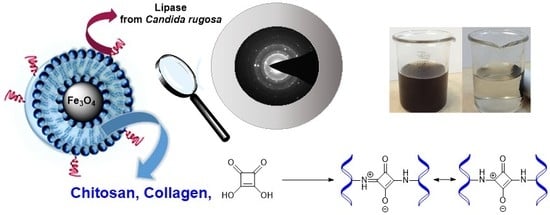
Chitosan–Collagen Coated Magnetic Nanoparticles for Lipase Immobilization—New Type of “Enzyme Friendly” Polymer Shell Crosslinking with Squaric Acid
Abstract
:1. Introduction
2. Results and Discussion
2.1. Magnetic Nanoparticles Synthesis and Characterization
2.2. Immobilization of Lipase from Candida rugosa
2.2.1. Lipase Activity and Recovery
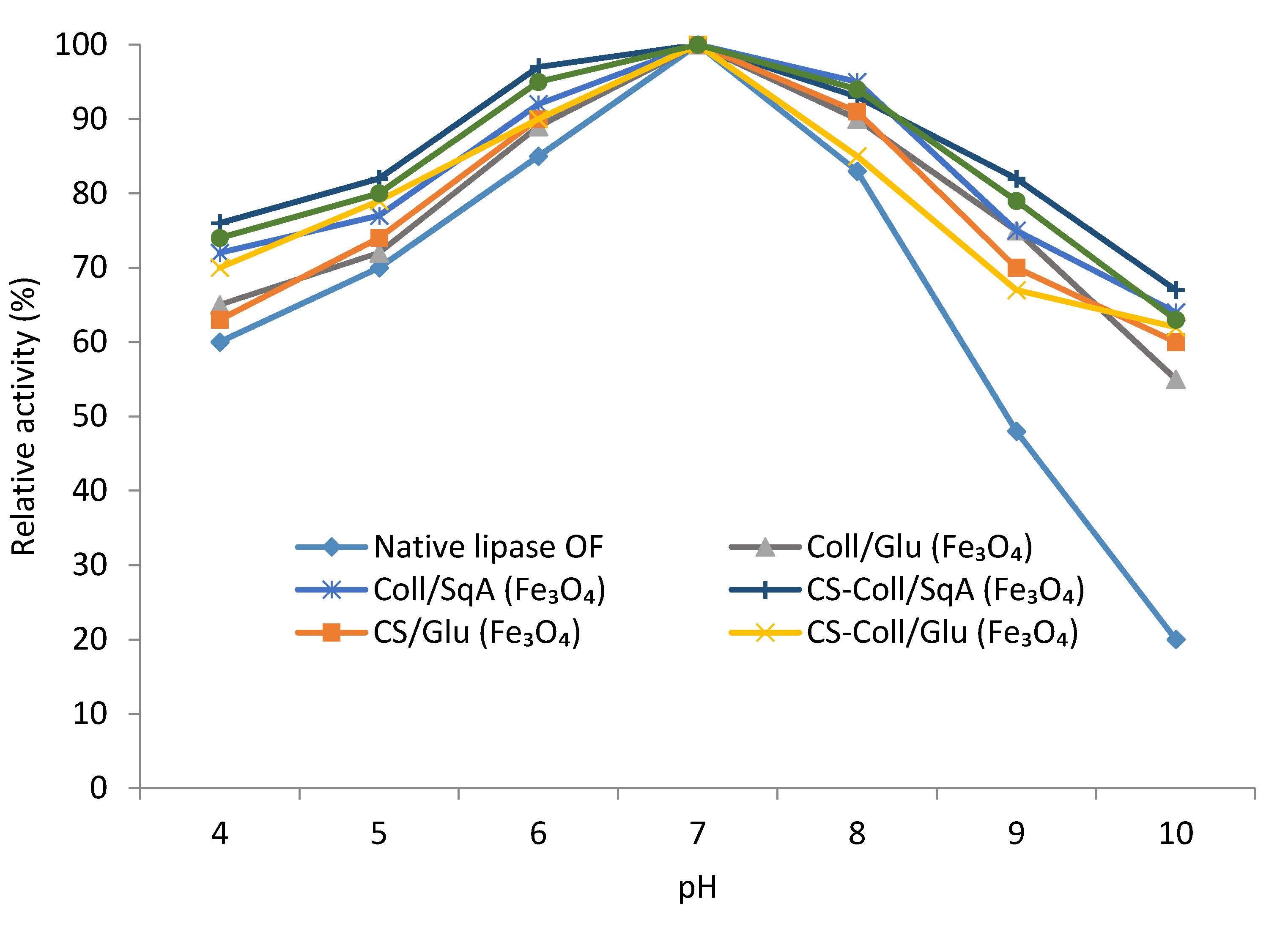
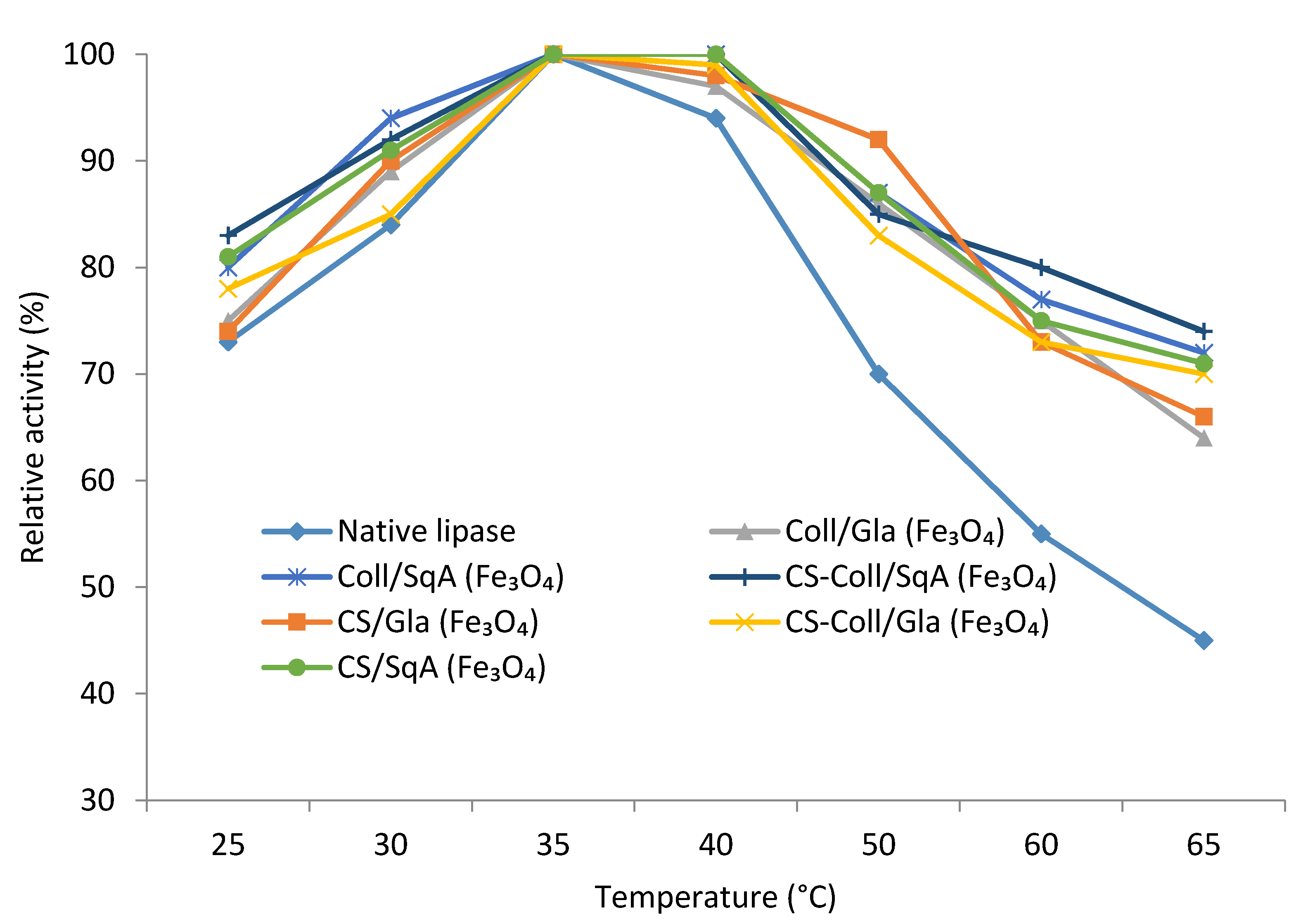
2.2.2. Immobilized Lipase Stability—Optimal pH and Temperature
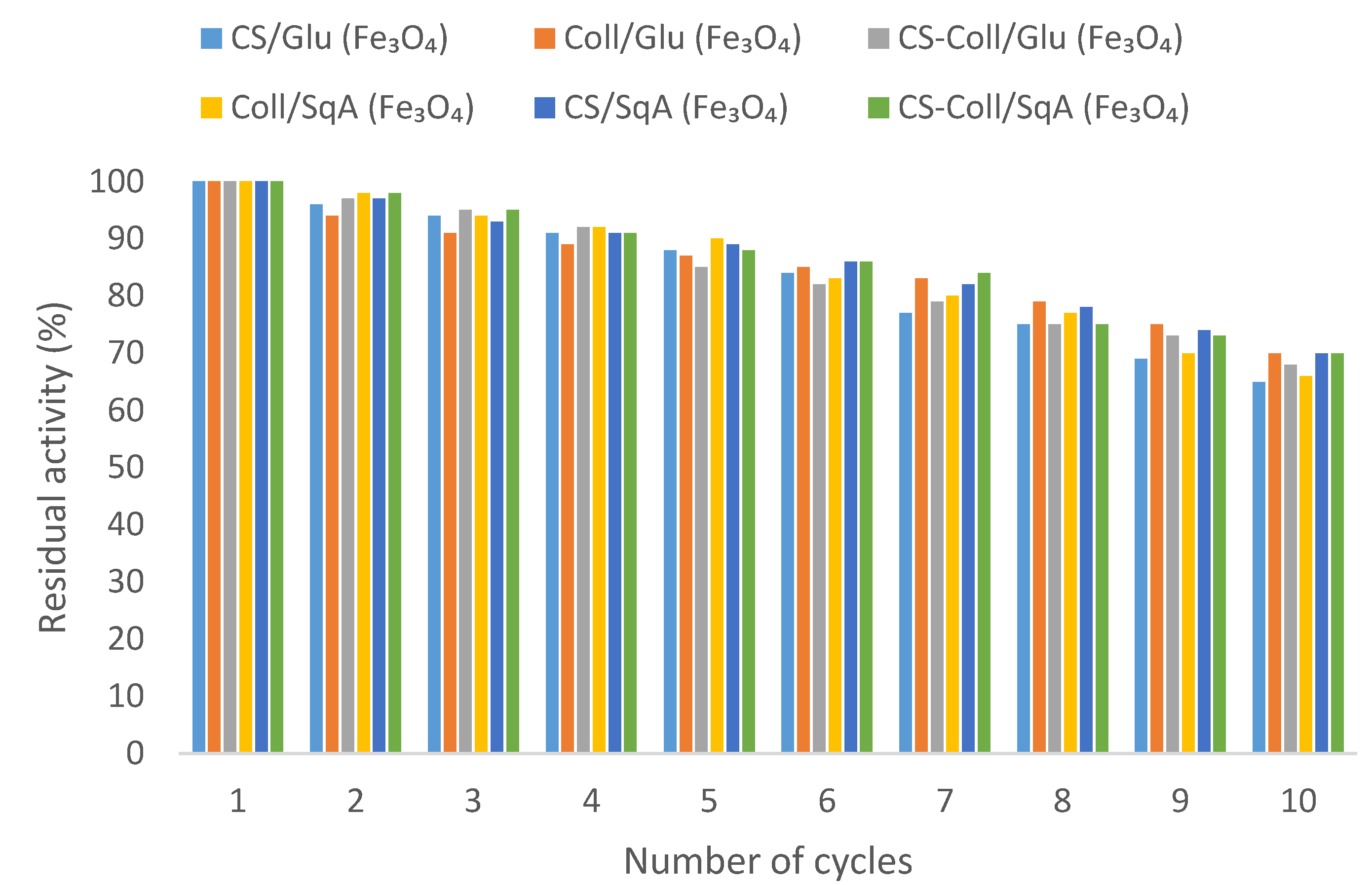
2.2.3. Reusability of Immobilized Lipase
3. Materials and Methods
3.1. Materials
3.2. Isolation of Collagen from Rats Tail Tendons
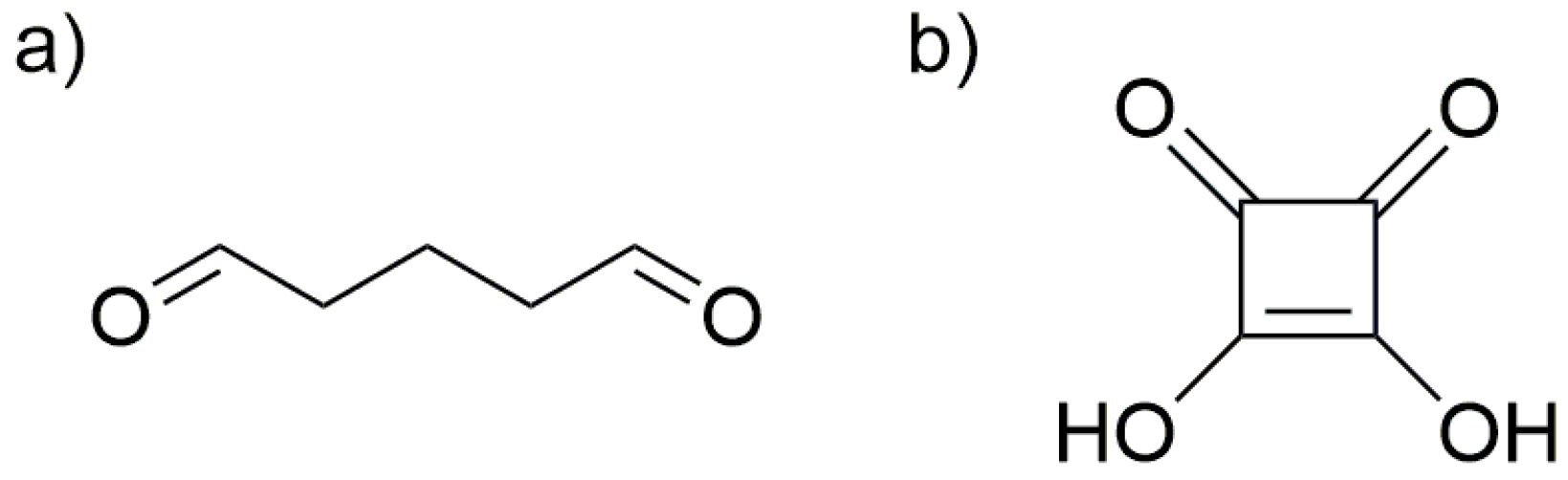
3.3. Synthesis of Collagen-Coated Nanoparticles Crosslinked with Glutaraldehyde Coll/Glu (Fe3O4)
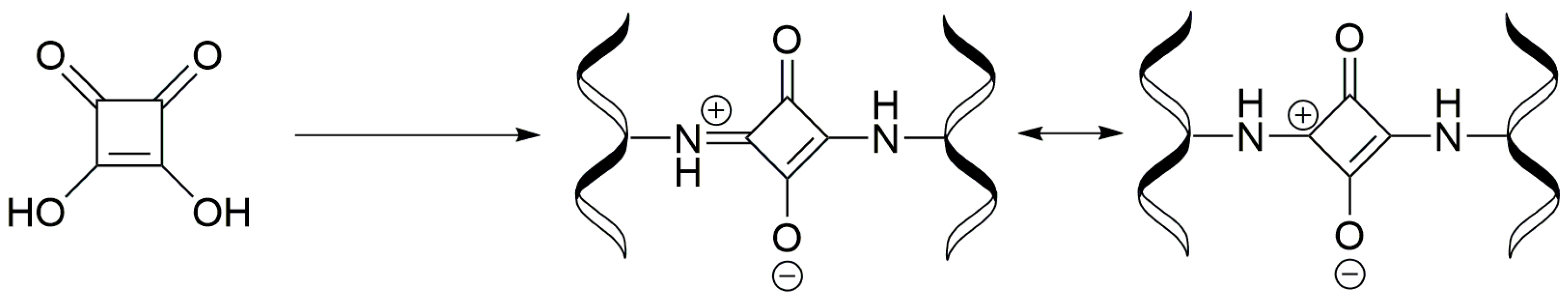
3.4. Synthesis of Collagen-Coated Nanoparticles Crosslinked with Squaric Acid Coll/SqA (Fe3O4)
3.5. Synthesis of Chitosan-Coated Nanoparticles Crosslinked with Glutaraldehyde CS/Glu (Fe3O4)
3.6. Synthesis of Chitosan-Coated Nanoparticles Crosslinked with Squaric Acid CS/SqA (Fe3O4)
3.7. Synthesis of Chitosan–Collagen (1:1) Coated Nanoparticles Crosslinked with Glutaraldehyde CS–Coll/Glu (Fe3O4)
3.8. Synthesis of Chitosan–Collagen (1:1) Coated Nanoparticles Crosslinked with Squaric Acid CS–Coll/SqA (Fe3O4)
3.9. Quantification of Available Primary Amino Groups on Magnetic Nanoparticles Surface
3.10. Immobilization of Lipase from Candida rugosa onto MNPs Surface
3.11. Assay of Immobilized Lipase Activity
3.12. Operational Stability (Reusability) of Lipase Immobilized onto MNPs
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Siódmiak, T.; Mangelings, D.; Heyden, Y.V.; Ziegler-Borowska, M.; Marszałł, M.P. High enantioselective novozym 435-catalyzed esterification of (R,S)-flurbiprofen monitored with a chiral stationary phase. Appl. Biochem. Biotechnol. 2015, 175, 2769–2785. [Google Scholar] [CrossRef] [PubMed]
- Rivero, C.; Palomo, J. Covalent immobilization of Candida rugosa lipase at alkaline pH and their application in the regioselective deprotection of per-O-acetylated thymidine. Catalysts 2016, 6, 115. [Google Scholar] [CrossRef]
- Grochulski, P.; Li, Y.; Schrag, J.D.; Bouthillier, F.; Smith, P.; Harrison, D.; Rubin, B.; Cygler, M. Insights into interfacial activation from an open structure of Candida rugosa lipase. J. Biol. Chem. 1993, 268, 12843–12847. [Google Scholar] [PubMed]
- Grochulski, P.; Li, Y.; Schrag, J.D.; Cygler, M. Two conformational states of Candida rugosa lipase. Protein Sci. 1994, 3, 82–91. [Google Scholar] [CrossRef] [PubMed]
- De Domínguez María, P.; Sánchez-Montero, J.M.; Sinisterra, J.V.; Alcántara, A.R. Understanding Candida rugosa lipases: An overview. Biotechnol. Adv. 2006, 24, 180–196. [Google Scholar] [CrossRef] [PubMed]
- De Albuquerque, T.L.; Rueda, N.; dos Santos, J.C.S.; Barbosa, O.; Ortiz, C.; Binay, B.; Özdemir, E.; Gonçalves, L.R.B.; Fernandez-Lafuente, R. Easy stabilization of interfacially activated lipases using heterofunctional divinyl sulfone activated-octyl agarose beads. Modulation of the immobilized enzymes by altering their nanoenvironment. Process Biochem. 2016, 51, 865–874. [Google Scholar] [CrossRef]
- Xu, L.; Ke, C.; Huang, Y.; Yan, Y. Immobilized Aspergillus niger lipase with SiO2 nanoparticles in sol-gel materials. Catalysts 2016, 6, 149. [Google Scholar] [CrossRef]
- Khoobi, M.; Motevalizadeh, S.F.; Asadgol, Z.; Forootanfar, H.; Shafiee, A.; Faramarzi, M.A. Synthesis of functionalized polyethylenimine-grafted mesoporous silica spheres and the effect of side arms on lipase immobilization and application. Biochem. Eng. J. 2014, 88, 131–141. [Google Scholar] [CrossRef]
- Garcia-Galan, C.; Berenguer-Murcia, Á.; Fernandez-Lafuente, R.; Rodrigues, R.C. Potential of different enzyme immobilization strategies to improve enzyme performance. Adv. Synth. Catal. 2011, 353, 2885–2904. [Google Scholar] [CrossRef]
- Mateo, C.; Palomo, J.M.; Fernandez-Lorente, G.; Guisan, J.M.; Fernandez-Lafuente, R. Improvement of enzyme activity, stability and selectivity via immobilization techniques. Enzyme Microb. Technol. 2007, 40, 1451–1463. [Google Scholar] [CrossRef]
- Rodrigues, R.C.; Ortiz, C.; Berenguer-Murcia, A.; Torres, R.; Fernandez-Lafuente, R. Modifying enzyme activity and selectivity by immobilization. Chem. Soc. Rev. 2013, 42, 6290–6307. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Lafuente, R. Stabilization of multimeric enzymes: Strategies to prevent subunit dissociation. Enzyme Microb. Technol. 2009, 45, 405–418. [Google Scholar] [CrossRef]
- Barbosa, O.; Ortiz, C.; Berenguer-Murcia, A.; Torres, R.; Rodrigues, R.C.; Fernandez-Lafuente, R. Glutaraldehyde in bio-catalysts design: A useful crosslinker and a versatile tool in enzyme immobilization. RSC Adv. 2014, 4, 1583–1600. [Google Scholar] [CrossRef]
- Badgujar, K.C.; Bhanage, B.M. Lipase immobilization on hyroxypropyl methyl cellulose support and its applications for chemo-selective synthesis of β-amino ester compounds. Process Biochem. 2016, 51, 1420–1433. [Google Scholar] [CrossRef]
- Sikora, A.; Chełminiak-Dudkiewicz, D.; Siódmiak, T.; Tarczykowska, A.; Sroka, W.D.; Ziegler-Borowska, M.; Marszałł, M.P. Enantioselective acetylation of (R,S)-atenolol: The use of Candida rugosa lipases immobilized onto magnetic chitosan nanoparticles in enzyme-catalyzed biotransformation. J. Mol. Catal. B 2016, 134(Pt. A), 43–50. [Google Scholar] [CrossRef]
- Karimi, M. Immobilization of lipase onto mesoporous magnetic nanoparticles for enzymatic synthesis of biodiesel. Biocatal. Agric. Biotechnol. 2016, 8, 182–188. [Google Scholar] [CrossRef]
- Motevalizadeh, S.F.; Khoobi, M.; Sadighi, A.; Khalilvand-Sedagheh, M.; Pazhouhandeh, M.; Ramazani, A.; Faramarzi, M.A.; Shafiee, A. Lipase immobilization onto polyethylenimine coated magnetic nanoparticles assisted by divalent metal chelated ions. J. Mol. Catal. B 2015, 120, 75–83. [Google Scholar] [CrossRef]
- Gupta, A.K.; Gupta, M. Synthesis and surface engineering of iron oxide nanoparticles for biomedical applications. Biomaterials 2005, 26, 3995–4021. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Bai, Y.X.; Li, Y.F.; Lei, L.; Chui, Y.J.; Xia, C.G. Characterization of Candida rugosa lipase immobilized onto magnetic microspheres with hydrophilicity. Precess Biochem. 2008, 43, 1179–1185. [Google Scholar] [CrossRef]
- Ren, Y.; Rivera, J.G.; He, L.; Kulkarni, H.; Lee, D.-K.; Messersmith, P.B. Facile, high efficiency immobilization of lipase enzyme on magnetic iron oxide nanoparticles via a biomimetic coating. BMC Biotechnol. 2011, 11, 63. [Google Scholar] [CrossRef] [PubMed]
- Marszałł, M.P.; Sroka, W.D.; Sikora, A.; Chełminiak, D.; Ziegler-Borowska, M.; Siódmiak, T.; Moaddel, R. Ligand fishing using new chitosan based functionalized Androgen receptor magnetic particles. J. Pharm. Biomed. Anal. 2016, 127, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Unsoy, G.; Yalcin, S.; Khodadust, R.; Gunduz, G.; Gunduz, U. Synthesis optimization and characterization of chitosan-coated iron oxide nanoparticles produced for biomedical applications. J. Nanopart. Res. 2012, 14, 964. [Google Scholar] [CrossRef]
- Pan, Y.; Du, X.; Zhao, F.; Xu, B. Magnetic nanoparticles for the manipulation of proteins and cells. Chem. Soc. Rev. 2012, 41, 2912–2942. [Google Scholar] [CrossRef] [PubMed]
- Skopinska-Wisniewska, J.; Olszewski, K.; Bajek, A.; Rynkiewicz, A.; Sionkowska, A. Dialysis as a method of obtaining neutral collagen gels. Mater. Sci. Eng. C 2014, 40, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.A.; Macedo, G.P.; Rodrigues, D.S.; Giordano, R.L.C.; Gonçalves, L.R.B. Immobilization of Candida antarctica lipase B by covalent attachment on chitosan-based hydrogels using different support activation strategies. Biochem. Eng. J. 2012, 60, 16–24. [Google Scholar] [CrossRef]
- Pandey, S.; Wilmer, E.N.; Morrell, D.S. Examining the efficacy and safety of squaric acid therapy for treatment of recalcitrant warts in children. Pediatr. Dermatol. 2015, 32, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Skopinska-Wisniewska, J.; Kuderko, J.; Bajek, A.; Maj, M.; Sionkowska, A.; Ziegler-Borowska, M. Collagen/elastin hydrogels cross-linked by squaric acid. Mater. Sci. Eng. C 2016, 60, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Sionkowska, A.; Kozłowska, J. Characterization of collagen/hydroxyapatite composite sponges as a potential bone substitute. Int. J. Biol. Macromol. 2010, 47, 483–487. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Xu, X.; Guo, S.; Han, W. Synthesis and self-assembly of chitosan-based copolymer with a pair of hydrophobic/hydrophilic grafts of polycaprolactone and poly(ethylene glycol). Carbohydr. Polym. 2009, 75, 401–407. [Google Scholar] [CrossRef]
- Marszałł, M.P.; Siódmiak, T. Immobilization of Candida rugosa lipase onto magnetic beads for kinetic resolution of (R,S)-ibuprofen. Catal. Commun. 2012, 24, 80–84. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Bakare, R.A.; Bhan, C.; Raghavan, D. Synthesis and characterization of collagen grafted poly(hydroxybutyrate-valerate) (PHBV) scaffold for loading of bovine serum albumin capped silver (AG/BSA) nanoparticles in the potential use of tissue engineering application. Biomacromolecules 2014, 15, 423–435. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, N.; Prakash, O.; Talat, M.; Hasan, S.H.; Pandey, R.K. α-Amylase immobilization on gelatin: Optimization of process variables. J. Genet. Eng. Biotechnol. 2012, 10, 161–167. [Google Scholar] [CrossRef]
- Moreno-Perez, S.; Ghattas, N.; Filice, M.; Guisan, J.M.; Fernandez-Lorente, G. Dramatic hyperactivation of lipase of Thermomyces lanuginosa by a cationic surfactant: Fixation of the hyperactivated form by adsorption on sulfopropyl-sepharose. J. Mol. Catal. B 2015, 122, 199–203. [Google Scholar] [CrossRef]
- Ghattas, N.; Filice, M.; Abidi, F.; Guisan, J.M.; Ben Salah, A. Purification and improvement of the functional properties of Rhizopus oryzae lipase using immobilization techniques. J. Mol. Catal. B 2014, 110, 111–116. [Google Scholar] [CrossRef]
- Shang, C.-Y.; Li, W.-X.; Zhang, R.-F. Immobilization of Candida rugosa lipase on ZnO nanowires/macroporous silica composites for biocatalytic synthesis of phytosterol esters. Mater. Res. Bull. 2015, 68, 336–342. [Google Scholar] [CrossRef]
- Hou, C.; Qi, Z.; Zhu, H. Preparation of core-shell magnetic polydopamine/alginate biocomposite for Candida rugosa lipase immobilization. Colloids Surf. B 2015, 128, 544–551. [Google Scholar] [CrossRef] [PubMed]
- Mahto, T.K.; Chowdhuri, A.R.; Sahoo, B.; Sahu, S.K. Polyaniline-functionalized magnetic mesoporous nanocomposite: A smart material for the immobilization of lipase. Polym. Compos. 2016, 37, 1152–1160. [Google Scholar] [CrossRef]
- Wang, J.; Zhao, G.; Jing, L.; Peng, X.; Li, Y. Facile self-assembly of magnetite nanoparticles on three-dimensional graphene oxide–chitosan composite for lipase immobilization. Biochem. Eng. J. 2015, 98, 75–83. [Google Scholar] [CrossRef]



| Nanoparticles Type | Size (nm) | Amount of NH2 Groups (mM/g) |
|---|---|---|
| CS/Glu (Fe3O4) | 16 | 3.73 |
| CS/SqA (Fe3O4) | 20 | 3.31 |
| Coll/Glu (Fe3O4) | 21 | 3.51 |
| Coll/SqA (Fe3O4) | 29 | 2.17 |
| CS–Coll/Glu (Fe3O4) | 24 | 2.05 |
| CS–Coll/SqA (Fe3O4) | 32 | 2.83 |
| Nanoparticles Type | Lipase Loading (mg/g) | Activity Recovery (%) | Lipase Activity (U) | Hydrolytic Activity (U/g Support) | Specific Activity (U/mg Lipase) |
|---|---|---|---|---|---|
| CS/Glu (Fe3O4) | 67 | 77 | 85.3 | 1706 | 25.5 |
| CS/SqA (Fe3O4) | 47 | 104 | 70.5 | 1410 | 30 |
| Coll/Glu (Fe3O4) | 63 | 85 | 82.1 | 1642 | 26 |
| Coll/SqA (Fe3O4) | 18 | 97 | 47 | 940 | 52.2 |
| CS–Coll/Glu (Fe3O4) | 74 | 94 | 90.7 | 1814 | 24.5 |
| CS–Coll/SqA (Fe3O4) | 27 | 112 | 60.1 | 1202 | 44.5 |
© 2017 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ziegler-Borowska, M.; Chelminiak-Dudkiewicz, D.; Siódmiak, T.; Sikora, A.; Wegrzynowska-Drzymalska, K.; Skopinska-Wisniewska, J.; Kaczmarek, H.; Marszałł, M.P. Chitosan–Collagen Coated Magnetic Nanoparticles for Lipase Immobilization—New Type of “Enzyme Friendly” Polymer Shell Crosslinking with Squaric Acid. Catalysts 2017, 7, 26. https://doi.org/10.3390/catal7010026
Ziegler-Borowska M, Chelminiak-Dudkiewicz D, Siódmiak T, Sikora A, Wegrzynowska-Drzymalska K, Skopinska-Wisniewska J, Kaczmarek H, Marszałł MP. Chitosan–Collagen Coated Magnetic Nanoparticles for Lipase Immobilization—New Type of “Enzyme Friendly” Polymer Shell Crosslinking with Squaric Acid. Catalysts. 2017; 7(1):26. https://doi.org/10.3390/catal7010026
Chicago/Turabian StyleZiegler-Borowska, Marta, Dorota Chelminiak-Dudkiewicz, Tomasz Siódmiak, Adam Sikora, Katarzyna Wegrzynowska-Drzymalska, Joanna Skopinska-Wisniewska, Halina Kaczmarek, and Michał P. Marszałł. 2017. "Chitosan–Collagen Coated Magnetic Nanoparticles for Lipase Immobilization—New Type of “Enzyme Friendly” Polymer Shell Crosslinking with Squaric Acid" Catalysts 7, no. 1: 26. https://doi.org/10.3390/catal7010026
APA StyleZiegler-Borowska, M., Chelminiak-Dudkiewicz, D., Siódmiak, T., Sikora, A., Wegrzynowska-Drzymalska, K., Skopinska-Wisniewska, J., Kaczmarek, H., & Marszałł, M. P. (2017). Chitosan–Collagen Coated Magnetic Nanoparticles for Lipase Immobilization—New Type of “Enzyme Friendly” Polymer Shell Crosslinking with Squaric Acid. Catalysts, 7(1), 26. https://doi.org/10.3390/catal7010026