin | 35 °C, pH 6.0 | faCOS |
| Chit42 | Chitosan QS1 | 35 °C, pH 6.0 | paCOS |
| Neutrase 0.8 L | Chitosan CHIT600 | 50 °C, pH 5.0 | fdCOS |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santos-Moriano, P.; Kidibule, P.; Míguez, N.; Fernández-Arrojo, L.; Ballesteros, A.O.; Fernández-Lobato, M.; Plou, F.J. Tailored Enzymatic Synthesis of Chitooligosaccharides with Different Deacetylation Degrees and Their Anti-Inflammatory Activity. Catalysts 2019, 9, 405. https://doi.org/10.3390/catal9050405
Santos-Moriano P, Kidibule P, Míguez N, Fernández-Arrojo L, Ballesteros AO, Fernández-Lobato M, Plou FJ. Tailored Enzymatic Synthesis of Chitooligosaccharides with Different Deacetylation Degrees and Their Anti-Inflammatory Activity. Catalysts. 2019; 9(5):405. https://doi.org/10.3390/catal9050405
Chicago/Turabian StyleSantos-Moriano, P., P. Kidibule, N. Míguez, L. Fernández-Arrojo, A.O. Ballesteros, M. Fernández-Lobato, and F.J. Plou. 2019. "Tailored Enzymatic Synthesis of Chitooligosaccharides with Different Deacetylation Degrees and Their Anti-Inflammatory Activity" Catalysts 9, no. 5: 405. https://doi.org/10.3390/catal9050405
APA StyleSantos-Moriano, P., Kidibule, P., Míguez, N., Fernández-Arrojo, L., Ballesteros, A. O., Fernández-Lobato, M., & Plou, F. J. (2019). Tailored Enzymatic Synthesis of Chitooligosaccharides with Different Deacetylation Degrees and Their Anti-Inflammatory Activity. Catalysts, 9(5), 405. https://doi.org/10.3390/catal9050405