Deconstruction of the CYP153A6 Alkane Hydroxylase System: Limitations and Optimization of In Vitro Alkane Hydroxylation
Abstract
1. Introduction
2. Results and Discussion
2.1. Heterologous Expression and Purification
2.2. Cofactor Regeneration Using Glucose Dehydrogenase
2.3. Stabilization of Reactions
2.4. Ratios of Three-Component Systems
2.5. Comparison of Glucose and Glycerol Dehydrogenase for Cofactor Regeneration
2.6. Biotransformations in 300 mL Stirred Bioreactors
3. Discussion
4. Materials and Methods
4.1. Expression Constructs
4.2. Heterologous Expression and Protein Purification
4.3. Biotransformations—1 mL Scale
4.4. Biotransformations on 60 mL Scale in Shake Flasks
4.5. Biotransformations on 120 mL Scale in Stirred Bioreactors
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ortiz de Montellano, P.R. Hydrocarbon hydroxylation by cytochrome P450 enzymes. Chem. Rev. 2010, 110, 932–948. [Google Scholar] [CrossRef] [PubMed]
- Girvan, H.M.; Munro, A.W. Applications of microbial cytochrome P450 enzymes in biotechnology and synthetic biology. Curr. Opin. Chem. Biol. 2016, 31, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Urlacher, V.B.; Girhard, M. Cytochrome P450 monooxygenases: An update on perspectives for synthetic application. Trends Biotechnol. 2012, 30, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Urlacher, V.B.; Eiben, S. Cytochrome P450 monooxygenases: Perspectives for synthetic application. Trends Biotechnol. 2006, 24, 324–330. [Google Scholar] [CrossRef] [PubMed]
- Roiban, G.-D.; Reetz, M.T. Expanding the toolbox of organic chemists: Directed evolution of P450 monooxygenases as catalysts in regio- and stereoselective oxidative hydroxylation. Chem. Commun. 2015, 51, 2208–2224. [Google Scholar] [CrossRef] [PubMed]
- Johnston, J.B.; Ouellet, H.; Podust, L.M.; Ortiz de Montellano, P.R. Structural control of cytochrome P450-catalyzed ω-hydroxylation. Arch. Biochem. Biophys. 2010, 2, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Malca, S.H.; Scheps, D.; Kühnel, L.; Venegas-Venegas, E.; Seifert, A.; Nestl, B.M.; Hauer, B. Bacterial CYP153A monooxygenases for the synthesis of omega-hydroxylated fatty acids. Chem. Commun. 2012, 48, 5115. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, S.M.; Danesh-Azari, H.R.; Spandolf, C.; Weissenborn, M.J.; Grogan, G.; Hauer, B. Structure-Guided Redesign of CYP153AM.aq for the Improved Terminal Hydroxylation of Fatty Acids. ChemCatChem 2016, 8, 3178. [Google Scholar] [CrossRef]
- Duan, Y.; Ba, L.; Gao, J.; Gao, X.; Zhu, D.; de Jong, R.M.; Mink, D.; Kaluzna, I.; Lin, Z. Semi-rational engineering of cytochrome CYP153A from Marinobacter aquaeolei for improved ω-hydroxylation activity towards oleic acid. Appl. Microbiol. Biotechnol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, S.M.; Weissenborn, M.J.; Gricman, Ł.; Notonier, S.; Pleiss, J.; Hauer, B. The Impact of Linker Length on P450 Fusion Constructs: Activity, Stability and Coupling. ChemCatChem 2016, 8, 1591–1597. [Google Scholar] [CrossRef]
- Hannemann, F.; Bichet, A.; Ewen, K.M.; Bernhardt, R. Cytochrome P450 systems—Biological variations of electron transport chains. Biochim. Biophys. Acta 2007, 1770, 330–344. [Google Scholar] [CrossRef] [PubMed]
- Kubota, M.; Nodate, M.; Yasumoto-Hirose, M.; Uchiyama, T.; Kagami, O.; Shizuri, Y.; Misawa, N. Isolation and functional analysis of cytochrome P450 CYP153A genes from various environments. Biosci. Biotechnol. Biochem. 2005, 69, 2421–2430. [Google Scholar] [CrossRef] [PubMed]
- Scheps, D.; Honda Malca, S.; Richter, S.M.; Marisch, K.; Nestl, B.M.; Hauer, B. Synthesis of ω-hydroxy dodecanoic acid based on an engineered CYP153A fusion construct. Microb. Biotechnol. 2013, 6, 694–707. [Google Scholar] [CrossRef] [PubMed]
- Gricman, Ł.; Weissenborn, M.J.; Hoffmann, S.M.; Borlinghaus, N.; Hauer, B.; Pleiss, J. Redox Partner Interaction Sites in Cytochrome P450 Monooxygenases: In Silico Analysis and Experimental Validation. ChemistrySelect 2016, 1, 1243–1251. [Google Scholar] [CrossRef]
- Jung, E.; Park, B.G.; Ahsan, M.M.; Kim, J.; Yun, H.; Choi, K.-Y.; Kim, B.-G. Production of ω-hydroxy palmitic acid using CYP153A35 and comparison of cytochrome P450 electron transfer system in vivo. Appl. Microbiol. Biotechnol. 2016, 100, 10375–10384. [Google Scholar] [CrossRef] [PubMed]
- Kohl, A.; Srinivasamurthy, V.; Böttcher, D.; Kabisch, J.; Bornscheuer, U.T. Co-expression of an alcohol dehydrogenase and a cyclohexanone monooxygenase for cascade reactions facilitates the regeneration of the NADPH cofactor. Enzyme Microb. Technol. 2018, 108, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Lundemo, M.T.; Notonier, S.; Striedner, G.; Hauer, B.; Woodley, J.M. Process limitations of a whole-cell P450 catalyzed reaction using a CYP153A-CPR fusion construct expressed in Escherichia coli. Appl. Microbiol. Biotechnol. 2016, 100, 1197–1208. [Google Scholar] [CrossRef] [PubMed]
- White, B.E.; Fenner, C.J.; Smit, M.S.; Harrison, S.T.L. Effect of cell permeability and dehydrogenase expression on octane activation by CYP153A6-based whole cell Escherichia coli catalysts. Microb. Cell Fact. 2017, 16, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Olaofe, O.A.; Fenner, C.J.; Gudiminchi, R.K.; Smit, M.S.; Harrison, S.T. The influence of microbial physiology on biocatalyst activity and efficiency in the terminal hydroxylation of n-octane using Escherichia coli expressing the alkane hydroxylase, CYP153A6. Microb. Cell Fact. 2013, 12, 8. [Google Scholar] [CrossRef] [PubMed]
- Van Beilen, J.B.; Holtackers, R.; Lüscher, D.; Bauer, U.; Witholt, B.; Duetz, W.A. Biocatalytic production of perillyl alcohol from limonene by using a novel Mycobacterium sp. cytochrome P450 alkane hydroxylase expressed in Pseudomonas putida. Appl. Environ. Microbiol. 2005, 71, 1737–1744. [Google Scholar] [CrossRef] [PubMed]
- Gudiminchi, R.K.; Randall, C.; Opperman, D.J.; Olaofe, O.A; Harrison, S.T.L.; Albertyn, J.; Smit, M.S. Whole-cell hydroxylation of n-octane by Escherichia coli strains expressing the CYP153A6 operon. Appl. Microbiol. Biotechnol. 2012, 96, 1507–1516. [Google Scholar] [CrossRef]
- Pennec, A.; Jacobs, C.L.; Opperman, D.J.; Smit, M.S. Revisiting Cytochrome P450-Mediated Oxyfunctionalization of Linear and Cyclic Alkanes. Adv. Synth. Catal. 2015, 357, 118–130. [Google Scholar] [CrossRef]
- Funhoff, E.G.; Bauer, U.; García-Rubio, I.; Witholt, B.; van Beilen, J.B. CYP153A6, a soluble P450 oxygenase catalyzing terminal-alkane hydroxylation. J. Bacteriol. 2006, 188, 5220–5227. [Google Scholar] [CrossRef] [PubMed]
- Van Beilen, J.B.; Funhoff, E.G.; van Loon, A.; Just, A.; Kaysser, L.; Bouza, M.; Holtackers, R.; Röthlisberger, M.; Li, Z.; Witholt, B. Cytochrome P450 alkane hydroxylases of the CYP153 family are common in alkane-degrading eubacteria lacking integral membrane alkane hydroxylases. Appl. Environ. Microbiol. 2006, 72, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Bordeaux, M.; Girval, D.; Rullaud, R.; Subileau, M.; Dubreucq, E.; Drone, J. High-cell-density cultivation of recombinant Escherichia coli, purification and characterization of a self-sufficient biosynthetic octane ω-hydroxylase. Appl. Microbiol. Biotechnol. 2014, 98, 6275–6283. [Google Scholar] [CrossRef] [PubMed]
- Weckbecker, A.; Hummel, W. Glucose dehydrogenase for the regeneration of NADPH and NADH. In Microbial Enzymes and Biotransformations; Barredo, J.L., Ed.; Humana Press: New York, NY, USA, 2005; Volume 17, pp. 225–238. [Google Scholar]
- Yamamoto, K.; Nagao, T.; Makino, Y.; Urabe, I.; Okada, H. Characterization of mutant glucose dehydrogenases with increasing stability. Ann. N. Y. Acad. Sci. 1990, 613, 362–365. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Chi, Y.T.; Toh, H.H.; Li, Z. Evolving P450pyr monooxygenase for highly regioselective terminal hydroxylation of n-butanol to 1,4-butanediol. Chem. Commun. 2015, 51, 914–917. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, Z. Evolving P450pyr Monooxygenase for Regio- and Stereoselective Hydroxylations. Chim. Int. J. Chem. 2015, 69, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Nebel, B.A.; Scheps, D.; Honda Malca, S.; Nestl, B.M.; Breuer, M.; Wagner, H.G.; Breitscheidel, B.; Kratz, D.; Hauer, B. Biooxidation of n-butane to 1-butanol by engineered P450 monooxygenase under increased pressure. J. Biotechnol. 2014, 191, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Liu, J.; Li, Z. Engineering of p450pyr hydroxylase for the highly regio- and enantioselective subterminal hydroxylation of alkanes. Angew. Chem. Int. Ed. Engl. 2014, 53, 3120–3124. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Liu, J.; Pham, S.Q.; Li, Z. Engineered P450pyr monooxygenase for asymmetric epoxidation of alkenes with unique and high enantioselectivity. Chem. Commun. 2013, 49, 11572. [Google Scholar] [CrossRef] [PubMed]
- Pham, S.Q.; Pompidor, G.; Liu, J.; Li, X.-D.; Li, Z. Evolving P450pyr hydroxylase for highly enantioselective hydroxylation at non-activated carbon atom. Chem. Commun. 2012, 48, 4618–4620. [Google Scholar] [CrossRef] [PubMed]
- Mohammadzadeh-K, A.; Feeney, R.E.; Samuels, R.B.; Smith, L.M. Solubility of alkanes in protein solutions. Biochim. Biophys. Acta–Protein Struct. 1967, 147, 583–589. [Google Scholar] [CrossRef]
- Piattoni, C.V.; Figueroa, C.M.; Asención Diez, M.D.; Parcerisa, I.L.; Antuña, S.; Comelli, R.A.; Guerrero, S.A.; Beccaria, A.J.; Iglesias, A.Á. Production and characterization of Escherichia coli glycerol dehydrogenase as a tool for glycerol recycling. Process Biochem. 2013, 48, 406–412. [Google Scholar] [CrossRef]
- Guengerich, F.P.; Martin, M.V.; Sohl, C.D.; Cheng, Q. Measurement of cytochrome P450 and NADPH-cytochrome P450 reductase. Nat. Protoc. 2009, 4, 1245–1251. [Google Scholar] [CrossRef] [PubMed]
- Schrewe, M.; Julsing, M.K.; Bühler, B.; Schmid, A. Whole-cell biocatalysis for selective and productive C–O functional group introduction and modification. Chem. Soc. Rev. 2013, 42, 6346–6377. [Google Scholar] [CrossRef] [PubMed]

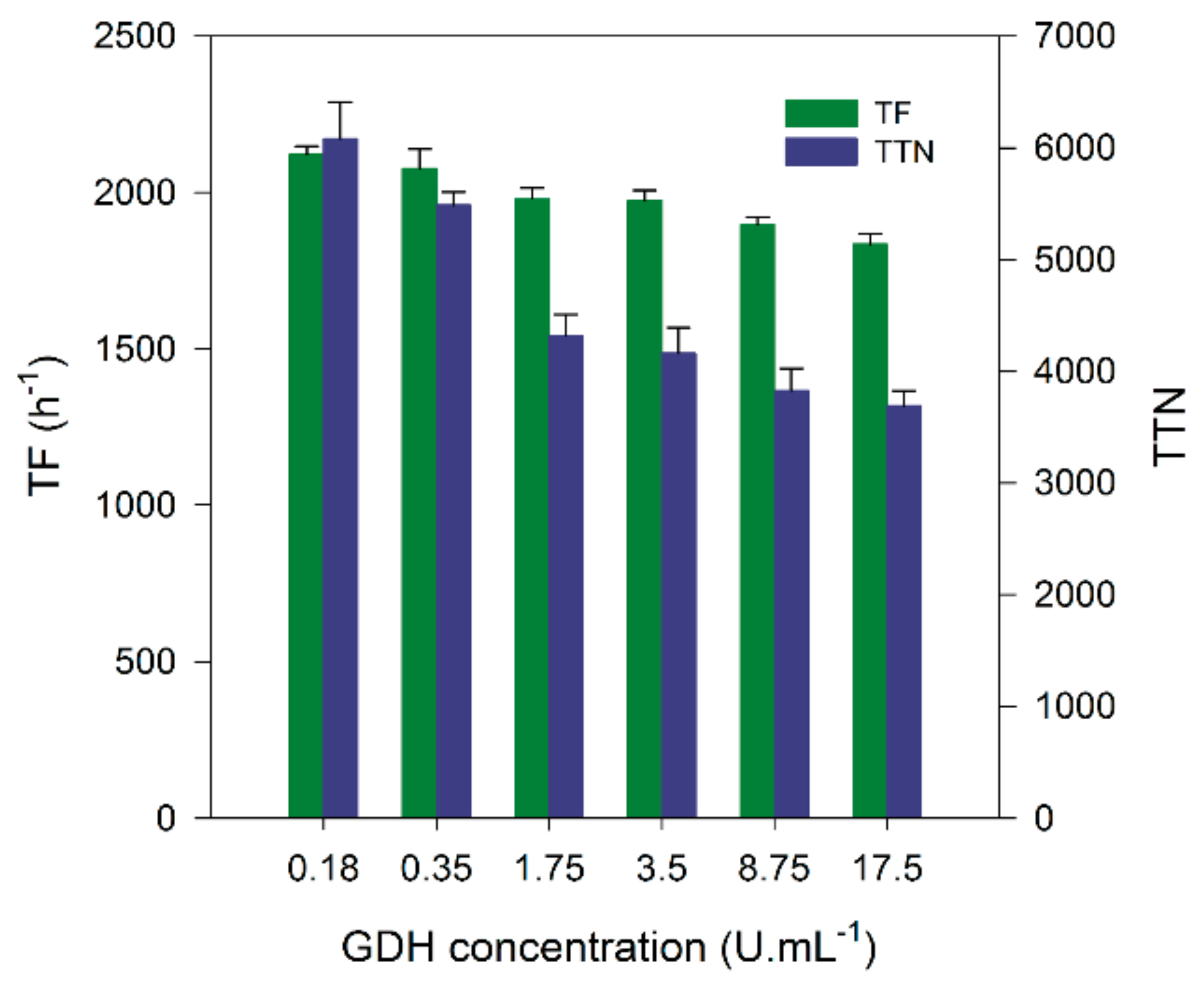

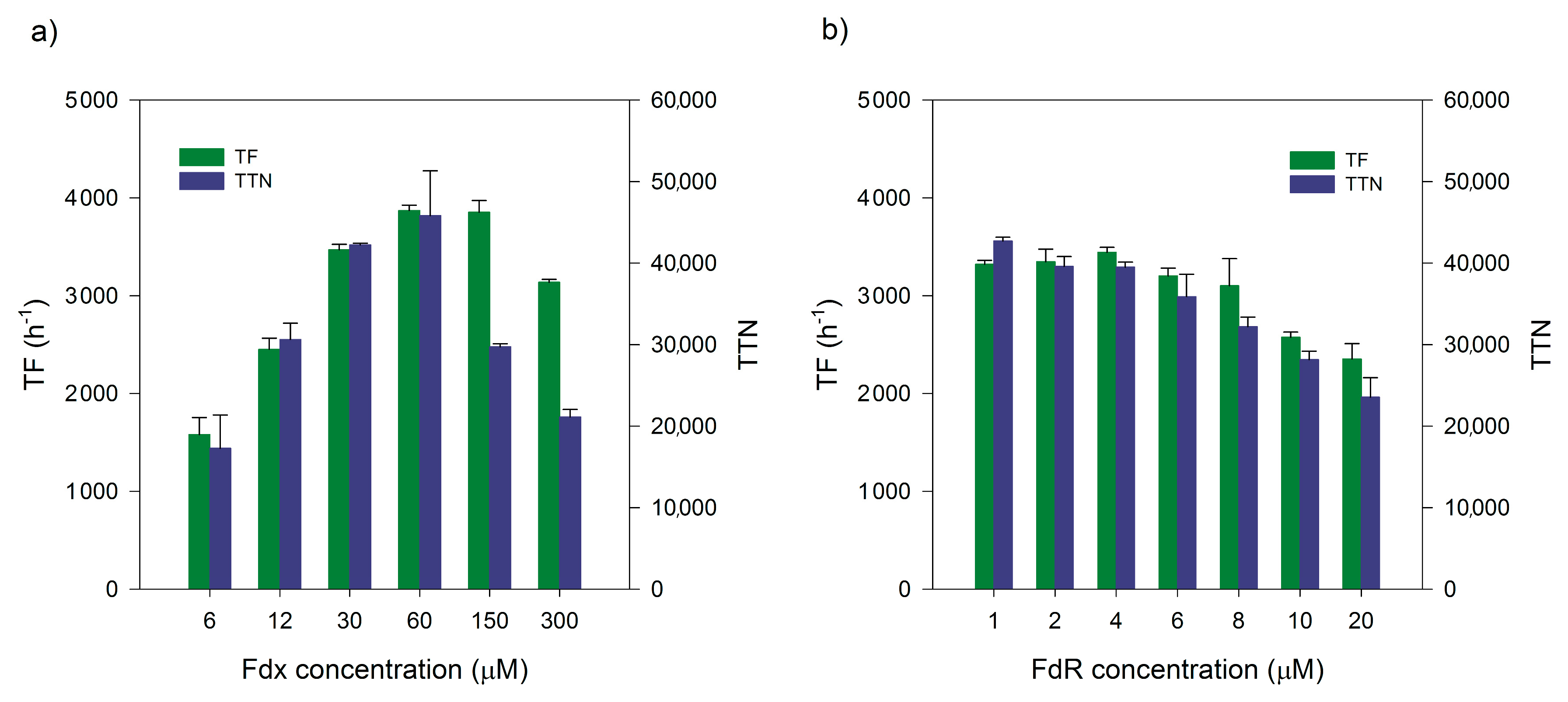
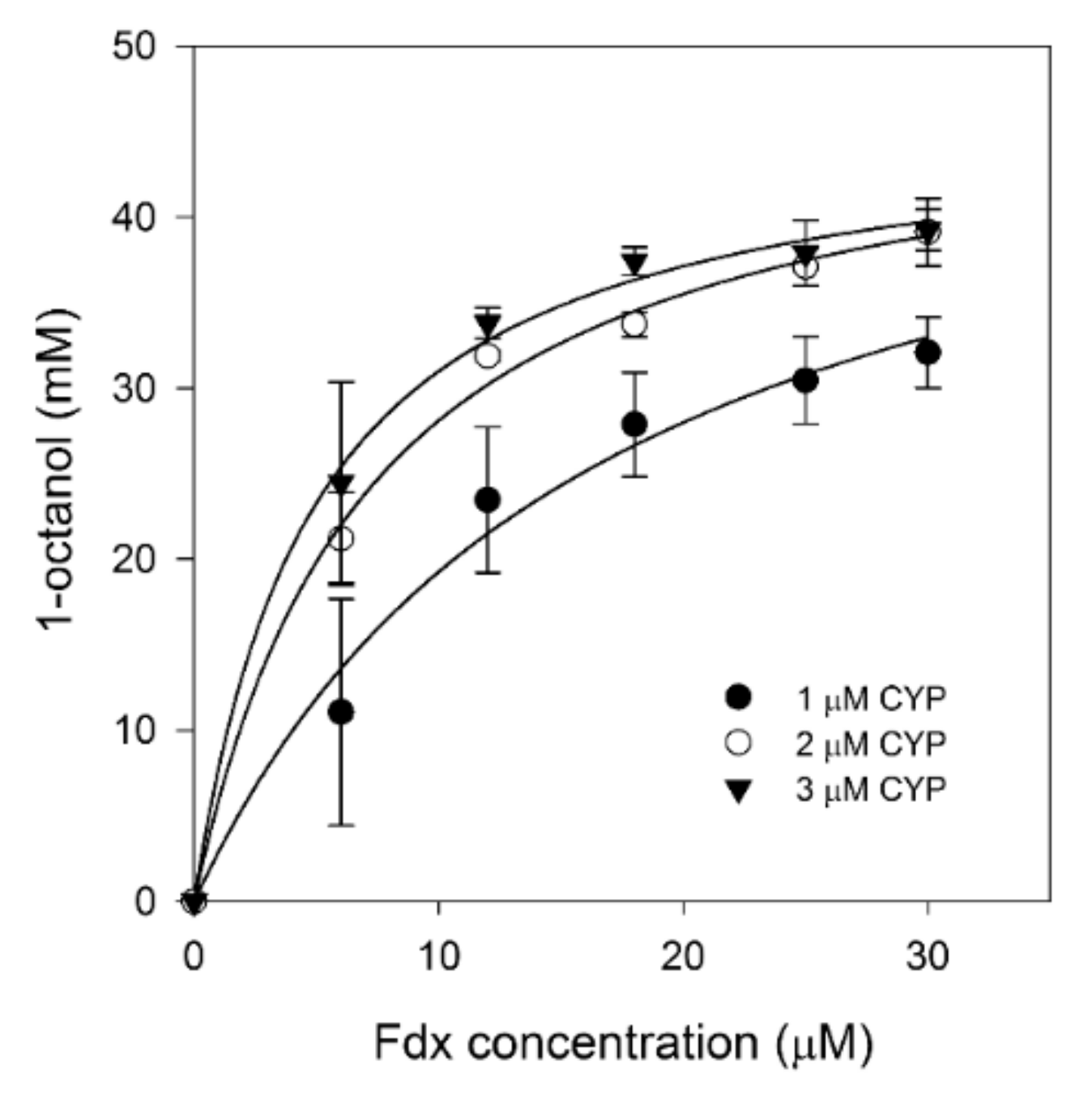
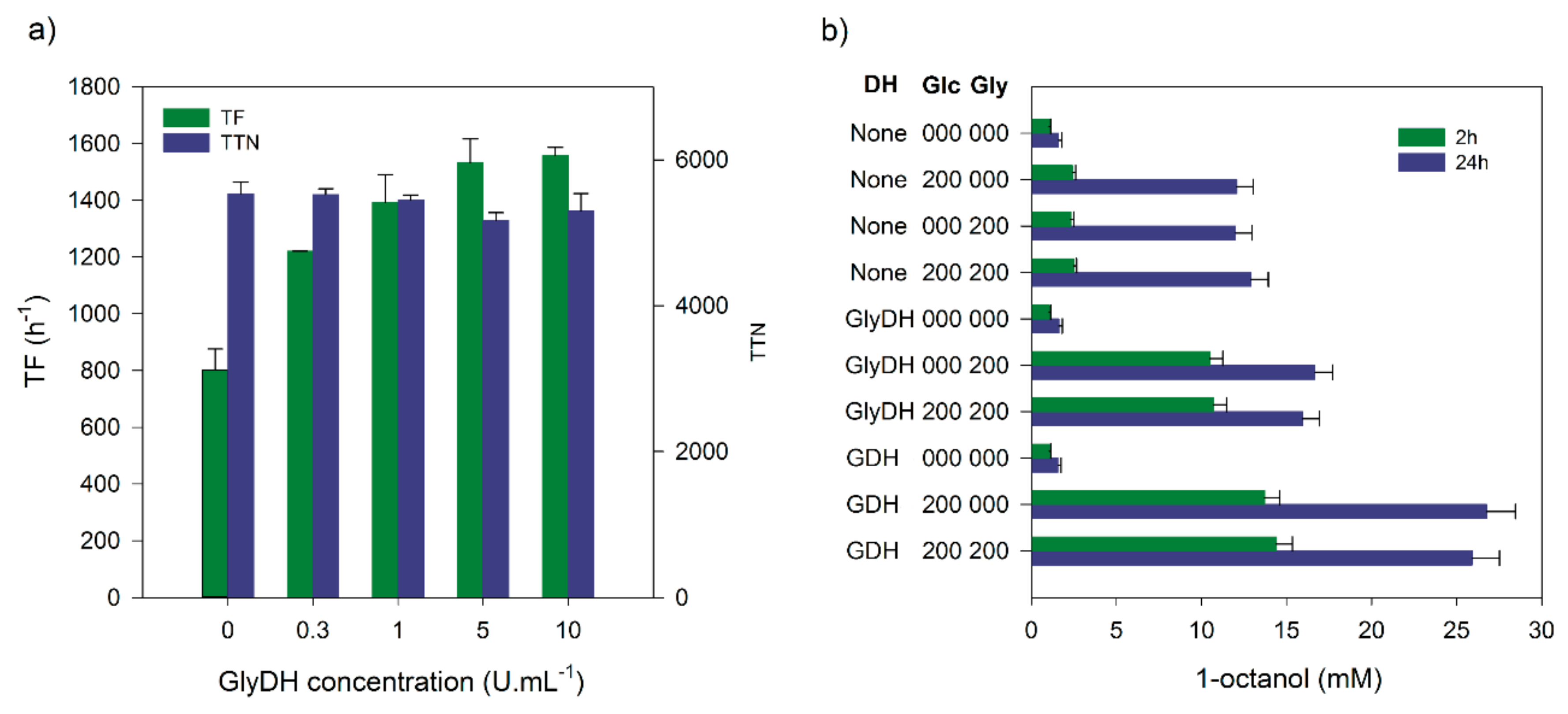

| Optimization Step | TF (h−1) | TTN | 1-Octanol (mM) |
|---|---|---|---|
| GDH Concentration | 2122 | 6078 1 | 6.1 |
| Addition of CFE | 1932 | 23,800 1 | 23.8 |
| Biocatalysts Ratios | |||
| CYP153A6:FdR:Fdx (2:1:30) | n.d. | 19,561 1 | 39.1 |
| (1:1:60) | 3872 | 45,828 2 | 45.8 |
| Reactor | Stirring (rpm) | pH Control | TF 1 (h−1) | TTN 2 | 1-Octanol (mM) 2 | CYP153A6 (µM) 3 | pH 3 |
|---|---|---|---|---|---|---|---|
| 1 | 600 | - | 690 | 4355 | 8.71 | n.d. | 6.4 |
| 2 | 200 | - | 181 | 3690 | 7.38 | 1.1 | 6.0 |
| 3 | 200 | + | 230 | 3505 | 7.01 | 0.4 | 7.7 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kochius, S.; Van Marwijk, J.; Ebrecht, A.C.; Opperman, D.J.; Smit, M.S. Deconstruction of the CYP153A6 Alkane Hydroxylase System: Limitations and Optimization of In Vitro Alkane Hydroxylation. Catalysts 2018, 8, 531. https://doi.org/10.3390/catal8110531
Kochius S, Van Marwijk J, Ebrecht AC, Opperman DJ, Smit MS. Deconstruction of the CYP153A6 Alkane Hydroxylase System: Limitations and Optimization of In Vitro Alkane Hydroxylation. Catalysts. 2018; 8(11):531. https://doi.org/10.3390/catal8110531
Chicago/Turabian StyleKochius, Svenja, Jacqueline Van Marwijk, Ana Cristina Ebrecht, Diederik Johannes Opperman, and Martha Sophia Smit. 2018. "Deconstruction of the CYP153A6 Alkane Hydroxylase System: Limitations and Optimization of In Vitro Alkane Hydroxylation" Catalysts 8, no. 11: 531. https://doi.org/10.3390/catal8110531
APA StyleKochius, S., Van Marwijk, J., Ebrecht, A. C., Opperman, D. J., & Smit, M. S. (2018). Deconstruction of the CYP153A6 Alkane Hydroxylase System: Limitations and Optimization of In Vitro Alkane Hydroxylation. Catalysts, 8(11), 531. https://doi.org/10.3390/catal8110531




