Abstract
Paenibacillus polymyxa exhibits remarkable hemicellulolytic activity. In the present study, 13 hemicellulose-degrading enzymes were identified from the secreted proteome of P. polymyxa KF-1 by liquid chromatography-tandem mass spectrometry analysis. α-L-arabinofuranosidase is an important member of hemicellulose-degrading enzymes. A novel α-L-arabinofuranosidase (PpAbf51b), belonging to glycoside hydrolase family 51, was identified from P. polymyxa. Recombinant PpAbf51b was produced in Escherichia coli BL21 (DE3) and was found to be a tetramer using gel filtration chromatography. PpAbf51b hydrolyzed neutral arabinose-containing polysaccharides, including sugar beet arabinan, linear-1,5-α-L-arabinan, and wheat arabinoxylan, with L-arabinose as the main product. The products from hydrolysis indicate that PpAbf51b functions as an exo-α-L-arabinofuranosidase. Combining PpAbf51b and Trichoderma longibrachiatum endo-1,4-xylanase produced significant synergistic effects for the degradation of wheat arabinoxylan. The α-L-arabinofuranosidase identified from the secretome of P. polymyxa KF-1 is potentially suitable for application in biotechnological industries.
1. Introduction
Lignocellulosic biomass is the most abundant renewable resource on earth, and lignocellulose shows potential for the production of biofuels and valuable chemicals [1]. Lignocellulose is composed of lignin, cellulose and hemicellulose [2]. The efficient conversion of lignocellulose to desirable target chemicals is challenging due to the complex structure of lignocellulosic materials [3,4]. Therefore, developing efficient lignocellulosic degradation approaches has attracted the attention of researchers. Many microorganisms exhibiting lignocellulosic biodegradation activities have been reported, with most species being filamentous fungi [5]. Research has shown that bacteria will play an increasingly important role in the bioconversion of lignocellulose due to their strong environmental adaptability and rich biochemical diversity [4]. However, compared with the numerous studies on fungi, there are only a few reports describing the degradation of lignocelluloses by bacteria [4]. Bacterial degradation of lignocellulose is not well understood, and lignocellulose-degrading enzymes from bacteria and the associated degradation pathways remain to be discovered.
Hemicellulose is one of the key components in lignocellulose [6]. Hemicellulose is mainly degraded by endoxylanase, β-xylosidase, α-L-arabinofuranosidase, mannanase, α-glucuronidase, and acetylxylan esterase [6,7]. α-L-arabinofuranosidase (Abf, EC. 3.2.1.55), which catalyzes the α-1,2-, α-1,3-, or α-1,5-linked L-arabinose or oligoarabinose side chains from the xylose backbone of hemicellulose [8], synergizes with other hemicelluloses to increase degradation efficiency. There has been growing interest in α-L-arabinofuranosidases due to their role in the degradation of hemicellulose, and their potential application in many biotechnological applications, such as the production of L-arabinose, improvement of wine flavors, clarification of fruit juices, and enhanced digestion of animal feedstuffs [8,9,10].
Paenibacillus polymyxa is a plant-growth-promoting rhizobacterium [11,12]. P. polymyxa was reported to use the major components of hemicellulose biomass and exhibited remarkable endoglucanase and α-L-arabinofuranosidase activities [13]. Genome sequencing of P. polymyxa revealed the presence of a diverse range of putative hemicellulose-degrading enzymes, signifying the potential of the bacterium for hemicellulose hydrolysis [11,14]. A glycoside hydrolase family 51 enzyme (PpAFase-1) from the fermentation broth of P. polymyxa was partially purified, identified, and overexpressed in Escherichia coli [13]. PpAFase-1 showed a synergistic effect with xylanase in hemicellulose degradation, but still has some disadvantages such as insufficient synergistic efficiency, which pushed us to explore robust α-L-arabinofuranosidases. In the present study, the secreted hemicellulose-degrading enzymes of P. polymyxa were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). An α-L-arabinofuranosidase belonging to the glycoside hydrolase (GH) family 51 was identified, heterologously expressed, and characterized, and its potential applications were explored.
2. Results
2.1. Measurement of the Extracellular Hemicellulytic Activities
The extracellular hemicellulytic activity of P. polymyxa KF-1 cultured in M9 minimal medium with arabinoxylan as the sole carbon source (10 g/L) at 30 °C for 36 h was measured. As shown in Table 1, P. polymyxa KF-1 showed high α-L-arabinofuranosidase activity and low levels of β-mannosidase, α-galactosidase, and β-xylosidase activities (Table 1). The specific activities against pNPαAraf, pNPβMan, pNPαGal, and pNPβXyl were 12.66, 1.22, 1.98, and 1.45 U/mg, respectively. P. polymyxa KF-1 secreted xylanase with a specific activity of 2.68 U/mg by using oat spelt xylan as a substrate.

Table 1.
Hemicellulytic activities of the enzyme extract of Paenibacillus polymyxa KF-1 on different substrates.
The hydrolytic activity of the enzyme extract toward wheat arabinoxylan was estimated by high-performance anion exchange chromatography (HPAEC). As shown in Figure 1, arabinoxylan was hydrolyzed by the enzyme extract of P. polymyxa with monosaccharides and oligosaccharides observed as products. The monosaccharide eluted at ~3.1 min was identified as L-arabinose. This result was confirmed by the reducing sugar assay. After 24 h, the reducing sugar released from arabinoxylan was 40.1 ± 3.7 mg/g.
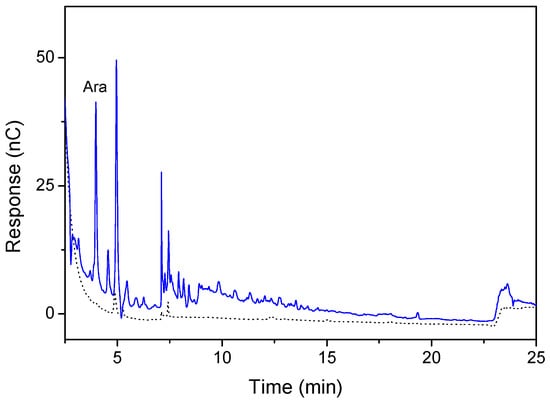
Figure 1.
High-performance anion exchange chromatography (HPAEC) analysis of the hydrolysis of arabinoxylan by Paenibacillus polymyxa enzyme extract. Reaction mixture containing 0.1 mg/mL enzyme extract and 5 mg/mL arabinoxylan was incubated at 37 °C for 24 h. The reaction was terminated by boiling the sample for 5 min, and the product was detected by HPAEC using a CarboPac PA-200 analytical column (3 × 250 mm). The black dotted line represents the substrate (wheat arabinoxylan) and the blue line represents the transformed product.
2.2. Hemicellulose-Degrading Enzymes Secreted by P. polymyxa
The complete degradation of hemicellulose requires a variety of enzymes, mainly endo-xylanase (EC 3.2.1.8), β-xylosidase (EC 3.2.1.37), and α-arabinofuranosidase (EC 3.2.1.55) [6]. Thirteen hemicellulose-degrading enzymes were identified by liquid chromatography-tandem mass spectrometry (LC-MS/MS). As listed in Table 2, four xylanases were assigned into the GH10 and GH11 families. Four GH43 enzymes were assigned, with two predicted as β-xylosidases, one as arabinoxylan arabinofuranohydrolase, and one as arabinan endo-1,5-α-L-arabinosidase. Two GH51 α-N-arabinofuranosidases with the α-L-AF_C superfamily domain were also identified as secreted enzymes. A GH27 α-galactosidase and a GH44 β-mannosidase were identified. The predicted pIs of the hemicellulases differed: the β-xylosidase, α-arabinofuranosidase, α-galactosidase, and β-mannosidase had weakly acidic pIs (pH 5.17–6.32). In addition to hemicellulases, other carbohydrate-degrading enzymes, including cellulose-degrading enzymes and pectin-degrading enzymes, were identified (Table S1).

Table 2.
Hemicellulose-degrading enzymes obtained by LC-MS/MS analysis.
2.3. Cloning and Sequence Analysis of α-L-Arabinofuranosidase
The gene encoding an α-L-arabinofuranosidase identified from the secreted proteomics analysis was cloned from the genome of P. polymyxa. The 1485-bp gene encoded a protein with a calculated molecular weight of 56.3 kDa. No signal peptide or non-classical signal was predicted. The protein, named PpAbf51b, showed high sequence similarity to GH51 α-L-arabinofuranosidase from Thermobacillus xylanilyticus (PDB ID: 2VRQ, identity 69%) [20]. However, PpAbf51b showed quite low identity to most published GH family 51 enzymes. The identities were approximately 26–28% to GH51 α-L-arabinofuranosidases from Thermotoga maritima (PDB ID: 3UG3), T. maritima MSB8 (PDB ID: 4ATW), Alicyclobacillus sp. A4 (KT781102), Geobacillus stearothermophilus T6 (PDB ID: 1PZ3), and Clostridium thermocellum (PDB ID: 2C7F) [20,21,22,23,24,25]. We noticed that the amino acid sequence similarities between the two α-L-arabinofuranosidases (WP_013368995.1 and WP_014599997.1) were only 27%. Previously, alignment analysis of the GH51 enzyme from T. xylanilyticus (PDB ID: 2VRQ) revealed that residue Glu176 in the conserved Gly-Asn-Glu sequence motif acted as the general acid/base and Glu298 in the conserved Asp-Glu-Trp domain acted as the catalytic nucleophile [20]. Alignment analysis indicated the presence of the conserved Gly-Asn-Glu and Asp-Glu-Trp motifs in PpAbf51b. Glu174 and Glu296 of PpAbf51b are hypothesized to act as the general acid/base and catalytic nucleophile, respectively (Figure 2).
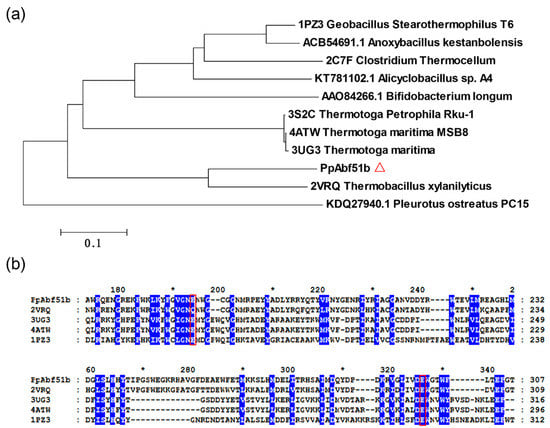
Figure 2.
(a) Phylogenetic analysis and (b) amino acid sequence alignment of PpAbf51b with GH51 arabinofuranosidases. (a) The phylogenetic tree was generated by MEGA6. (Δ) PpAbf51b from P. polymyxa KF-1; (b) Arabinoifuranosidases from G. stearothermophilus T6 (PDB: 1PZ3), T. xylanilyticus D3 (PDB: 2VRK), T. maritima MSB8 (PDB: 3UG3), and T. maritima MSB8 (4ATW) were aligned with PpAbf51b using the Clustal Omega program [26]. The conserved residues are labeled with a blue background. The catalytic glutamate residues are indicated by a red box.
The homology model of PpAbf51b was obtained using the GH51 α-L-arabinofuranosidase from T. xylanilyticus (PDB ID: 2VRQ, identity 69%) as the template [20]. The overall structure contains two characteristic GH51 domains: a (β/α)8-barrel catalytic domain and a C-terminal jelly roll architecture domain (Figure 3), which is similar to published GH51 structures.
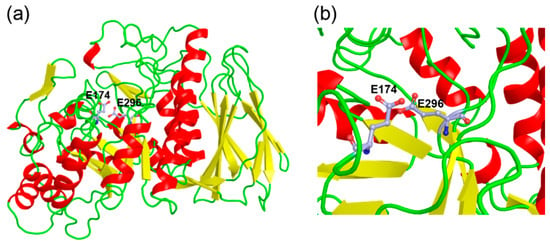
Figure 3.
The three-dimensional structure model of PpAbf51b predicted by SWISS-MODEL. (a) Amino acids from position 5 to position 493 of PpAbf51b were covered by the model; (b) E174 and E296, labeled in light blue, are predicted to be putative catalytic residues. The root-mean-square deviation (RMSD) was determined to be 0.070 between the homology model of PpAbf51b and the α-L-arabinofuranosidase from T. xylanilyticus (PDB ID: 2VRQ).
2.4. Expression and Purification of PpAbf51b
The recombinant protein was purified by Ni-NTA column chromatography, and 15.3 mg of purified protein was obtained from a 200-mL culture. A specific activity of 57.5 U/mg was obtained with pNPαAraf (1 mM) as a substrate. A single protein band with an estimated molecular weight (Mw) of 56.4 kDa was observed by SDS-PAGE analysis, which is consistent with the calculated Mr (Figure 4a). The native Mr was determined by gel filtration chromatography performed on Superdex 200 10/300 GL column (Figure 4b). The native Mr was calculated to be approximately 229.5 kDa, which indicated PpAbf51b existed as a tetramer.
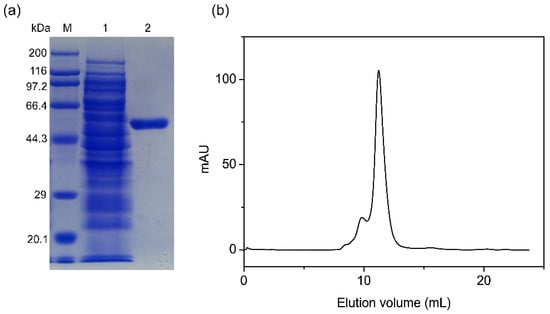
Figure 4.
Molecular weight of PpAbf51b determined by (a) SDS-PAGE and (b) gel filtration chromatography. (a) SDS-PAGE analysis (M), protein molecular weight markers (TaKaRa, code number 3597Q) (1), supernatant of cell lysis from recombinant Escherichia coli BL21 (DE3) cells harboring pET-28a-ppabf51b plasmid, 2, recombinant PpAbf51b purified from Ni-NTA agarose column; (b) Gel filtration chromatography analysis of purified PpAbf51b by Superdex 200 10/300 GL column.
2.5. Characterization of Purified PpAbf51b
PpAbf51b was incubated at different pH values (2.0–11.0) and showed the highest activity at pH 6.5 (Figure 5a). PpAbf51b was quite stable over the pH range of 4.0 to 11.0. More than 75% activity was retained at pH 4.0 and 11.0 after preincubation of the enzyme for 1 hour at 40 °C (Figure 5c). The optimal temperature was determined to be 40 °C. Enzymatic activity was near maximal over a relatively wide temperature range of 30 to 70 °C (Figure 5b). The enzyme was stable at temperatures lower than 60 °C. At 60 °C, the activity was still greater than 70% after 1 hour’s incubation (Figure 5d). In subsequent studies, the pH and temperature for hydrolyzing arabinose-containing polysaccharides by PpAbf51b were chosen to be pH 6.5 and 40 °C, respectively.
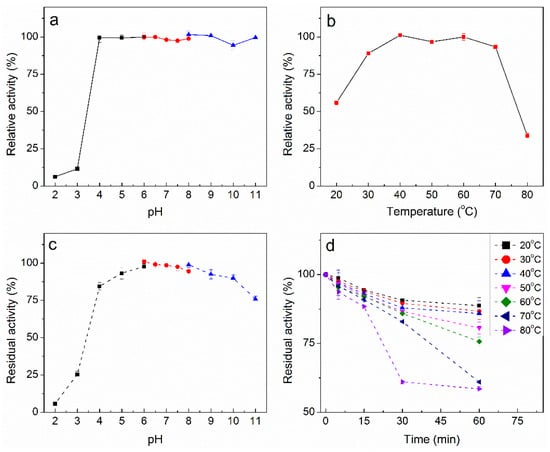
Figure 5.
Effect of pH and temperature on activity and stability of PpAbf51b. (a) Optimal pH and (c) pH stability were determined by incubating samples at various pH values for 15 min at 40 °C. (b) Optimal temperature and (d) temperature stability were determined by incubating samples at various temperatures at pH 6.5 for 15 min. The enzyme activity was detected by using pNPαAraf as a substrate (1 mM). The following buffers were used: 50 mM acetate buffer, pH 2.0–6.0; 50 mM phosphate buffer, pH 6.0–8.0; and 50 mM glycine sodium buffer, pH 8.0–11.0.
The effect of metal ions and chemicals on the activity of PpAbf51b was examined. FeCl2, at a concentration of 10 mM, enhanced PpAbf51b activity by more than 30%. CuCl2 and Triton X-100 showed slightly positive effects, leading to an activity increase of more than 10%. AlCl3 inhibited the activity of PpAbf51b with 72.4% of activity retained. For other tested ions and chemicals, inhibitory effects of around 10–20% were detected. The chemicals DTT, SDS, and Tween 40/60/80 did not significantly affect the activity of PpAbf51b (Table 3).

Table 3.
Effect of various metal ions and chemicals on the activity of PpAbf51b.
The effect of monosaccharides on the activity of PpAbf51b was determined. The residual activity of PpAbf51b was 85.4% and 78.8% when incubated with 100 mM L-arabinose or D-xylose, respectively. At a concentration of 500 mM, L-arabinose and D-xylose partially inhibited enzyme activity, with inhibition percentages of 26.1% and 36.9%, respectively (Figure 6).
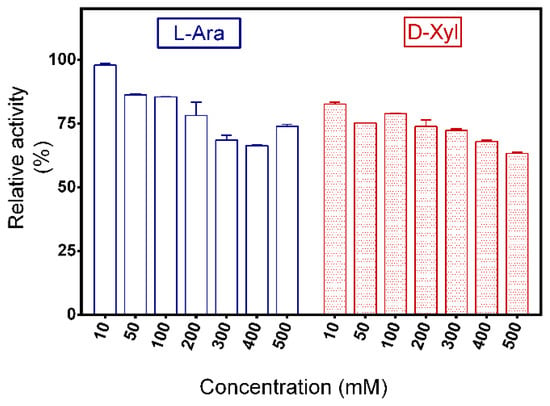
Figure 6.
Effect of L-arabinose and D-xylose on the activity of PpAbf51b. Values represent the mean ± SD (n = 3).
2.6. Substrate Specificity of PpAbf51b
Activities of PpAbf51b on 10 p-nitrophenyl–linked glycosides were determined. PpAbf51b exhibited high activity toward pNPαAraf and relative low activity toward pNPαGal. No activity was observed on other pNP glycosides (Table 4). These results demonstrate that PpAbf51b is an α-L-arabinofuranosidase. Using pNPαAraf as a substrate, the kinetic values Km, Vmax, and kcat were determined to be 0.2 mM, 58.1 μmol/min/mg, and 54.7 s−1, respectively. The catalytic efficiency kcat/Km was calculated to be 273.3 s−1mM−1.

Table 4.
Hydrolytic activity of PpAbf51b on different p-nitrophenyl linked glycosides.
The hydrolytic activity toward various arabinose-containing polysaccharides was examined. As shown in Figure 7, PpAbf51b was active against sugar beet arabinan, with L-arabinose detected by HPAEC after 16 h incubation. Similar enzyme activity was observed on linear-1,5-α-arabinan and wheat arabinoxylan. The degradation of sugar beet arabinan generated the highest amount of arabinose, followed by arabinoxylan and linear-1,5-α-arabinan. PpAbf51b showed no activity toward arabinogalactan or xylan.
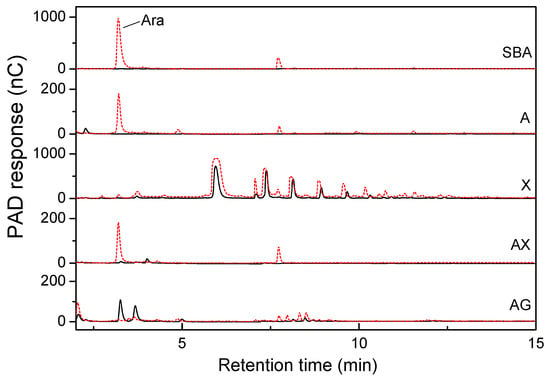
Figure 7.
HPAEC analysis of the hydrolysis products of neutral arabinose-containing polysaccharides by PpAbf51b. SBA, sugar beet arabinan; A, Linear-1, 5-α-arabinan; X, oat spelt xylan; AX, wheat arabinoxylan; AG, larch wood arabinogalactan. The black solid line represents the substrate and the red dashed line represents the hydrolysis product.
2.7. Synergistic Hydrolysis of Wheat Arabinoxylan by PpAbf51b and Tl-Xyn
The complete degradation of arabinoxylan requires the synergistic catalysis of endoxylanase and arabinofuranosidase. The synergistic effect of Trichoderma longibrachiatum endo-1,4-xylanase (Tl-Xyn) and PpAbf51b was detected by HPAEC with wheat arabinoxylan as the substrate (Figure 8). Using Tl-Xyn or PpAbf51b alone, Tl-Xyn liberated xylose (45.5 mg/g) and xylobiose (117.9 mg/g), whereas PpAbf51b released only arabinose (29.4 mg/g). All the simultaneous reactions and sequential enzyme combinations had significant synergistic effects on the degradation of wheat arabinoxylan, releasing substantially more arabinose, xylose, and xylobiose (Table 5). The degree of synergy for the simultaneous reaction was 1.38. For sequential enzyme combinations, the degree of synergy was 1.43 for PpAbf51b/Tl-Xyn (first addition of PpAbf51b) and 1.67 for Tl-Xyn/PpAbf51b (first addition of Tl-Xyn). Initial addition of Tl-Xyn followed by PpAbf51b liberated the most reducing sugar (321.9 mg/g). The molecular weight distribution of the hydrolytic products was determined by high-performance gel permeation chromatography (HPGPC) (Figure 9). The wheat arabinoxylan (retention time 9.67 min, ~330 kDa) was degraded to smaller molecular weight species after the simultaneous and sequential enzyme combinations.
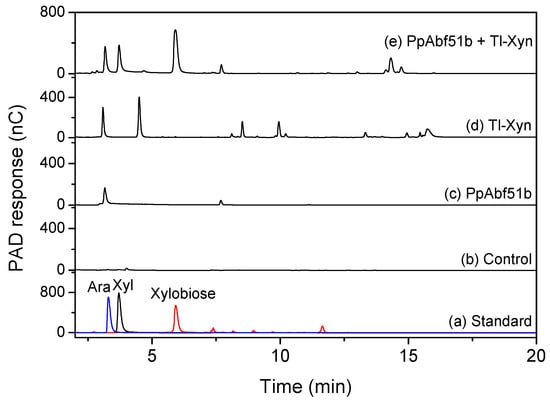
Figure 8.
HPAEC analysis of the hydrolysis products of wheat arabinoxylan. (a) Standard, arabinose, xylose, and xylobiose; (b) Wheat arabinoxylan (control) was hydrolyzed by (c) PpAbf51b; (d) Tl-Xyn, or (e) the combination of PpAbf51b and Tl-Xyn.

Table 5.
Synergistic effect of PpAbf51b and T. longibrachiatum endo-1, 4-β-Xylanase Tl-Xyn.
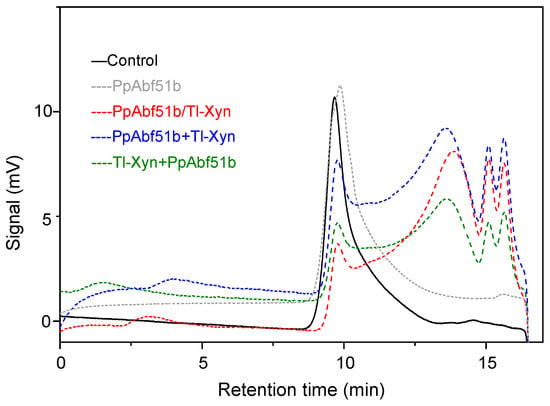
Figure 9.
HPGPC analysis of the hydrolysis products of wheat arabinoxylan. Wheat arabinoxylan was hydrolyzed by PpAbf51b, PpAbf51b/Tl-Xyn (the simultaneous reaction), PpAbf51b and Tl-Xyn (sequential enzyme combination with PpAbf51b added firstly), or Tl-Xyn and PpAbf51b (sequential enzyme combination with Tl-Xyn added firstly). Control: wheat arabinoxylan.
3. Discussion
The degradation products of hemicellulose are used widely in the production of pulp, paper, chemicals, and biofuels [6]. Degradation can be achieved using different types of processing, such as thermal, mechanical, chemical, and biological [6,7]. Among these methods, the specific action of hemicellulytic-degrading enzymes produced by microorganisms is considered to be the most promising approach [4,5].
P. polymyxa, previously known as Bacillus polymyxa, has a rich xylanolytic machinery, which makes the bacterium a potentially suitable candidate for the hydrolysis of hemicellulose [13]. Several alkaline endoxylanases, acidic endoxylanase, xyloglucanase, and α-L-arabinofuranosidase from this bacterium have been purified or cloned [13,27]. Genome sequence analysis of P. polymyxa revealed a number of genes coding for a variety of hemicellulases, which signifies the potential use of this bacterium in the general hydrolysis of hemicellulose (Table S2) [11,14]. A glycoside hydrolase family 51 enzyme (PpAFase-1) from the fermentation broth of P. polymyxa was partially purified, identified, and overexpressed in E. coli [13]. PpAFase-1 showed a synergistic effect with xylanase in hemicellulose degradation, but still has some disadvantages, such as insufficient synergistic efficiency, which pushed us to explore robust α-L-arabinofuranosidases.
In this report, the secreted hemicellulose-degrading enzymes of P. polymyxa KF-1 were identified by LC-MS/MS. As shown in Table 2, 13 proteins identified from the secreted proteomics analysis were assigned to hemicellulose-degrading enzymes. According to the activity assays, P. polymyxa displayed endoxylanase, β-mannosidase, β-xylosidase, α-galactosidase, and α-L-arabinofuranosidase activities (Table 1), with the α-L-arabinofuranosidase activity being measured as the highest. Therefore, P. polymyxa is a good candidate for obtaining an α-L-arabinofuranosidase.
Four proteins predicted as arabinofuranosidases were identified from the enzyme extract. Two were assigned to GH43 and the other two were assigned to GH51 α-N-arabinofuranosidases. Previously, the GH51 α-L-arabinofuranosidase (WP_013368995.1) from the enzyme extract of P. polymyxa was cloned, expressed, and named PpAFase-1 [13]. Here, another GH51 α-L-arabinofuranosidase PpAbf51b (WP_014599997.1) was cloned and expressed. PpAbf51b was found to show only 27% identity with PpAFase-1, so the functional diversity between the two GH51 enzymes was studied. PpAbf51b showed low sequence identity to several characterized bacterial GH51 α-L-arabinofuranosidases with identities of approximately 26–28% [21,28]. Although the identities were low, the sequence alignment showed that all sequences have the conserved general acid/base and catalytic nucleophile motifs, which are features of the GH family 51 (Figure 2). A structural model of PpAbf51b was obtained by SWISS-MODEL (Figure 3). The overall structure of PpAbf51b is similar to that of GH51 arabinofuranosidase from T. xylanilyticus (PDB ID: 2VRQ) [20], which confirmed that PpAbf51b belonged to the GH family 51. Previous studies showed that GH51 α-L-arabinofuranosidases function as monomers, tetramers, and hexamers. For example, GH51 α-L-arabinofuranosidases from G. stearothermophilus T6 (1PZ3), T. petrophila RKU-1 (3S2C), and T. maritima (4ATW) are hexamers, whereas AbfATK4 from G. caldoxylolyticus TK4, AbfAC26Sari from Anoxybacillus kestanbolensis AC26Sari, and Abf from Bifidobacterium longum B667 are tetramers [22,25,28,29,30,31]. The GH51 α-L-arabinofuranosidase from Pleurotus ostreatus is a monomer. For PpAbf51b, the native molecular mass was determined to be 229.5 kDa by gel filtration chromatography, which confirmed that the protein is a tetramer.
Most bacterial GH51 α-L-arabinofuranosidases are optimally active under neutral pH (pH 6.0–7.0) and mesophilic (30–50 °C) conditions [23]. Similarly, PpAbf51b was most active at pH 6.5 and 40 °C. Previously, two GH51 α-L-arabinofuranosidases from Paenibacillus sp. were characterized, which had higher optimal temperatures, 75 °C for THSAbf from Paenibacillus sp. THS1 and 60 °C for rAbfA from Paenibacillus sp. DG-22 [24,32]. PpAbf51b showed better stability than other published bacterial GH51 α-L-arabinofuranosidases. PpAbf51b showed stability over a broad pH range (pH 4.0–11.0), which was similar to that of the GH51 α-L-arabinofuranosidase Tx-Abf from T. xylanilyticus [33], but better than that of PpAFase-1. Since some endo-xylanases usually exhibit maximal activity at alkaline conditions, PpAbf51b, with excellent alkali tolerance, would be suitable for synergistic catalysis with endoxylanases and β-xylosidases. PpAbf51b showed good resistance to various metal ions and chemicals at a concentration of 10 mM, such as Co2+, Ag+, and Hg2+, which have been shown to inhibit GH51 enzymes [29,30]. The resistance against metal ions and chemicals by PpAbf51b was stronger than that of PpAFase-1, which was significantly inhibited by Fe2+, Cu2+, Mg2+, EDTA, SDS, Tween-40, Tween-60, Tween-80, and TritonX-100 at 10 mM [13]. For some glycoside hydrolases, the product inhibits the enzyme activity and becomes a limiting step in enzymatic technologies [34]. PpAbf51b showed good tolerance toward L-arabinose and D-xylose even at 500 mM. Previously, the GH51 enzyme PpAFase-1 was found to show tolerance to L-arabinose and D-xylose at concentrations lower than 50 mM. The excellent tolerance to monosaccharides and good stability makes PpAbf51b more suitable for biotechnological applications.
Besides α-L-arabinofuranosidase activity, endoglucanase, endoxylanase, and β-xylosidase activities are also included in the GH family 51. For example, Ac-Abf51A from Alicyclobacillus sp. A4, ArfB from C. stercorarium, and AF from B. stearothermophilus T-6 were reported to have relative weak β-xylosidase activity [23,25,35]; however, no hydrolytic activity of PpAbf51b was observed toward pNPXyl. Therefore, PpAbf51b displays rather narrow substrate specificity when compared with other GH51 α-L-arabinofuranosidases. Previously, Alicyclobacillus sp. enzyme Ac-Abf51A was reported to release arabinose and xylo-oligosaccharides from wheat arabinoxylan [23]. In contrast, PpAbf51b yielded L-arabinose as the main product from arabinoxylan, which indicated that PpAbf51b was an exo-acting enzyme and was quite specific against α-linked non-reducing terminal L-arabinofuranose residues. Wheat arabinoxylan is a β-1,4-linked xylan backbone substituent with an L-arabinose residue at the C-2 or C-3 position [36]. The hydrolysis action of PpAbf51b on wheat arabinoxylan suggested that PpAbf51b had hydrolytic activity against α-1,2- and/or α-1,3-linked non-reducing terminal L-arabinofuranose residues, but no activity toward β-1,4-linked xylose. This substrate specificity makes PpAbf51b a potentially useful enzyme for hemicellulose degradation.
Previous studies reported that the level of reducing sugar liberated from hemicellulose improved significantly due to the synergistic effect of GH51 arabinofuranosidase and endoxylanase [23,37]. For example, Ac-Abf51A from Alicyclobacillus sp. A4 exhibited a synergistic effect with endo-xylanase XynBE18 on the degradation of water-soluble wheat arabinoxylan. Maximum sugar liberation was observed when both enzymes were used simultaneously [37]. More than five commercially available endoxylanases were detected for their degradation efficiency with arabinoxylan as a substrate. Tl-Xyn exhibited maximal degradation efficiency (data not shown). Therefore, Tl-xyn was chosen for synergistic catalysis. When PpAbf51b was incubated in combination with T. longibrachiatum endo-1, 4-β-xylanase Tl-Xyn sequentially or simultaneously, more reducing sugars were released from wheat arabinoxylan. In contrast with the results obtained for Ac-Abf51A and XynBE18, the maximal synergistic effect was observed when PpAbf51b was added after Tl-Xyn. The results suggest that the initial addition of an endo-1,4-β-xylanase promotes branch removal by PpAbf51b. The substrate specificity and biochemical stability of PpAbf51b make this protein a potential candidate for hemicellulose degradation on an industrial scale.
4. Materials and Methods
4.1. Reagents
p-Nitrophenyl (pNP) linked glycosides including pNP-β-D-xylopyranoside (pNPβXyl), pNP-α-L-arabinofuranoside (pNPαAraf), pNP-α-L-arabinopyranoside (pNPαArap), pNP-α-D-mannopyranoside (pNPαMan), pNP-β-D-mannopyranoside (pNPβMan), pNP-α-D-glucuronide (pNPαGlcA), pNP-α-D-galactopyranoside (pNPαGal), pNP-β-D-galactopyranoside (pNPβGal), pNP-α-D-glucopyranoside (pNPαGlc), pNP-β-D-glucopyranoside (pNPβGlc), pNP-α-L-rhamnopranoside (pNPαRha), oat spelt xylan, and locust bean gum were obtained from Sigma-Aldrich (St. Louis, MO, USA). Sugar beet arabinan (P-ARAB), linear 1,5-α-L-arabinan (P-LARB), wheat arabinoxylan (P-WAXYL), larch wood arabinogalactan (P-ARGAL), and the GH11 endo-1,4-β-xylanase M3 from Trichoderma longibrachiatum (Tl-Xyn, E-XYTR3) were purchased from Megazyme (Wicklow, Ireland). E. coli DH5α cells were used as the cloning strain, and BL21 (DE3) cells and pET-28a (+) were chosen as the expression strain and expression vector, respectively (Novagen, Madison, WI, USA).
4.2. Strains and Culture Conditions
The P. polymyxa KF-1 (CCTCC AB 2018146) strain was cultured on LB agar (10% tryptone, 5% yeast extract, 10% NaCl, 2% agar, pH 7.0) at 30 °C in darkness. After incubation for 12 h, bacterial colonies were inoculated in 100 mL M9 minimal medium with arabinoxylan as the sole carbon source (Na2HPO4·7H2O 12.8 g/L, KH2PO4 3 g/L, NaCl 0.5 g/L, NH4Cl 1 g/L, MgSO4 2 mM, CaCl2 0.1 mM, wheat arabinoxylan 10 g/L, pH 7.2). The cells were grown at 30 °C and shaking at 180 rpm for 36 h. The fermentation broth was centrifuged at 5000 rpm and 4 °C for 10 min, the supernatant was filtrated by 0.22 μm microporous membrane to remove the bacteria, and used as the enzyme extract for the enzymatic activity assay and LC-MS/MS analysis.
4.3. Hemicellulytic Activities of the P. polymyxa KF-1 Enzyme Extract
The endo-1,4-xylanase and endo-1,4-mannanase activities of the P. polymyxa KF-1 enzyme extract were determined with oat spelt xylan and locust bean gum as the substrates, respectively. Reaction mixtures containing 20 μL enzyme extract (final concentration 0.1 mg/mL), 180 μL phosphate buffer (pH 6.5), and 1 mg/mL substrate were incubated at 37 °C for 1 h. The reactions were terminated by boiling samples for 5 min, and the released reducing sugar was measured by the DNS method [38]. The activities of β-xylosidase, α-L-arabinofuranosidase, β-mannosidase, α-glucuronidase and α-galactosidase activities were measured with pNPβXyl, pNPαAraf, pNPβMan, pNPαGlcA, and pNPαGal as substrates, respectively. Reaction mixtures containing 20 μL enzyme extract (final concentration 0.1 mg/mL), 180 μL phosphate buffer (pH 6.5), and 1 mM substrate were incubated at 37 °C for 1 h, and the absorbance of the samples at 405 nm were read [13]. One unit of enzyme activity was determined as the enzyme required to release 1 μmol of reducing sugar/p-nitrophenol from the substrate per minute under the above assay conditions.
The activity of the P. polymyxa KF-1 enzyme extract toward wheat arabinoxylan was detected by HPAEC [36]. Reaction mixtures containing 100 μL enzyme extract (final concentration 0.1 mg/mL) and 5 mg/mL substrate in 1 mL of phosphate buffer (pH 6.5) were incubated at 37 °C for 24 h. The reaction was terminated by boiling samples for 5 min, and the product was detected by HPAEC using a CarboPac PA-200 analytical column (3 × 250 mm) [7,36]. The reducing sugar released was determined by the DNS method, the experiment was done in triplicate [38].
4.4. LC-MS/MS Analysis
The secreted proteins from the fermentation supernatant of P. polymyxa KF-1 were analyzed by LC-MS/MS by Beijing BiotechPack Scientific (Beijing, China) [39]. The experiment was performed in triplicate. The fermentation supernatant was filtered by 10 kDa ultrafiltration membrane, precipitated with five volumes of cool acetone containing 10% TCA (w/v), and resolved in 1 mL of resolving solution (9 M urea, 4% CHAPS, 1% IPG buffer, and 1% DTT). The protein solution was digested with trypsin (150 ng, diluted by 25 mM NH4HCO3) at 4 °C for 40 min. Subsequently, the sample was extracted twice with 100 μL of 5% TFA-50% acetonitrile-45% H2O at 37 °C for 1 h. The solution was sonicated and centrifuged, and the extracts were combined and vacuum dried. The peptides were fractionated by Acclaim PepMap and Acclaim PepMap RSLC C18 columns. The peptides were eluted at 300 nL/min by Q Exactive mass spectrometer (Thermo Fisher, Waltham, MA, USA) with the following gradient program: 0–5 min, 4%–10% solvent B; 5–85 min, 10%–22% solvent B; 85–110 min, 22%–40% solvent B; 111–120 min, 95% solvent B, with solvent A consisting of 0.1% formic acid and solvent B consisting of 0.1% formic acid-80% acetonitrile. The obtained raw data were screened in Uniprot against P. polymyxa using the MaxQuant software [40,41]. The defaulting searching parameters for MaxQuant were as follows: fixed modifications = carbamidomethyl (C), variable modifications = oxidation (M), enzyme = trypsin, maximum missed cleavages = 2, peptide mass tolerance = 20 ppm, fragment mass tolerance = 0.6 Da, and mass values = monoisotopic, significance threshold = 0.05. The protein group score was reported by MaxQuant MS/MS Ion Search, which indicated the individual posterior error probabilities (PEPs) of the peptides of a protein group [18]. The false discovery rate (FDR) used for both peptides and proteins levels were 0.01 (1%). Identification of proteins is considered acceptable if at least one unique peptide was identified [18,19,41].
4.5. Gene Cloning, Protein Expression, and Purification
The genomic DNA of P. polymyxa KF-1 was extracted using the E.Z.N.A. Bacterial DNA Kit (D3350-01, Omega Bio-tek). The α-L-arabinofuranosidase gene was amplified with genomic DNA as the template. The oligonucleotides used for PCR were: 5′-CGGGATCCATGGTGAAGGGTTCTATTAT-3′ and 5′-CCGCTCGAGTTAAGGAGCGATTGTCAGCA-3′ (BamHI and XhoI sites are underlined). The PCR product and vector pET-28a were digested with BamHI/XhoI, and ligated to form the recombinant plasmid pET-28a-ppabf51b. The recombinant plasmid was used to transform E. coli DH5α cells. The recombinant plasmid with the correct coding sequence was used to transform E. coli BL21 (DE3) cells [42].
E. coli BL21 (DE3) cells harboring the recombinant pET-28a-ppabf51b plasmid were inoculated in LB broth at 37 °C and 180 rpm for 3 h with an inoculum volume of 1%. Protein expression was initiated by the addition of isopropyl β-D-1-thiogalactopyranoside to a final concentration of 0.5 mM and cells were grown for a further 12 h at 25 °C. Cells were harvested by centrifugation and the intracellular recombinant protein was released from the cells by sonication. The recombinant protein, PpAbf51b, which included an N-terminal His6-tag, was purified by Ni-NTA agarose (Qiagen, Hilden, Germany) using a linear gradient of 10–200 mM imidazole (in 20 mM Tris-HCl, pH 7.0). The α-L-arabinofuranosidase activity of each fraction was determined with pNPαAraf (1 mM) as a substrate [13]. The fractions with high α-L-arabinofuranosidase activity were combined, dialyzed to remove imidazole, and the protein concentration was measured by the BCA method [43].
The purified PpAbf51b was analyzed by SDS-PAGE using a 10% separation gel and 3.9% stacking gel [44]. The native molecular mass was detected by gel filtration chromatography performed on Superdex 200 10/300 GL column (GE Healthcare, Little Chalfont, UK). The column was pre-equilibrated and eluted by 50 mM phosphate buffer (pH 7.0) at a flow rate of 0.4 mL/min. The elution was monitored by UV-vis detector at 280 nm. Apoferritin (443 kDa), alcohol dehydrogenase (150 kDa), albumin (66 kDa), and carbonic anhydrase (29 kDa) were used to draw the standard curve.
4.6. Sequence Analysis and Protein Structure Prediction
The amino acid sequence was analyzed by BLASTp and aligned with published GH51 enzymes by Clustal Omega [26]. The phylogenetic tree was generated by MEGA6 [45]. The signal peptide and non-classical signal were predicted by the SignalP 4.1 Server [15] and the SecretomeP 2.0 server [16]. The protein structure was modeled by SWISS-MODEL [46] using GH51 α-L-arabinofuranosidase from T. xylanilyticus (PDB ID: 2VRQ, identity 69%) as the template [20].
4.7. Characterization of Recombinant PpAbf51b
The effect of pH on PpAbf51b activity was determined over the pH range of 2.0 to 11.0 using pNPαAraf (1 mM) as the substrate. The effect of pH on enzyme stability was determined by incubating samples at various pH values for 15 min at 40 °C. Residual arabinofuranosidase activity was measured under standard assay conditions. The effect of temperature on PpAbf51b activity was investigated by measuring the arabinofuranosidase activity at temperatures ranging from 20 to 80 °C at pH 6.5 for 15 min. The effect of temperature on enzyme stability was determined by incubating the enzyme at temperatures between 20 and 80 °C for up to 1 h. Residual arabinofuranosidase activity was measured under standard assay conditions (pH 6.5, 40 °C, reaction time 15 min). The initial activity measured prior to incubation was set to 100% [13].
The effects of metal ions and chemicals on the activity of PpAbf51b were determined by incubating 1 μg purified enzyme with each metal ion or chemical (10 mM) for 1 h at 40 °C and pH 6.5. The residual activity was then determined under standard assay conditions. The initial activity measured prior to incubation was set as 100% [13].
The effect of monosaccharides D-xylose and L-arabinose on PpAbf51b activity was assayed by preincubation of 1μg purified enzyme with each monosaccharide (10–500 mM) at 40 °C and pH 6.5 for 1 h. Enzymatic activity was then determined under standard assay conditions [13].
The standard assay conditions were as following: 2 μg purified enzyme and 1 mM pNPαAraf were incubated in 200 μL phosphate buffer (pH 6.5) at 40 °C for 15 min, then the absorbance at 405 nm were recorded [13].
4.8. Substrate Specificity of PpAbf51b
Activity of PpAbf51b against 10 p-nitrophenyl–linked glycosides was determined by reading the absorbance at 405 nm after 15-min reaction at 40 °C. The reaction mixture consisted of 2 μg recombinant enzyme, 200 μL phosphate buffer (pH 6.5), and 1 mM substrate. The kinetics values of PpAbf51b on pNPAraf were determined by the 15 min-incubation (substrate concentration 0.1–5 mM) with 2 μg purified enzyme at pH 6.5 and 40 °C. Km and Vmax values were calculated by Lineweaver-Burk plots [47].
The hydrolytic activity of PpAbf51b on arabinose-containing polymers, including sugar beet arabinan, linear-1,5-α-L-arabinan, oat spelt xylan, wheat arabinoxylan, and larch wood arabinogalactan, were detected. Reaction mixtures containing 140 μL 50 mM phosphate buffer (pH 6.5), 50 μL 4 mg/mL substrate, and 10 μL purified PpAbf51b (10 μg) were incubated at 40 °C for up to 16 h. The enzymatic products were analyzed by HPAEC [7].
The synergistic effect of PpAbf51b and T. longibrachiatum endo-1,4-β-xylanase (Tl-Xyn) on the degradation of wheat arabinoxylan was detected. The reaction mixture of 100 μL containing 50 μL 4 mg/mL arabinoxylan and 50 μL Tl-Xyn and/or PpAbf51b (1 U for each) was incubated in 50 mM phosphate buffer (pH 6.5) at 40 °C. After 12 h, the reaction mixture was precipitated by 80% ethanol, dried, and re-dissolved in 400 μL distilled water, and analyzed by HPAEC [7,36]. For sequential reactions, the substrate was incubated with the first enzyme for 4 h and the second enzyme was added. The degree of synergy was calculated as the ratio between the reducing sugar released from the combination reaction (Tl-Xyn and PpAbf51b) and the sum of the reducing sugar released from each enzyme separately. The molecular weight distribution of the hydrolytic product was determined by HPGPC using a TSK-gel G-3000PWXL column (7.8 × 300 mm, TOSOH, Japan). The column was pre-calibrated using standard dextrans (1, 5, 12, 25, and 50 kDa) [48].
5. Conclusions
Soil bacterium P. polymyxa KF-1 produces various CAZymes including hemicellulases. In this report, 13 hemicellulose-degrading enzymes were identified from LC-MS/MS analysis of secreted proteins by P. polymyxa. A novel α-L-arabinofuranosidase PpAbf51b identified from the secretome was heterologously expressed and characterized. The enzyme showed excellent pH (4.0–11.0) and temperature (≤60°C) stability and was active in the presence of metal ions (10 mM), chemicals (10 mM), and monosaccharides (500 mM). The enzyme acted as an exo-type α-L-arabinofuranosidase and showed a synergistic effect with endo-1,4-β-xylanase. Thus, PpAbf51b is a hemicellulase potentially suitable for biotechnological applications.
Supplementary Materials
The following are available online at http://www.mdpi.com/2073-4344/8/12/589/s1: Table S1: Other carbohydrate degrading enzymes identified by LC-MS/MS from P. polymyxa; Table S2: Other predicted hemicellulose-degrading enzymes in the genome sequence of P. polymyxa.
Author Contributions
Conceptualization, J.G.; Data curation, Y.H. and Y.Z.; Formal analysis, S.T.; Funding acquisition, Q.L.; Methodology, Y.Z. and Y.H.; Software, Y.Z., G.-Z. and J.G.; Validation, Y.L.; Visualization, J.G.; Writing—Original draft, Q.L. and J.G.; Writing—Review & editing, J.G.
Funding
We thank the Natural Science Foundation of China (31770852, 31741007) for supporting this study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Paulova, L.; Patakova, P.; Branska, B.; Rychtera, M.; Melzoch, K. Lignocellulosic ethanol: Technology design and its impact on process efficiency. Biotechnol. Adv. 2015, 33, 1091–1107. [Google Scholar] [CrossRef] [PubMed]
- Fen, L.; Xuwei, Z.; Nanyi, L.; Puyu, Z.; Shuang, Z.; Xue, Z.; Pengju, L.; Qichao, Z.; Haiping, L. Screening of lignocellulose-degrading superior mushroom strains and determination of their CMCase and laccase activity. Sci. World J. 2014, 2014, 763108. [Google Scholar] [CrossRef] [PubMed]
- Lopes, A.M.; Ferreira Filho, E.X.; Moreira, L.R.S. An update on enzymatic cocktails for lignocellulose breakdown. J. Appl. Microbiol. 2018, 125, 632–645. [Google Scholar] [CrossRef] [PubMed]
- De Gonzalo, G.; Colpa, D.I.; Habib, M.H.M.; Fraaije, M.W. Bacterial enzymes involved in lignin degradation. J. Biotechnol. 2016, 236, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.K.; Arora, D.S. Fungal degradation of lignocellulosic residues: An aspect of improved nutritive quality. Crit. Rev. Microbiol. 2015, 41, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Saha, B.C. Hemicellulose bioconversion. J. Ind. Microbiol. Biotechnol. 2003, 30, 279–291. [Google Scholar] [CrossRef] [PubMed]
- Goldbeck, R.; Damasio, A.R.L.; Goncalves, T.A.; Machado, C.B.; Paixao, D.A.A.; Wolf, L.D.; Mandelli, F.; Rocha, G.J.M.; Ruller, R.; Squina, F.M. Development of hemicellulolytic enzyme mixtures for plant biomass deconstruction on target biotechnological applications. Appl. Microbiol. Biotechnol. 2014, 98, 8513–8525. [Google Scholar] [CrossRef] [PubMed]
- Wilkens, C.; Andersen, S.; Dumon, C.; Berrin, J.-G.; Svensson, B. GH62 arabinofuranosidases: Structure, function and applications. Biotechnol. Adv. 2017, 35, 792–804. [Google Scholar] [CrossRef] [PubMed]
- Seri, K.; Sanai, K.; Matsuo, N.; Kawakubo, K.; Xue, C.; Inoue, S. L-arabinose selectively inhibits intestinal sucrase in an uncompetitive manner and suppresses glycemic response after sucrose ingestion in animals. Metabolism 1996, 45, 1368–1374. [Google Scholar] [CrossRef]
- Lagaert, S.; Pollet, A.; Courtin, C.M.; Volckaert, G. beta-xylosidases and alpha-L-arabinofuranosidases: Accessory enzymes for arabinoxylan degradation. Biotechnol. Adv. 2014, 32, 316–332. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, Q.; Li, Y.; Gao, J.; Fan, X. Draft Genome Sequence of Paenibacillus polymyxa KF-1, an Excellent Producer of Microbicides. Genome Announc. 2016, 4. [Google Scholar] [CrossRef] [PubMed]
- Naghmouchi, K.; Baah, J.; Cudennec, B.; Drider, D. Required characteristics of Paenibacillus polymyxa JB-0501 as potential probiotic. Arch. Microbiol. 2013, 195, 537–543. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Zhao, Y.; Zhang, G.; Li, Y.; Li, Q. Production optimization, purification, expression, and characterization of a novel α-l-arabinofuranosidase from Paenibacillus polymyxa. Electron. J. Biotechnol. 2018, 36, 24–33. [Google Scholar] [CrossRef]
- Rybakova, D.; Wetzlinger, U.; Muller, H.; Berg, G. Complete Genome Sequence of Paenibacillus polymyxa Strain Sb3-1, a Soilborne Bacterium with Antagonistic Activity toward Plant Pathogens. Genome Announc. 2015, 3. [Google Scholar] [CrossRef] [PubMed]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Bendtsen, J.D.; Kiemer, L.; Fausboll, A.; Brunak, S. Non-classical protein secretion in bacteria. BMC Microbiol. 2005, 5, 58. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [PubMed]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef] [PubMed]
- McManus, F.P.; Lamoliatte, F.; Thibault, P. Identification of cross talk between SUMOylation and ubiquitylation using a sequential peptide immunopurification approach. Nat. Protoc. 2017, 12, 2342–2358. [Google Scholar] [CrossRef] [PubMed]
- Paes, G.; Skov, L.K.; O’Donohue, M.J.; Remond, C.; Kastrup, J.S.; Gajhede, M.; Mirza, O. The structure of the complex between a branched pentasaccharide and Thermobacillus xylanilyticus GH-51 arabinofuranosidase reveals xylan-binding determinants and induced fit. Biochemistry 2008, 47, 7441–7451. [Google Scholar] [CrossRef] [PubMed]
- Dumbrepatil, A.; Park, J.-M.; Jung, T.Y.; Song, H.-N.; Jang, M.-U.; Han, N.S.; Kim, T.-J.; Woo, E.J. Structural analysis of alpha-L-arabinofuranosidase from Thermotoga maritima reveals characteristics for thermostability and substrate specificity. J. Microbiol. Biotechnol. 2012, 22, 1724–1730. [Google Scholar] [CrossRef] [PubMed]
- Souza, T.A.C.B.; Santos, C.R.; Souza, A.R.; Oldiges, D.P.; Ruller, R.; Prade, R.A.; Squina, F.M.; Murakami, M.T. Structure of a novel thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1. Protein Sci. 2011, 20, 1632–1637. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Bai, Y.; Yang, P.; Luo, H.; Huang, H.; Meng, K.; Shi, P.; Wang, Y.; Yao, B. A novel bifunctional GH51 exo-alpha-l-arabinofuranosidase/endo-xylanase from Alicyclobacillus sp. A4 with significant biomass-degrading capacity. Biotechnol. Biofuels 2015, 8, 197. [Google Scholar] [CrossRef] [PubMed]
- Bouraoui, H.; Desrousseaux, M.-L.; Ioannou, E.; Alvira, P.; Manai, M.; Remond, C.; Dumon, C.; Fernandez-Fuentes, N.; O’Donohue, M.J. The GH51 alpha-l-arabinofuranosidase from Paenibacillus sp. THS1 is multifunctional, hydrolyzing main-chain and side-chain glycosidic bonds in heteroxylans. Biotechnol. Biofuels 2016, 9, 140. [Google Scholar] [CrossRef] [PubMed]
- Shallom, D.; Belakhov, V.; Solomon, D.; Shoham, G.; Baasov, T.; Shoham, Y. Detailed kinetic analysis and identification of the nucleophile in alpha-L-arabinofuranosidase from Geobacillus stearothermophilus T-6, a family 51 glycoside hydrolase. J. Biol. Chem. 2002, 277, 43667–43673. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Higgins, D.G. Clustal omega. Curr. Protoc. Bioinform. 2014, 48, 3–13. [Google Scholar] [CrossRef]
- Morales, P.; Sendra, J.M.; Perez-Gonzalez, J.A. Purification and characterization of an arabinofuranosidase from Bacillus polymyxa expressed in Bacillus subtilis. Appl. Microbiol. Biotechnol. 1995, 44, 112–117. [Google Scholar] [CrossRef] [PubMed]
- Im, D.-H.; Kimura, K.; Hayasaka, F.; Tanaka, T.; Noguchi, M.; Kobayashi, A.; Shoda, S.; Miyazaki, K.; Wakagi, T.; Fushinobu, S. Crystal structures of glycoside hydrolase family 51 alpha-L-arabinofuranosidase from Thermotoga maritima. Biosci. Biotechnol. Biochem. 2012, 76, 423–428. [Google Scholar] [CrossRef] [PubMed]
- Canakci, S.; Belduz, A.O.; Saha, B.C.; Yasar, A.; Ayaz, F.A.; Yayli, N. Purification and characterization of a highly thermostable alpha-L-Arabinofuranosidase from Geobacillus caldoxylolyticus TK4. Appl. Microbiol. Biotechnol. 2007, 75, 813–820. [Google Scholar] [CrossRef] [PubMed]
- Canakci, S.; Kacagan, M.; Inan, K.; Belduz, A.O.; Saha, B.C. Cloning, purification, and characterization of a thermostable alpha-L-arabinofuranosidase from Anoxybacillus kestanbolensis AC26Sari. Appl. Microbiol. Biotechnol. 2008, 81, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Margolles, A.; de los Reyes-Gavilan, C.G. Purification and functional characterization of a novel alpha-L-arabinofuranosidase from Bifidobacterium longum B667. Appl. Environ. Microbiol. 2003, 69, 5096–5103. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Lee, Y.-E. Cloning, expression, and characterization of a thermostable GH51 alpha-L-arabinofuranosidase from Paenibacillus sp. DG-22. J. Microbiol. Biotechnol. 2014, 24, 236–244. [Google Scholar] [CrossRef] [PubMed]
- Debeche, T.; Cummings, N.; Connerton, I.; Debeire, P.; O’Donohue, M.J. Genetic and biochemical characterization of a highly thermostable alpha-L-arabinofuranosidase from Thermobacillus xylanilyticus. Appl. Environ. Microbiol. 2000, 66, 1734–1736. [Google Scholar] [CrossRef] [PubMed]
- De Giuseppe, P.O.; Souza, T.D.A.; Souza, F.H.M.; Zanphorlin, L.M.; Machado, C.B.; Ward, R.J.; Jorge, J.A.; Furriel, R.P.M.; Murakami, M.T. Structural basis for glucose tolerance in GH1 beta-glucosidases. Acta Crystallogr. D Biol. Crystallogr. 2014, 70, 1631–1639. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, W.H.; Bronnenmeier, K.; Krause, B.; Lottspeich, F.; Staudenbauer, W.L. Debranching of arabinoxylan: Properties of the thermoactive recombinant alpha-L-arabinofuranosidase from Clostridium stercorarium (ArfB). Appl. Microbiol. Biotechnol. 1995, 43, 856–860. [Google Scholar] [CrossRef] [PubMed]
- Guilloux, K.; Gaillard, I.; Courtois, J.; Courtois, B.; Petit, E. Production of arabinoxylan-oligosaccharides from flaxseed (Linum usitatissimum). J. Agric. Food Chem. 2009, 57, 11308–11313. [Google Scholar] [CrossRef] [PubMed]
- Shi, P.; Chen, X.; Meng, K.; Huang, H.; Bai, Y.; Luo, H.; Yang, P.; Yao, B. Distinct actions by Paenibacillus sp. strain E18 alpha-L-arabinofuranosidases and xylanase in xylan degradation. Appl. Environ. Microbiol. 2013, 79, 1990–1995. [Google Scholar] [CrossRef] [PubMed]
- McCleary, B.V.; McGeough, P. A Comparison of Polysaccharide Substrates and Reducing Sugar Methods for the Measurement of endo-1,4-beta-Xylanase. Appl. Biochem. Biotechnol. 2015, 177, 1152–1163. [Google Scholar] [CrossRef] [PubMed]
- Talamantes, T.; Ughy, B.; Domonkos, I.; Kis, M.; Gombos, Z.; Prokai, L. Label-free LC-MS/MS identification of phosphatidylglycerol-regulated proteins in Synechocystis sp. PCC6803. Proteomics 2014, 14, 1053–1057. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Carlson, A.; Sinitcyn, P.; Mann, M.; Cox, J. Visualization of LC-MS/MS proteomics data in MaxQuant. Proteomics 2015, 15, 1453–1456. [Google Scholar] [CrossRef] [PubMed]
- Sonnett, M.; Gupta, M.; Nguyen, T.; Wuhr, M. Quantitative Proteomics for Xenopus Embryos II, Data Analysis. Methods Mol. Biol. 2018, 1865, 195–215. [Google Scholar] [CrossRef] [PubMed]
- Lessard, J.C. Molecular cloning. Methods Enzymol. 2013, 529, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Walker, J.M. The bicinchoninic acid (BCA) assay for protein quantitation. Methods Mol. Biol. 1994, 32, 5–8. [Google Scholar] [CrossRef] [PubMed]
- Brunelle, J.L.; Green, R. One-dimensional SDS-polyacrylamide gel electrophoresis (1D SDS-PAGE). Methods Enzymol. 2014, 541, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Gallo Cassarino, T.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014, 42, W252–W258. [Google Scholar] [CrossRef] [PubMed]
- Ghim, Y.S.; Chang, H.N. Diffusional falsification of kinetic constants on Lineweaver-Burk plots. J. Theor. Biol. 1983, 105, 91–102. [Google Scholar] [CrossRef]
- Wu, D.; Cui, L.; Yang, G.; Ning, X.; Sun, L.; Zhou, Y. Preparing rhamnogalacturonan II domains from seven plant pectins using Penicillium oxalicum degradation and their structural comparison. Carbohydr. Polym. 2018, 180, 209–215. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).