RNA-Cleaving DNAzymes: Old Catalysts with New Tricks for Intracellular and In Vivo Applications
Abstract
1. Introduction
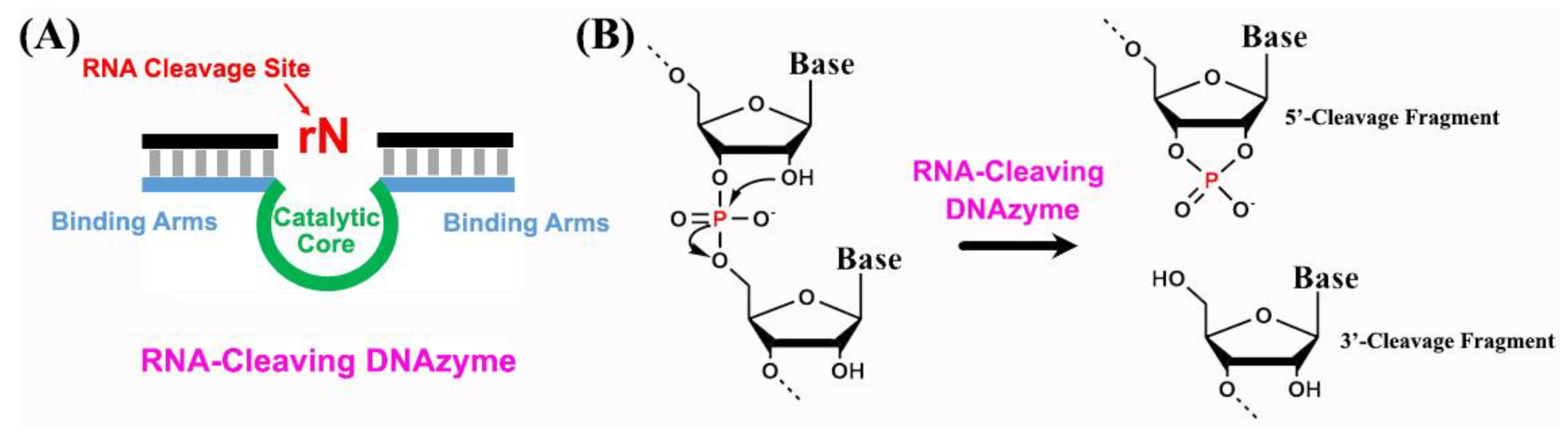
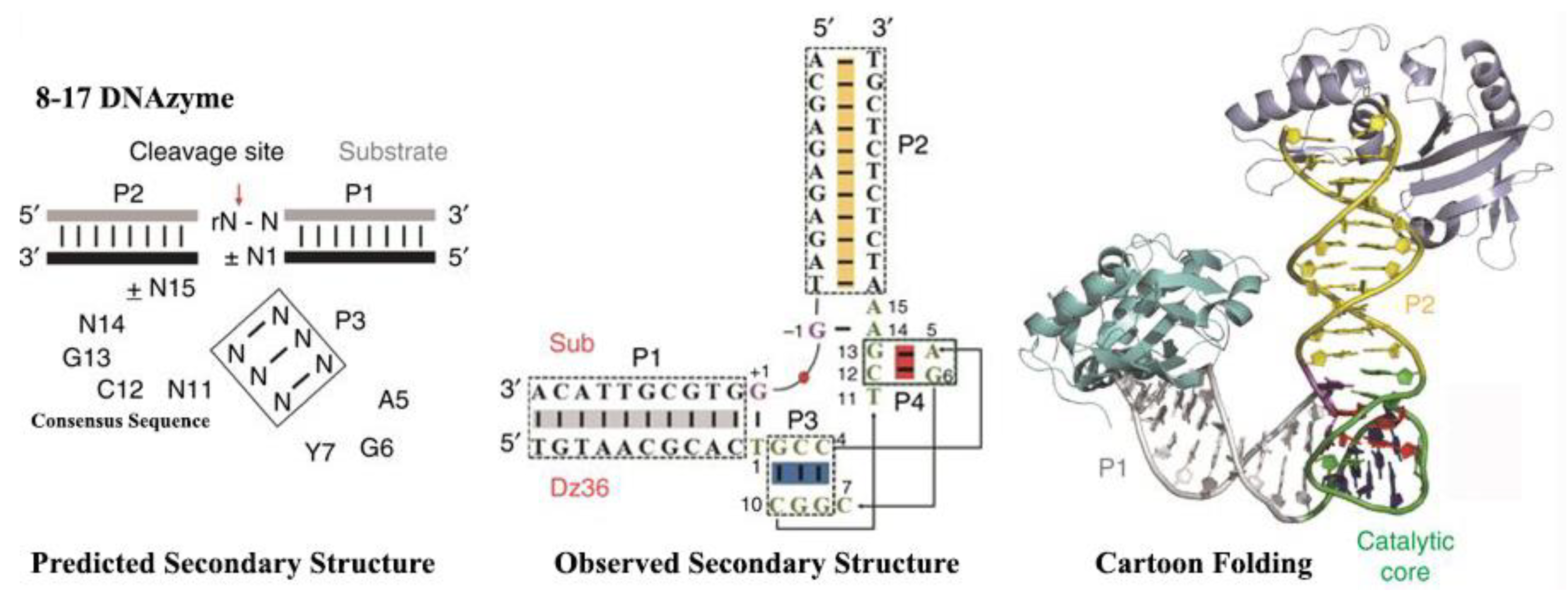
2. Structural Characterization of RNA-Cleaving DNAzymes
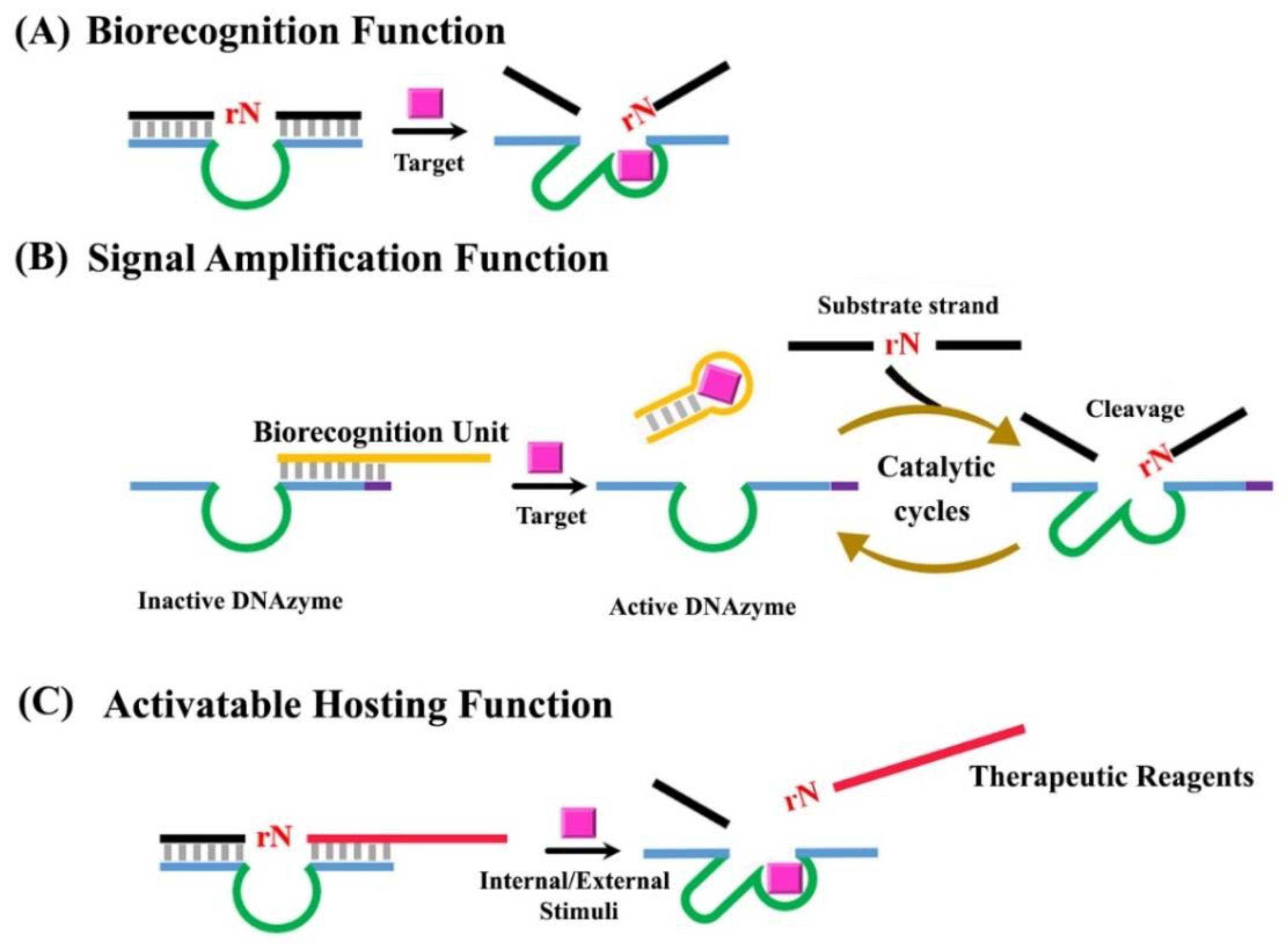
3. The General Concept of Translating RNA-Cleaving DNAzymes into Functional Probes
4. RNA-Cleaving DNAzymes-Based Functional Probes for Intracellular Applications
4.1. Intracellular Sensing and Imaging of Metal Ions
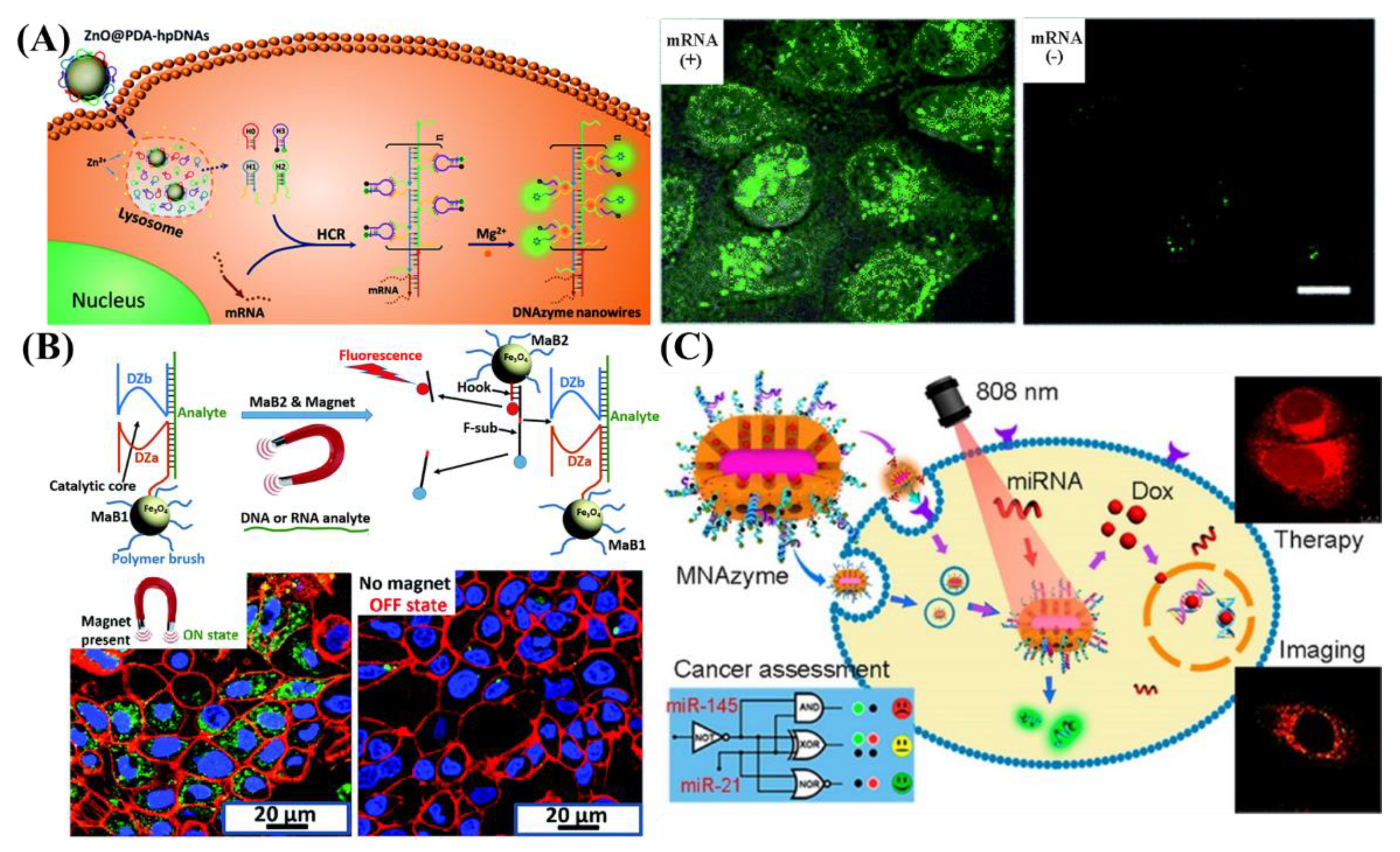
4.2. Intracellular Sensing and Imaging of RNAs
4.3. Intracellular Sensing and Imaging of Adenosine 5′-Triphosphate
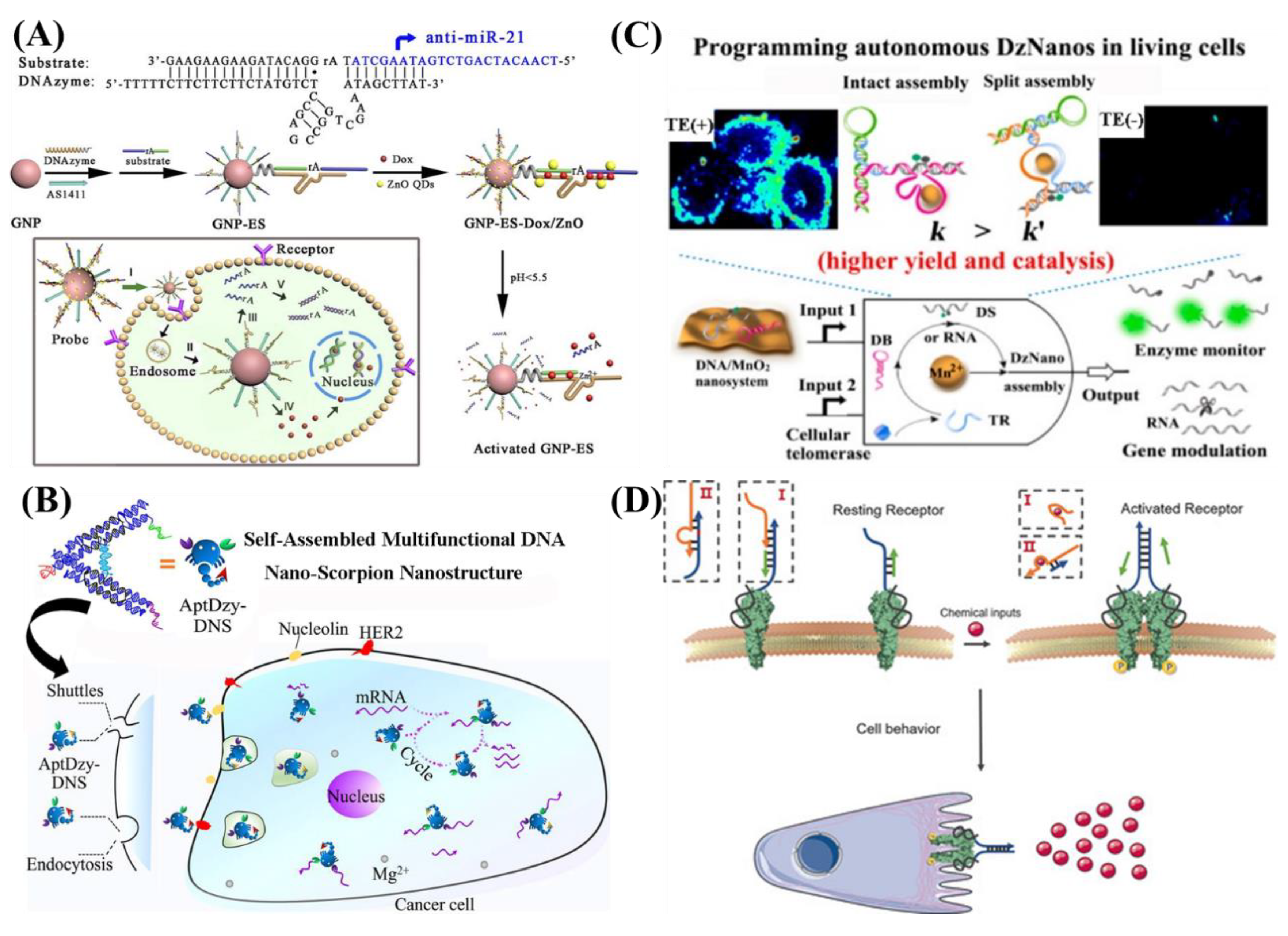
4.4. Intracellular Gene Regulation
4.5. Other Intracellular Applications
5. RNA-Cleaving DNAzymes-Based Functional Probes for In Vivo Applications
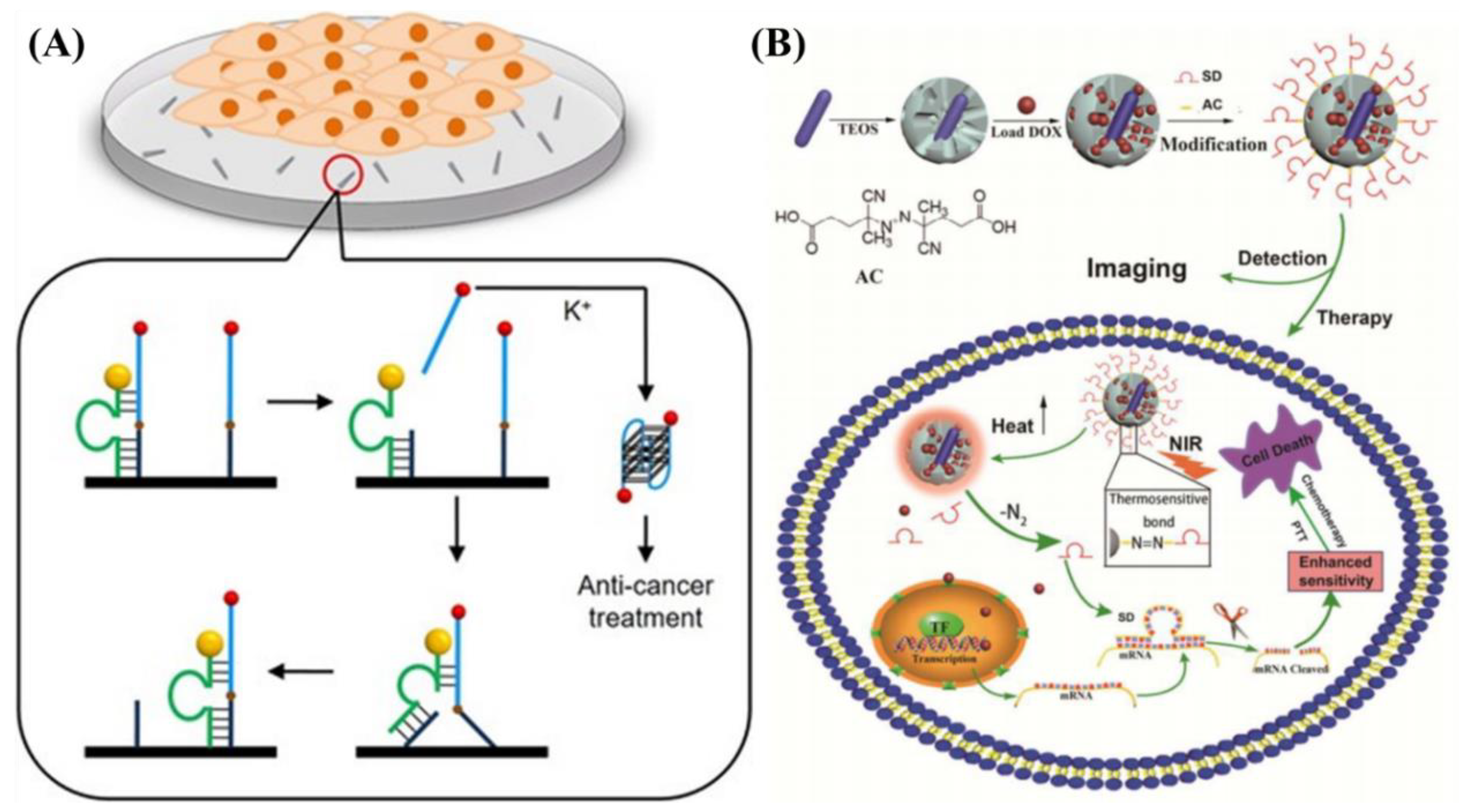
5.1. In Vivo Cancer Therapy
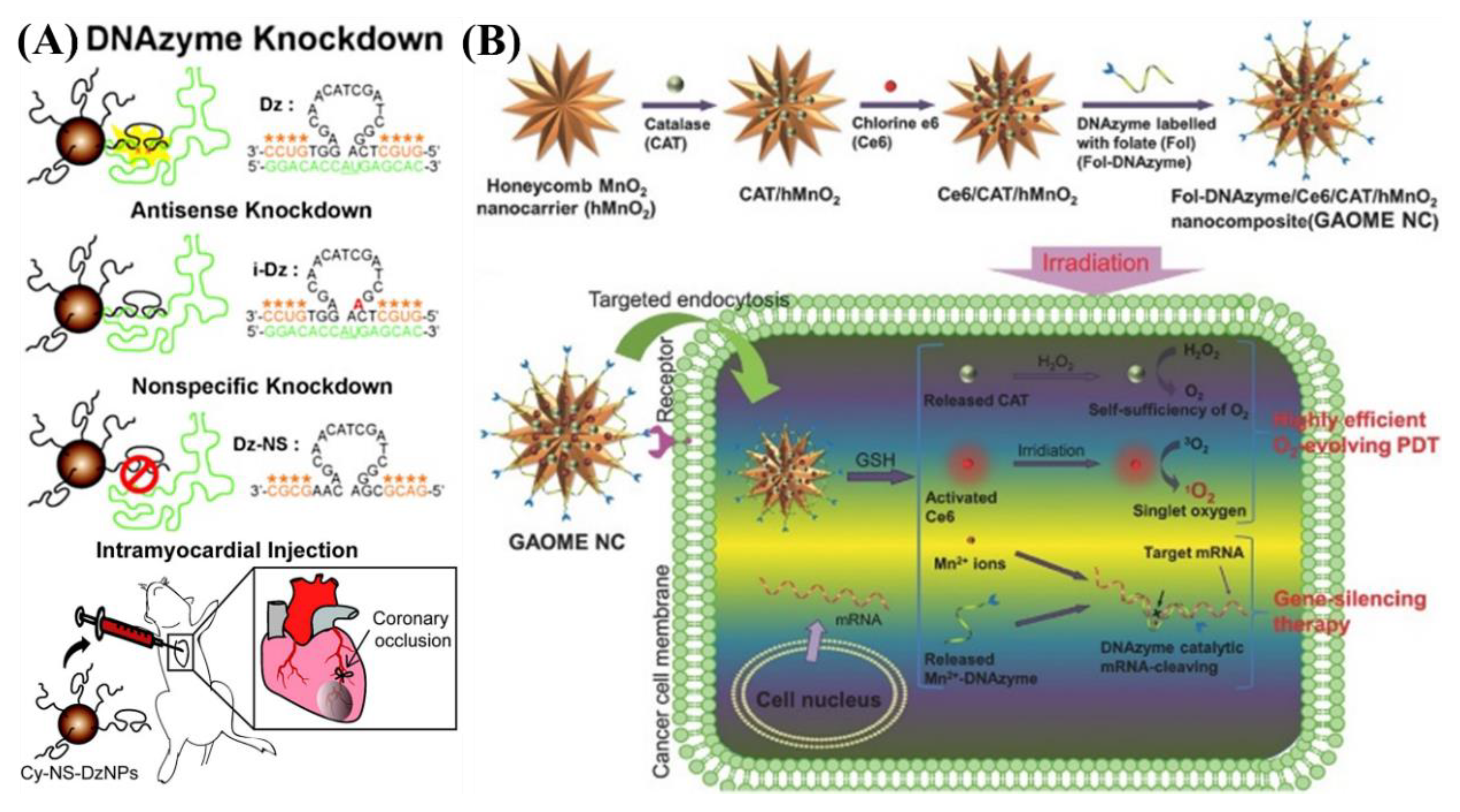
5.2. In Vivo Gene Regulation
5.3. Other In Vivo Applications
6. Clinical Trials of RNA-Cleaving DNAzymes
7. Conclusions and Perspectives
Funding
Acknowledgments
Conflicts of Interest
References
- Ward, W.L.; Plakos, K.; DeRose, V.J. Nucleic acid catalysis: Metals, nucleobases, and other cofactors. Chem. Rev. 2014, 114, 4318–4342. [Google Scholar] [CrossRef] [PubMed]
- Breaker, R.R.; Joyce, G.F. A DNA enzyme that cleaves RNA. Chem. Biol. 1994, 1, 223–229. [Google Scholar] [CrossRef]
- Santoro, S.W.; Joyce, G.F. A general purpose RNA-cleaving DNA enzyme. Proc. Natl. Acad. Sci. USA 1997, 94, 4262–4266. [Google Scholar] [CrossRef] [PubMed]
- Silverman, S.K. Deoxyribozymes: Selection design and serendipity in the development of DNA catalysts. Acc. Chem. Res. 2009, 42, 1521–1531. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Cao, Z.; Lu, Y. Functional nucleic acid sensors. Chem. Rev. 2009, 109, 1948–1998. [Google Scholar] [CrossRef] [PubMed]
- Silverman, S.K. Catalytic DNA: Scope, Applications, and Biochemistry of Deoxyribozymes. Trends Biochem. Sci. 2016, 41, 595–609. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Lu, Y. Biocomputing for Portable, Resettable, and Quantitative Point-of-Care Diagnostics: Making the Glucose Meter a Logic-Gate Responsive Device for Measuring Many Clinically Relevant Targets. Angew. Chem. 2018, 57, 9702–9706. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.J.; Xing, H.; Lu, Y. Translating molecular detections into a simple temperature test using a target-responsive smart thermometer. Chem. Sci. 2018, 9, 3906–3910. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Newbigging, A.M.; Wang, Z.; Tao, J.; Deng, W.; Le, X.C.; Zhang, H. DNAzyme-Mediated Assays for Amplified Detection of Nucleic Acids and Proteins. Anal. Chem. 2018, 90, 190–207. [Google Scholar] [CrossRef] [PubMed]
- Baum, D.A.; Silverman, S.K. Deoxyribozymes: Useful DNA catalysts in vitro and in vivo. Cell. Mol. Life Sci. 2008, 65, 2156–2174. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Ding, J.; Liu, J. Theranostic DNAzymes. Theranostics 2017, 7, 1010–1025. [Google Scholar] [CrossRef] [PubMed]
- Potaczek, D.P.; Unger, S.D.; Zhang, N.; Taka, S.; Michel, S.; Akdag, N.; Lan, F.; Helfer, M.; Hudemann, C.; Eickmann, M.; et al. Development and characterization of DNAzyme candidates demonstrating significant efficiency against human rhinoviruses. J. Allergy Clin. Immunol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Morrison, D.; Rothenbroker, M.; Li, Y.F. DNAzymes: Selected for Applications. Small Methods 2018, 2, 1700319. [Google Scholar] [CrossRef]
- Hwang, K.; Hosseinzadeh, P.; Lu, Y. Biochemical and Biophysical Understanding of Metal Ion Selectivity of DNAzymes. Inorg. Chim. Acta 2016, 452, 12–24. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Chang, D.; Li, Y. Discovery and Biosensing Applications of Diverse RNA-Cleaving DNAzymes. Acc. Chem. Res. 2017, 50, 2273–2283. [Google Scholar] [CrossRef] [PubMed]
- Hollenstein, M. DNA Catalysis: The Chemical Repertoire of DNAzymes. Molecules 2015, 20, 20777–20804. [Google Scholar] [CrossRef] [PubMed]
- Gong, L.; Zhao, Z.; Lv, Y.F.; Huan, S.Y.; Fu, T.; Zhang, X.B.; Shen, G.L.; Yu, R.Q. DNAzyme-based biosensors and nanodevices. Chem. Commun. 2015, 51, 979–995. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Liu, J. Multi-metal-dependent nucleic acid enzymes. Metallomics 2018, 10, 30–48. [Google Scholar] [CrossRef] [PubMed]
- Wiraja, C.; Yeo, D.C.; Lio, D.; Zheng, M.; Xu, C. Functional Imaging with Nucleic Acid-based Sensors: Technology, Application and Future Healthcare Prospects. Chembiochem 2018. [Google Scholar] [CrossRef] [PubMed]
- Müller, S.; Appel, B.; Balke, D.; Hieronymus, R.; Nübel, C. Thirty-five years of research into ribozymes and nucleic acid catalysis: Where do we stand today? F1000Research 2016, 5, 1511. [Google Scholar] [CrossRef] [PubMed]
- Lan, T.; Lu, Y. Metal Ion-Dependent DNAzymes and Their Applications as Biosensors. In Interplay between Metal Ions and Nucleic Acids; Sigel, A., Sigel, H., Sigel, R.K.O., Eds.; Springer: Dordrecht, The Netherlands, 2012; pp. 217–248. [Google Scholar]
- Nowakowski, J.; Shim, P.J.; Joyce, G.F.; Stout, C.D. Crystallization of the 10–23 DNA enzyme using a combinatorial screen of paired oligonucleotides. Acta Crystallogr. Sect. D Biol. Crystallogr. 1999, 55, 1885–1892. [Google Scholar] [CrossRef]
- Nowakowski, J.; Shim, P.J.; Prasad, G.S.; Stout, C.D.; Joyce, G.F. Crystal structure of an 82-nucleotide RNA-DNA complex formed by the 10–23 DNA enzyme. Nat. Struct. Biol. 1999, 6, 151–156. [Google Scholar] [PubMed]
- Parkinson, G.N.; Lee, M.P.H.; Neidle, S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature 2002, 417, 876–880. [Google Scholar] [CrossRef] [PubMed]
- Kondo, J.; Yamada, T.; Hirose, C.; Okamoto, I.; Tanaka, Y.; Ono, A. Crystal Structure of Metallo DNA Duplex Containing Consecutive Watson-Crick-like T-Hg-II-T Base Pairs. Angew. Chem. 2014, 53, 2385–2388. [Google Scholar] [CrossRef] [PubMed]
- Ponce-Salvatierra, A.; Wawrzyniak-Turek, K.; Steuerwald, U.; Hobartner, C.; Pena, V. Crystal structure of a DNA catalyst. Nature 2016, 529, 231–234. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.H.; Yu, X.; Chen, Y.Q.; Zhang, J.; Wu, B.X.; Zheng, L.N.; Haruehanroengra, P.; Wang, R.; Li, S.H.; Lin, J.Z.; et al. Crystal structure of an RNA-cleaving DNAzyme. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Wirmer-Bartoschek, J.; Schwalbe, H. Understanding How DNA Enzymes Work. Angew. Chem. 2016, 55, 5376–5377. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.J.; Cheng, F.F.; Zheng, T.T.; Zhu, J.J. Versatile aptasensor for electrochemical quantification of cell surface glycan and naked-eye tracking glycolytic inhibition in living cells. Biosens. Bioelectron. 2017, 89, 937–945. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, C.; Liu, X.; Zhou, X.; Wang, F. Construction of an enzyme-free concatenated DNA circuit for signal amplification and intracellular imaging. Chem. Sci. 2018, 9, 5842–5849. [Google Scholar] [CrossRef] [PubMed]
- Giannetti, A.; Tombelli, S.; Baldini, F. Oligonucleotide optical switches for intracellular sensing. Anal. Bioanal. Chem. 2013, 405, 6181–6196. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.J.; Cheng, F.F.; Li, J.J.; Zhu, J.J.; Lu, Y. Fluorescent nanoprobes for sensing and imaging of metal ions: Recent advances and future perspectives. Nano Today 2016, 11, 309–329. [Google Scholar] [CrossRef] [PubMed]
- Dean, K.M.; Qin, Y.; Palmer, A.E. Visualizing metal ions in cells: An overview of analytical techniques, approaches, and probes. Biochim. Biophys. Acta 2012, 1823, 1406–1415. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Hwang, K.; Lan, T.; Lu, Y. A DNAzyme-gold nanoparticle probe for uranyl ion in living cells. J. Am. Chem. Soc. 2013, 135, 5254–5257. [Google Scholar] [CrossRef] [PubMed]
- Zhai, T.T.; Ye, D.; Shi, Y.; Zhang, Q.W.; Qin, X.; Wang, C.; Xia, X.H. Plasmon Coupling Effect-Enhanced Imaging of Metal Ions in Living Cells Using DNAzyme Assembled Core-Satellite Structures. ACS Appl. Mater. Interfaces 2018, 10, 33966–33975. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Zhao, D.X.; Huang, J.X.; Li, J.; Zuo, X.L.; Wang, L.H.; Fan, C.H. Protein-mimicking nanoparticle (Protmin)-based nanosensor for intracellular analysis of metal ions. Nucl. Sci. Technol. 2018, 29, 5. [Google Scholar] [CrossRef]
- Zhang, L.; Yin, Q.-H.; Li, J.-M.; Huang, H.-Y.; Wu, Q.; Mao, Z.-W. Functionalization of dendritic polyethylene with cationic poly(p-phenylene ethynylene) enables efficient siRNA delivery for gene silencing. J. Mater. Chem. B 2013, 1, 2245–2251. [Google Scholar] [CrossRef]
- Li, L.; Feng, J.; Fan, Y.; Tang, B. Simultaneous imaging of Zn(2+) and Cu(2+) in living cells based on DNAzyme modified gold nanoparticle. Anal. Chem. 2015, 87, 4829–4835. [Google Scholar] [CrossRef] [PubMed]
- Si, H.; Sheng, R.; Li, Q.; Feng, J.; Li, L.; Tang, B. Highly Sensitive Fluorescence Imaging of Zn(2+) and Cu(2+) in Living Cells with Signal Amplification Based on Functional DNA Self-Assembly. Anal. Chem. 2018, 90, 8785–8792. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Liang, W.; Li, D.; Yuan, R.; Xiang, Y. Dual-color encoded DNAzyme nanostructures for multiplexed detection of intracellular metal ions in living cells. Biosens. Bioelectron. 2016, 85, 573–579. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Yin, X.; Huan, S.Y.; Chen, L.; Hu, X.X.; Xiong, M.Y.; Chen, K.; Zhang, X.B. Two-Photon DNAzyme-Gold Nanoparticle Probe for Imaging Intracellular Metal Ions. Anal. Chem. 2018, 90, 3118–3123. [Google Scholar] [CrossRef] [PubMed]
- Hong, C.Y.; Wu, S.X.; Li, S.H.; Liang, H.; Chen, S.; Li, J.; Yang, H.H.; Tan, W. Semipermeable Functional DNA-Encapsulated Nanocapsules as Protective Bioreactors for Biosensing in Living Cells. Anal. Chem. 2017, 89, 5389–5394. [Google Scholar] [CrossRef] [PubMed]
- Hwang, K.; Wu, P.W.; Kim, T.; Lei, L.; Tian, S.L.; Wang, Y.X.; Lu, Y. Photocaged DNAzymes as a General Method for Sensing Metal Ions in Living Cells. Angew. Chem. 2014, 53, 13798–13802. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Satyavolu, N.S.R.; Wu, Z.; Zhang, J.R.; Zhu, J.J.; Lu, Y. Near-Infrared Photothermally Activated DNAzyme-Gold Nanoshells for Imaging Metal Ions in Living Cells. Angew. Chem. 2017, 56, 6798–6802. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Feng, M.; Xiao, L.; Tong, A.; Xiang, Y. Postsynthetic Modification of DNA Phosphodiester Backbone for Photocaged DNAzyme. ACS Chem. Biol. 2016, 11, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Fan, H.; Satyavolu, N.S.R.; Wang, W.; Lake, R.; Jiang, J.H.; Lu, Y. Imaging Endogenous Metal Ions in Living Cells Using a DNAzyme-Catalytic Hairpin Assembly Probe. Angew. Chem. 2017, 56, 8721–8725. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Wu, X.; Krueger, C.J.; Chen, A.K. Engineering Novel Molecular Beacon Constructs to Study Intracellular RNA Dynamics and Localization. Genom. Proteom. Bioinform. 2017, 15, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Pitchiaya, S.; Heinicke, L.A.; Custer, T.C.; Walter, N.G. Single molecule fluorescence approaches shed light on intracellular RNAs. Chem. Rev. 2014, 114, 3224–3265. [Google Scholar] [CrossRef] [PubMed]
- Santangelo, P.J. Molecular beacons and related probes for intracellular RNA imaging. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2010, 2, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Bai, M.; Zhao, Y.; Cao, K.; Cao, X.; Zhao, Y. MnO2-Nanosheet-Powered Protective Janus DNA Nanomachines Supporting Robust RNA Imaging. Anal. Chem. 2018, 90, 2271–2276. [Google Scholar] [CrossRef] [PubMed]
- He, D.; He, X.; Yang, X.; Li, H.W. A smart ZnO@polydopamine-nucleic acid nanosystem for ultrasensitive live cell mRNA imaging by the target-triggered intracellular self-assembly of active DNAzyme nanostructures. Chem. Sci. 2017, 8, 2832–2840. [Google Scholar] [CrossRef] [PubMed]
- Bakshi, S.F.; Guz, N.; Zakharchenko, A.; Deng, H.; Tumanov, A.V.; Woodworth, C.D.; Minko, S.; Kolpashchikov, D.M.; Katz, E. Nanoreactors based on DNAzyme-functionalized magnetic nanoparticles activated by magnetic field. Nanoscale 2018, 10, 1356–1365. [Google Scholar] [CrossRef] [PubMed]
- Makarova, J.A.; Shkurnikov, M.U.; Wicklein, D.; Lange, T.; Samatov, T.R.; Turchinovich, A.A.; Tonevitsky, A.G. Intracellular and extracellular microRNA: An update on localization and biological role. Prog. Histochem. Cytochem. 2016, 51, 33–49. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, R.; Orbay, H.; Cai, W. Molecular Imaging Strategies for In Vivo Tracking of MicroRNAs: A Comprehensive Review. Curr. Med. Chem. 2013, 20, 3594–3603. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; He, Z.; Wang, C.; Chen, J.; Zhao, J.; Zhu, X.; Li, C.Z.; Min, Q.; Zhu, J.J. In situ amplification of intracellular microRNA with MNAzyme nanodevices for multiplexed imaging, logic operation, and controlled drug release. ACS Nano 2015, 9, 789–798. [Google Scholar] [CrossRef] [PubMed]
- Belter, A.; Rolle, K.; Piwecka, M.; Fedoruk-Wyszomirska, A.; Naskret-Barciszewska, M.Z.; Barciszewski, J. Inhibition of miR-21 in glioma cells using catalytic nucleic acids. Sci. Rep. 2016, 6, 24516. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Huang, J.; Yang, X.; Yang, Y.; Quan, K.; Xie, N.; Li, J.; Ma, C.; Wang, K. Gold Nanoparticle Loaded Split-DNAzyme Probe for Amplified miRNA Detection in Living Cells. Anal. Chem. 2017, 89, 8377–8383. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.T.; Chen, T.T.; Huo, J.; Chu, X. Nanoscale Zeolitic Imidazolate Framework-8 for Ratiometric Fluorescence Imaging of MicroRNA in Living Cells. Anal. Chem. 2017, 89, 12351–12359. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Cui, M.; Zhou, H.; Yang, W. DNAzyme Based Nanomachine for in Situ Detection of MicroRNA in Living Cells. ACS Sens. 2017, 2, 1847–1853. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Huang, J.; Yang, X.; He, X.; Quan, K.; Xie, N.; Ou, M.; Wang, K. Gold Nanoparticle Based Hairpin-Locked-DNAzyme Probe for Amplified miRNA Imaging in Living Cells. Anal. Chem. 2017, 89, 5850–5856. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Li, X.F.; Zhang, H.; Le, X.C. A microRNA-initiated DNAzyme motor operating in living cells. Nat. Commun. 2017, 8, 14378. [Google Scholar] [CrossRef] [PubMed]
- Mahdiannasser, M.; Karami, Z. An innovative paradigm of methods in microRNAs detection: Highlighting DNAzymes, the illuminators. Biosens. Bioelectron. 2018, 107, 123–144. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.T.; Zhao, M.P. In-vivo fluorescence imaging of adenosine 5’-triphosphate. TrAC Trensd Anal. Chem. 2016, 80, 190–203. [Google Scholar] [CrossRef]
- Yang, Y.; Huang, J.; Yang, X.; Quan, K.; Wang, H.; Ying, L.; Xie, N.; Ou, M.; Wang, K. Aptazyme-Gold Nanoparticle Sensor for Amplified Molecular Probing in Living Cells. Anal. Chem. 2016, 88, 5981–5987. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.L.; Wu, J.; Yao, Y.; Zhang, Y.; Liao, X.J.; Geng, D.Q.; Tang, D.Q. Proximity hybridization triggered strand displacement and DNAzyme assisted strand recycling for ATP fluorescence detection in vitro and imaging in living cells. RSC Adv. 2018, 8, 28161–28171. [Google Scholar] [CrossRef]
- Fan, H.; Zhang, X.; Lu, Y. Recent advances in DNAzyme-based gene silencing. Sci. China Chem. 2017, 60, 591–601. [Google Scholar] [CrossRef]
- Young, D.D.; Lively, M.O.; Deiters, A. Activation and Deactivation of DNAzyme and Antisense Function with Light for the Photochemical Regulation of Gene Expression in Mammalian Cells. J. Am. Chem. Soc. 2010, 132, 6183–6193. [Google Scholar] [CrossRef] [PubMed]
- Yehl, K.; Joshi, J.R.; Greene, B.L.; Dyer, R.B.; Nahta, R.; Salaita, K. Catalytic Deoxyribozyme-Modified Nanoparticles for RNAi-Independent Gene Regulation. ACS Nano 2012, 6, 9150–9157. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, A.K.; Cairns-Gibson, D.F.; Santiana, J.J.; Tolentino, M.Q.; Barber, H.M.; Rouge, J.L. Enzymatically Ligated DNA-Surfactants: Unmasking Hydrophobically Modified DNA for Intracellular Gene Regulation. ChemBioChem 2018, 19, 1734–1739. [Google Scholar] [CrossRef] [PubMed]
- Awino, J.K.; Gudipati, S.; Hartmann, A.K.; Santiana, J.J.; Cairns-Gibson, D.F.; Gomez, N.; Rouge, J.L. Nucleic Acid Nanocapsules for Enzyme-Triggered Drug Release. J. Am. Chem. Soc. 2017, 139, 6278–6281. [Google Scholar] [CrossRef] [PubMed]
- Zokaei, E.; Badoei-Dalfrad, A.; Ansari, M.; Karami, Z.; Eslaminejad, T.; Nematollahi-Mahani, S.N. Therapeutic Potential of DNAzyme Loaded on Chitosan/Cyclodextrin Nanoparticle to Recovery of Chemosensitivity in the MCF-7 Cell Line. Appl. Biochem. Biotechnol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Zhao, Z.; Yan, G.; Zhang, X.; Yang, C.; Meng, H.; Chen, Z.; Liu, H.; Tan, W. A smart DNAzyme-MnO(2) nanosystem for efficient gene silencing. Angew. Chem. 2015, 54, 4801–4805. [Google Scholar] [CrossRef] [PubMed]
- He, Z.M.; Zhang, P.H.; Li, X.; Zhang, J.R.; Zhu, J.J. A Targeted DNAzyme-Nanocomposite Probe Equipped with Built-in Zn(2+) Arsenal for Combined Treatment of Gene Regulation and Drug Delivery. Sci. Rep. 2016, 6, 22737. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Mo, F.; Wu, J.; Huang, Y.; Zhou, H.; Ding, S.; Chen, W. A multifunctional DNA nano-scorpion for highly efficient targeted delivery of mRNA therapeutics. Sci. Rep. 2018, 8, 10196. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Bai, M.; Cao, K.; Zhao, Y.; Cao, X.W.; Wei, J.; Wu, N.; Li, J.; Wang, L.H.; Fan, C.H.; et al. Programming Enzyme-Initiated Autonomous DNAzyme Nanodevices in Living Cells. ACS Nano 2017, 11, 11908–11914. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, M.; Shi, T.H.; Yang, S.H.; Zhang, J.H.; Wang, H.H.; Nie, Z. A DNA-Mediated Chemically Induced Dimerization (D-CID) Nanodevice for Nongenetic Receptor Engineering To Control Cell Behavior. Angew. Chem. 2018, 57, 10226–10230. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Bai, M.; Cao, K.; Zhao, Y.; Wei, J.; Zhao, Y.X. Fabricating MnO2 Nanozymes as Intracellular Catalytic DNA Circuit Generators for Versatile Imaging of Base-Excision Repair in Living Cells. Adv. Funct. Mater. 2017, 27, 1702748. [Google Scholar] [CrossRef]
- Chen, X.; Zhao, J.; Chen, T.; Gao, T.; Zhu, X.; Li, G. Nondestructive Analysis of Tumor-Associated Membrane Protein Integrating Imaging and Amplified Detection in situ Based on Dual-Labeled DNAzyme. Theranostics 2018, 8, 1075–1083. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.J.; Zheng, T.T.; Cheng, F.F.; Zhang, J.R.; Zhu, J.J. Toward the Early Evaluation of Therapeutic Effects: An Electrochemical Platform for Ultrasensitive Detection of Apoptotic Cells. Anal. Chem. 2011, 83, 7902–7909. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.P.; Zhang, J.J.; Cheng, F.F.; Zheng, T.T.; Wang, C.M.; Zhu, J.J. Green and facile synthesis of highly biocompatible graphene nanosheets and its application for cellular imaging and drug delivery. J. Mater. Chem. 2011, 21, 12034–12040. [Google Scholar] [CrossRef]
- Zhang, J.J.; Gu, M.M.; Zheng, T.T.; Zhu, J.J. Synthesis of Gelatin-Stabilized Gold Nanoparticles and Assembly of Carboxylic Single-Walled Carbon Nanotubes/Au Composites for Cytosensing and Drug Uptake. Anal. Chem. 2009, 81, 6641–6648. [Google Scholar] [CrossRef] [PubMed]
- Li, F.R.; Cha, T.G.; Pan, J.; Ozcelikkale, A.; Han, B.; Choi, J.H. DNA Walker-Regulated Cancer Cell Growth Inhibition. ChemBioChem 2016, 17, 1138–1141. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Li, Z.H.; Liu, H.F.; Chen, S.Z.; Wang, F.; Wang, L.; Li, N.; Ge, K.; Yang, X.J.; Liang, X.J.; et al. Biodegradable, multifunctional DNAzyme nanoflowers for enhanced cancer therapy. NPG Asia Mater. 2017, 9, e365. [Google Scholar] [CrossRef]
- Jin, Y.; Liang, L.N.; Sun, X.J.; Yu, G.S.; Chen, S.Z.; Shi, S.T.; Liu, H.F.; Li, Z.H.; Ge, K.; Liu, D.D.; et al. Deoxyribozyme-nanosponges for improved photothermal therapy by overcoming thermoresistance. NPG Asia Mater. 2018, 10, 373–384. [Google Scholar] [CrossRef]
- Sun, X.; Jin, Y.; Wang, H.; Feng, N.; Li, Z.; Liu, D.; Ge, K.; Liu, H.; Zhang, J.-C.; Yang, X. A NIR-light activated nanoplatform for sensitizing triple negative breast cancer against therapeutic resistance to enhance treatment effect. J. Mater. Chem. B 2018. [Google Scholar] [CrossRef]
- Yu, L.; Chen, Y.; Lin, H.; Gao, S.; Chen, H.; Shi, J. Magnesium-Engineered Silica Framework for pH-Accelerated Biodegradation and DNAzyme-Triggered Chemotherapy. Small 2018, 14, e1800708. [Google Scholar] [CrossRef] [PubMed]
- Ryoo, S.R.; Jang, H.; Kim, K.S.; Lee, B.; Kim, K.B.; Kim, Y.K.; Yeo, W.S.; Lee, Y.; Kim, D.E.; Min, D.H. Functional delivery of DNAzyme with iron oxide nanoparticles for hepatitis C virus gene knockdown. Biomaterials 2012, 33, 2754–2761. [Google Scholar] [CrossRef]
- Somasuntharam, I.; Yehl, K.; Carroll, S.L.; Maxwell, J.T.; Martinez, M.D.; Che, P.L.; Brown, M.E.; Salaita, K.; Davis, M.E. Knockdown of TNF-alpha by DNAzyme gold nanoparticles as an anti-inflammatory therapy for myocardial infarction. Biomaterials 2016, 83, 12–22. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Li, Y.; Gao, X.; Yu, Z.; Pan, W.; Tang, B. Multiplexed gene silencing in living cells and in vivo using a DNAzymes-CoOOH nanocomposite. Chem. Commun. 2017, 53, 4962–4965. [Google Scholar] [CrossRef] [PubMed]
- He, D.; Hai, L.; He, X.; Yang, X.; Li, H.-W. Glutathione-Activatable and O2/Mn2+-Evolving Nanocomposite for Highly Efficient and Selective Photodynamic and Gene-Silencing Dual Therapy. Adv. Funct. Mater. 2017, 27, 1704089. [Google Scholar] [CrossRef]
- Popp, V.; Gerlach, K.; Mott, S.; Turowska, A.; Garn, H.; Atreya, R.; Lehr, H.A.; Ho, I.C.; Renz, H.; Weigmann, B.; et al. Rectal Delivery of a DNAzyme That Specifically Blocks the Transcription Factor GATA3 and Reduces Colitis in Mice. Gastroenterology 2017, 152, 176–192. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Cho, E.A.; Li, Y.; Sockler, J.; Parish, C.R.; Chong, B.H.; Edwards, J.; Dodds, T.J.; Ferguson, P.M.; Wilmott, J.S.; et al. Melanoma protective antitumor immunity activated by catalytic DNA. Oncogene 2018, 37, 5115–5126. [Google Scholar] [CrossRef] [PubMed]
- Dass, C.R. Deoxyribozymes: Cleaving a path to clinical trials. Trends Pharmacol. Sci. 2004, 25, 395–397. [Google Scholar] [CrossRef] [PubMed]
- Monteleone, G.; Fantini, M.C.; Onali, S.; Zorzi, F.; Sancesario, G.; Bernardini, S.; Calabrese, E.; Viti, F.; Monteleone, I.; Biancone, L.; et al. Phase I Clinical Trial of Smad7 Knockdown Using Antisense Oligonucleotide in Patients With Active Crohn’s Disease. Mol. Ther. 2012, 20, 870–876. [Google Scholar] [CrossRef] [PubMed]
- Krug, N.; Hohlfeld, J.M.; Buhl, R.; Renz, J.; Garn, H.; Renz, H. Blood eosinophils predict therapeutic effects of a GATA3-specific DNAzyme in asthma patients. J. Allergy Clin. Immunol. 2017, 140, 625–628. [Google Scholar] [CrossRef] [PubMed]
- Homburg, U.; Renz, H.; Timmer, W.; Hohlfeld, J.M.; Seitz, F.; Luer, K.; Mayer, A.; Wacker, A.; Schmidt, O.; Kuhlmann, J.; et al. Safety and tolerability of a novel inhaled GATA3 mRNA targeting DNAzyme in patients with T(H)2-driven asthma. J. Allergy Clin. Immunol. 2015, 136, 797–800. [Google Scholar] [CrossRef] [PubMed]
- Sel, S.; Wegmann, M.; Dicke, T.; Sel, S.; Henke, W.; Yildirim, A.O.; Renz, H.; Garn, H. Effective prevention and therapy of experimental allergic asthma using a GATA-3-specific DNAzyme. J. Allergy Clin. Immunol. 2008, 121, 910–916. [Google Scholar] [CrossRef] [PubMed]
- Turowska, A.; Librizzi, D.; Baumgartl, N.; Kuhlmann, J.; Dicke, T.; Merkel, O.; Homburg, U.; Hoffken, H.; Renz, H.; Garn, H. Biodistribution of the GATA-3-specific DNAzyme hgd40 after inhalative exposure in mice, rats and dogs. Toxicol. Appl. Pharmacol. 2013, 272, 365–372. [Google Scholar] [CrossRef] [PubMed]
- Sulaiman, I.; Lim, J.C.W.; Soo, H.L.; Stanslas, J. Molecularly targeted therapies for asthma: Current development, challenges and potential clinical translation. Pulm. Pharmacol. Ther. 2016, 40, 52–68. [Google Scholar] [CrossRef] [PubMed]
- Garn, H.; Renz, H. GATA-3-specific DNAzyme—A novel approach for stratified asthma therapy. Eur. J. Immunol. 2017, 47, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Krug, N.; Hohlfeld, J.M.; Kirsten, A.M.; Kornmann, O.; Beeh, K.M.; Kappeler, D.; Korn, S.; Ignatenko, S.; Timmer, W.; Rogon, C.; et al. Allergen-Induced Asthmatic Responses Modified by a GATA3-Specific DNAzyme. N. Engl. J. Med. 2015, 372, 1987–1995. [Google Scholar] [CrossRef] [PubMed]
- Greulich, T.; Hohlfeld, J.M.; Neuser, P.; Lueer, K.; Klemmer, A.; Schade-Brittinger, C.; Harnisch, S.; Garn, H.; Renz, H.; Homburg, U.; et al. A GATA3-specific DNAzyme attenuates sputum eosinophilia in eosinophilic COPD patients: A feasibility randomized clinical trial. Respir. Res. 2018, 19, 55. [Google Scholar] [CrossRef] [PubMed]
- Cho, E.A.; Moloney, F.J.; Cai, H.; Au-Yeung, A.; China, C.; Scolyer, R.A.; Yosufi, B.; Raftery, M.J.; Deng, J.Z.; Morton, S.W.; et al. Safety and tolerability of an intratumorally injected DNAzyme, Dz13, in patients with nodular basal-cell carcinoma: A phase 1 first-in-human trial (DISCOVER). Lancet 2013, 381, 1835–1843. [Google Scholar] [CrossRef]
- Li, Z.H.; Wang, J.; Li, Y.X.; Liu, X.W.; Yuan, Q. Self-assembled DNA nanomaterials with highly programmed structures and functions. Mat. Chem. Front. 2018, 2, 423–436. [Google Scholar] [CrossRef]
- Tack, F.; Noppe, M.; Van Dijck, A.; Dekeyzer, N.; Van Der Leede, B.J.; Bakker, A.; Wouters, W.; Janicot, M.; Brewster, M.E. Delivery of a DNAzyme targeting c-myc to HT29 colon carcinoma cells using a gold nanoparticulate approach. Die Pharm. 2008, 63, 221–225. [Google Scholar]
- Xing, Z.; Gao, S.; Duan, Y.; Han, H.; Li, L.; Yang, Y.; Li, Q. Delivery of DNAzyme targeting aurora kinase A to inhibit the proliferation and migration of human prostate cancer. Int. J. Nanomed. 2015, 10, 5715–5727. [Google Scholar]
- Hu, Q.; Li, H.; Wang, L.; Gu, H.; Fan, C. DNA Nanotechnology-Enabled Drug Delivery Systems. Chem. Rev. 2018. [Google Scholar] [CrossRef] [PubMed]


© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J. RNA-Cleaving DNAzymes: Old Catalysts with New Tricks for Intracellular and In Vivo Applications. Catalysts 2018, 8, 550. https://doi.org/10.3390/catal8110550
Zhang J. RNA-Cleaving DNAzymes: Old Catalysts with New Tricks for Intracellular and In Vivo Applications. Catalysts. 2018; 8(11):550. https://doi.org/10.3390/catal8110550
Chicago/Turabian StyleZhang, JingJing. 2018. "RNA-Cleaving DNAzymes: Old Catalysts with New Tricks for Intracellular and In Vivo Applications" Catalysts 8, no. 11: 550. https://doi.org/10.3390/catal8110550
APA StyleZhang, J. (2018). RNA-Cleaving DNAzymes: Old Catalysts with New Tricks for Intracellular and In Vivo Applications. Catalysts, 8(11), 550. https://doi.org/10.3390/catal8110550




