Engineering Malic Enzyme CO2 Fixation Activity via a Structure–Sequence–SCANNER (3S) Co-Evolution Strategy
Abstract
1. Introduction
2. Results and Discussion
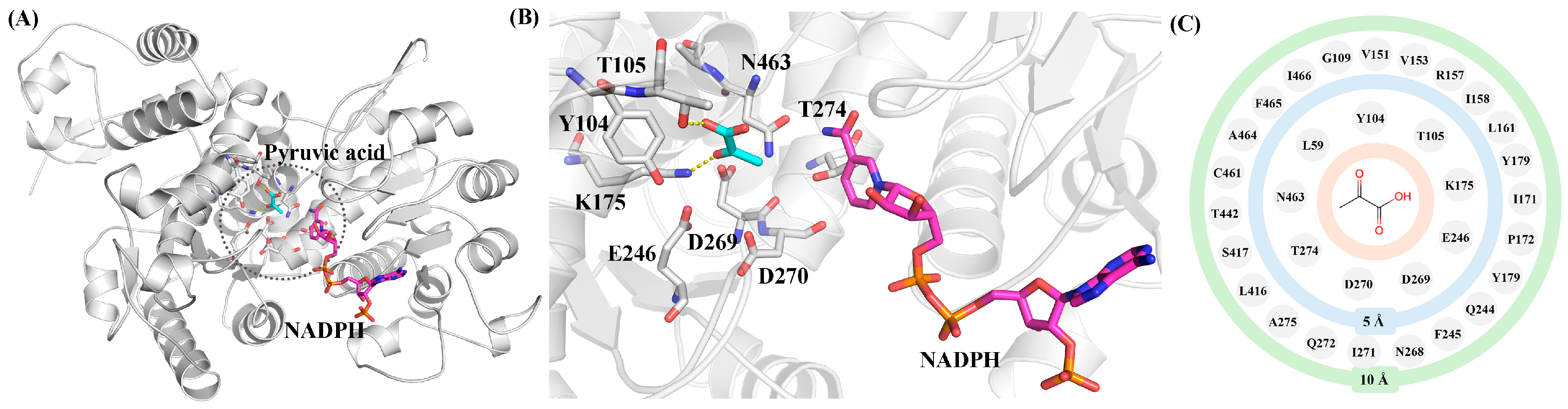
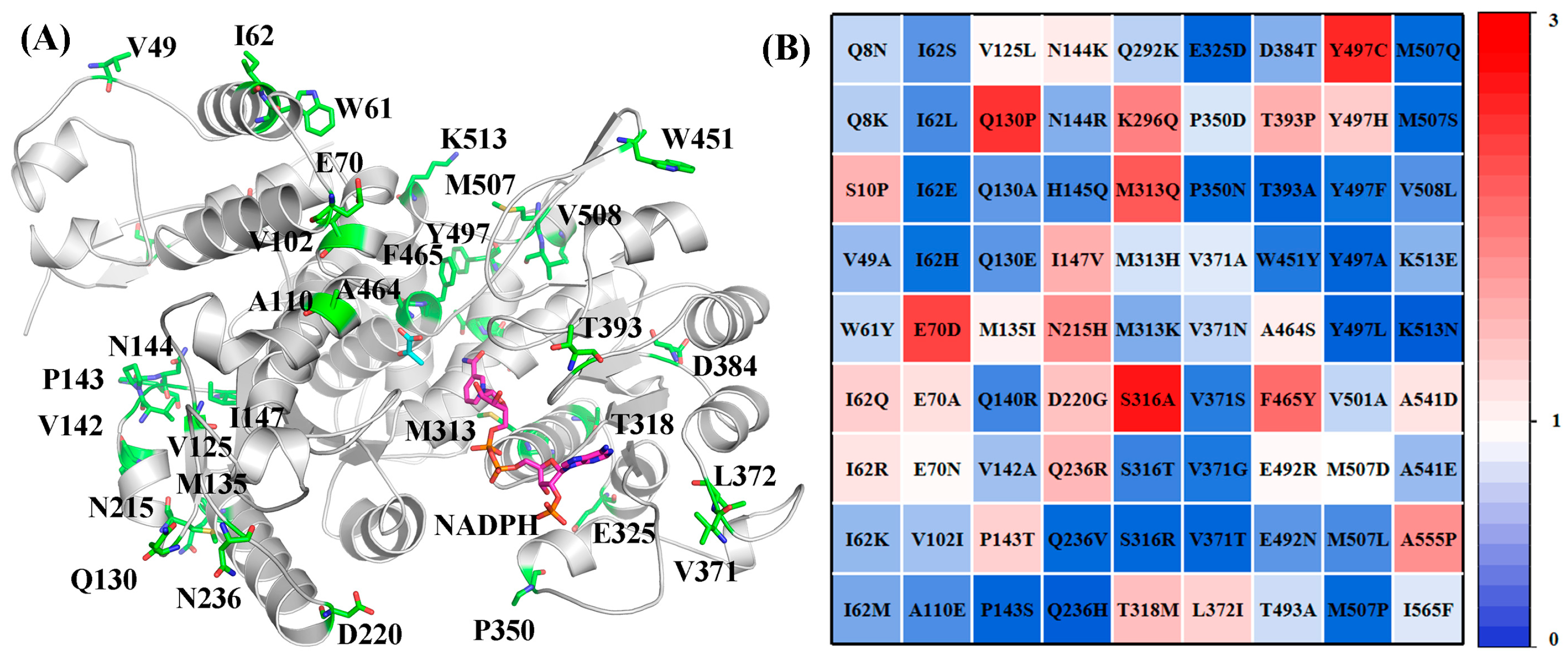
2.1. Structure-Guided Construction of ME Variant Libraries
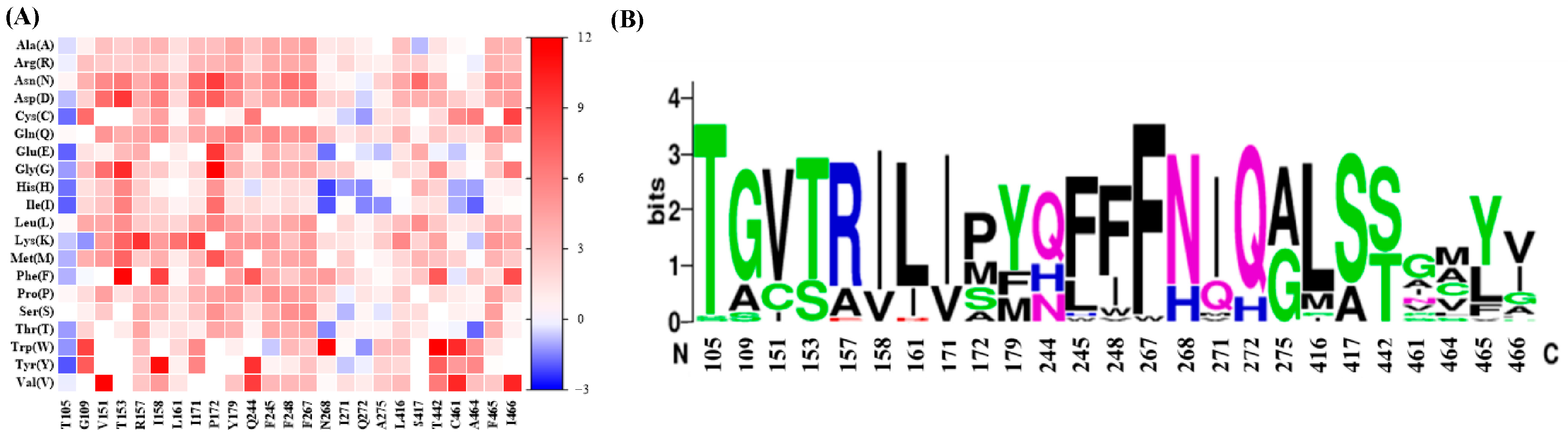
2.2. Sequence-Guided Construction of ME Variant Libraries
2.3. SCANNER-Guided Construction of ME Variant Libraries
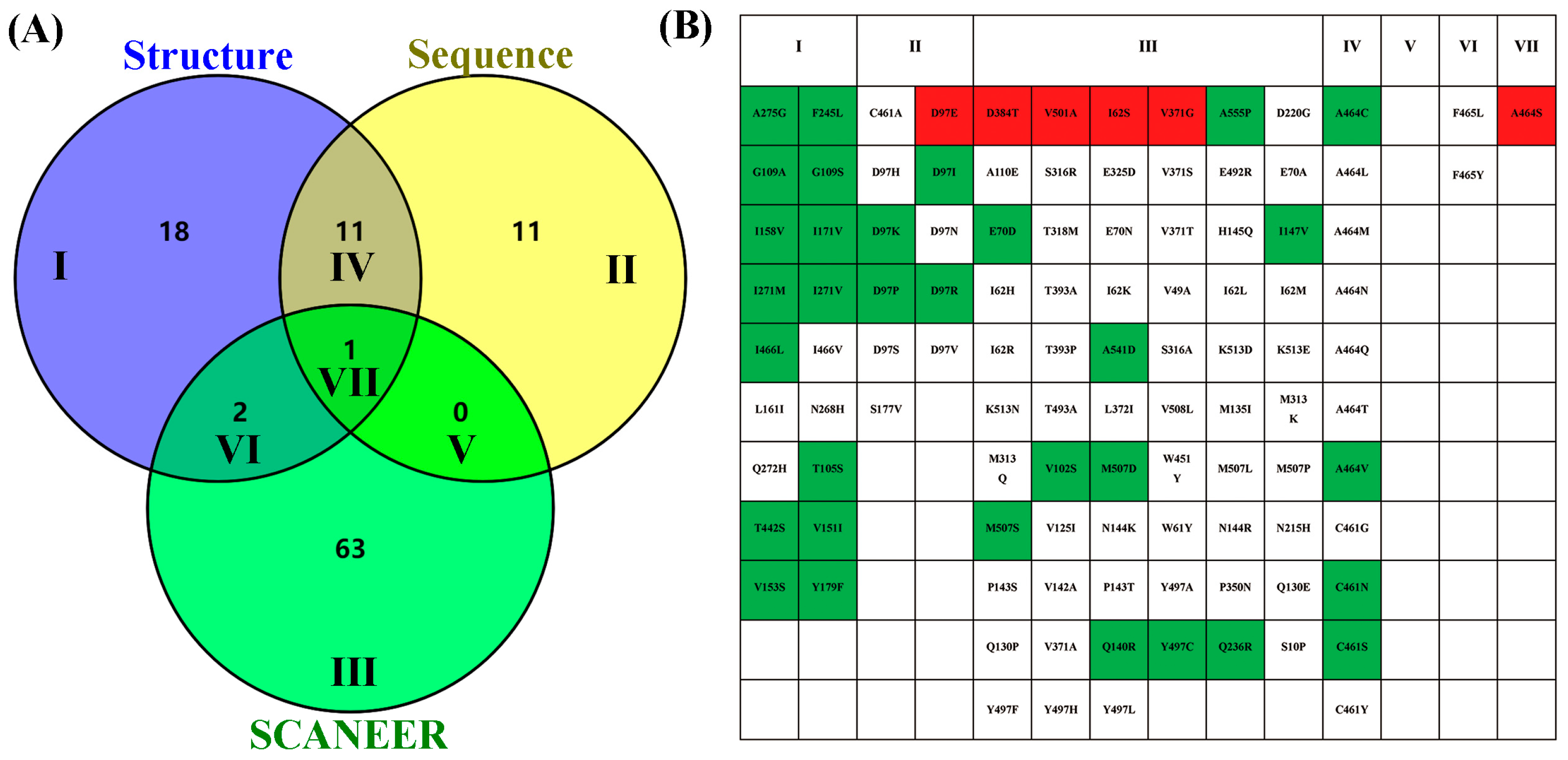
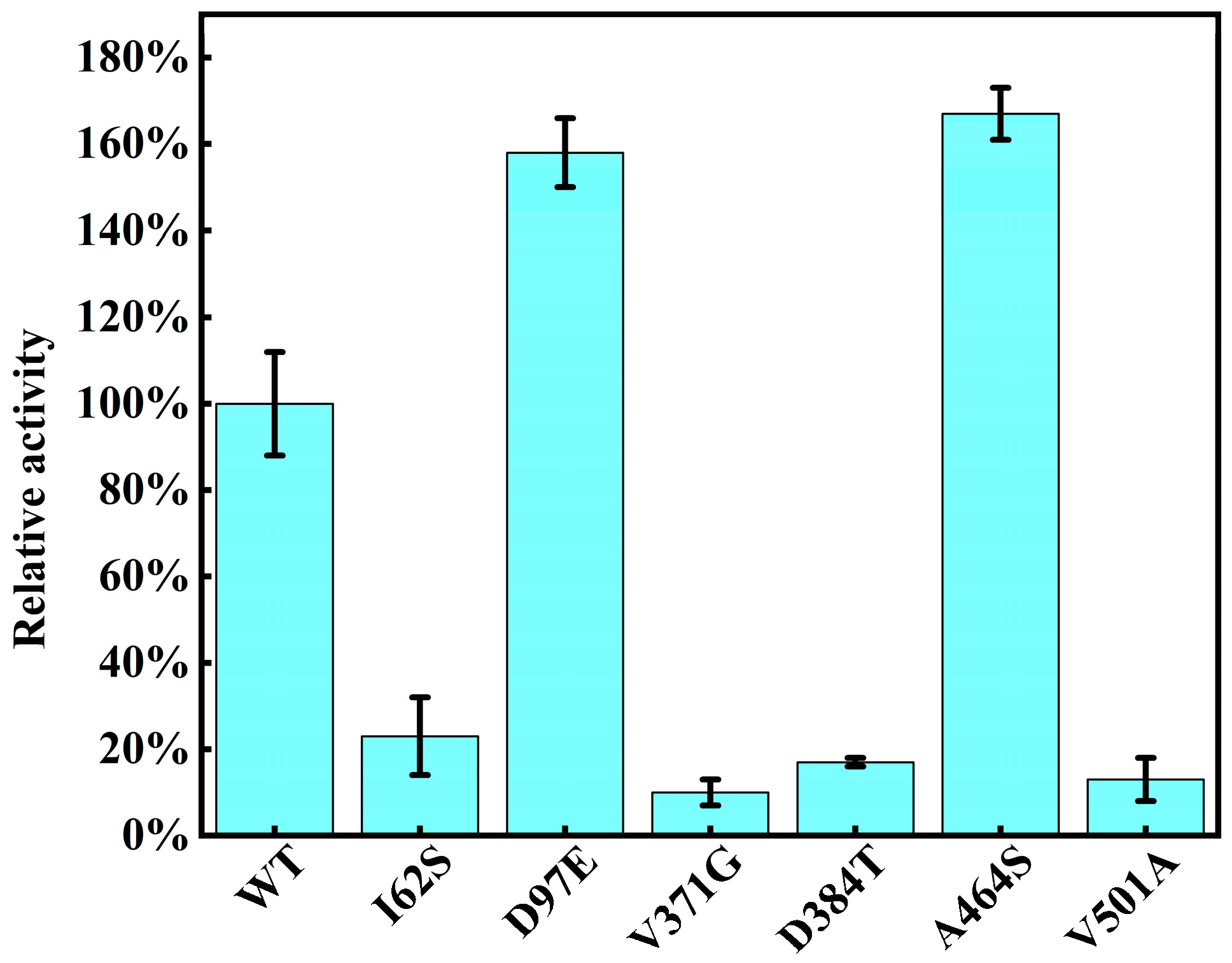
2.4. Preliminary Screening of ME Variant Libraries
2.5. Purified Enzyme Re-Screening and Kinetic Characterization
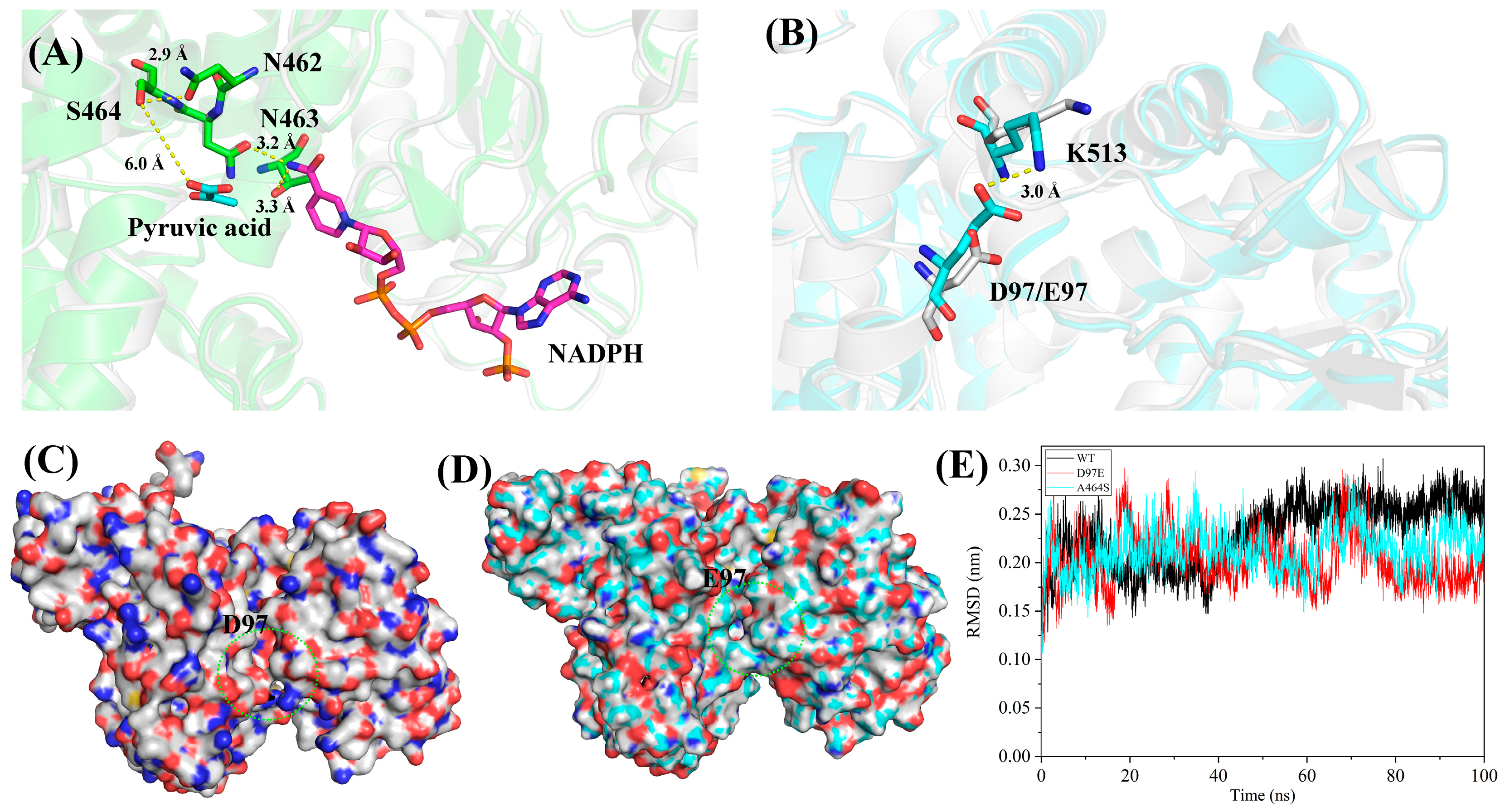
2.6. Mechanistic Analysis of the Enhanced Catalytic Activity of the Variants
3. Materials and Methods
3.1. Materials
3.2. Structural Prediction and Molecular Docking
3.3. Multiple Sequence Alignment
3.4. SCANNER-Based Multiple Sequence Alignment and Co-Evolution Analysis
3.5. FoldX Calculation of Gibbs Free Energy Changes
3.6. Construction of Mutant Libraries
3.7. Expression of Wild-Type and Mutant Enzymes
3.8. Purification of the Enzymes
3.9. Library Screening
3.10. Determination of Kinetic Parameters
3.11. Molecular Dynamics Simulations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Yang, C.; Mao, L.; Chen, Y.X.; Zhou, Y.H.; Zhang, R.F.; Yi, Z.W.; Zhang, D.C.; Zhang, G.Y. Ancestral carbonic anhydrase with significantly enhanced stability and activity for CO2 capture and utilization. Bioresour. Technol. 2025, 419, 132054. [Google Scholar] [CrossRef]
- Karishma, S.; Kamalesh, R.; Saravanan, A.; Deivayanai, V.C.; Yaashikaa, P.R.; Vickram, A.S. A review on recent advancements in biochemical fixation and transformation of CO2 into constructive products. Biochem. Eng. J. 2024, 208, 109366. [Google Scholar] [CrossRef]
- Lee, J.; Yu, H.E.; Lee, S.Y. Metabolic engineering of microorganisms for carbon dioxide utilization. Curr. Opin. Biotech. 2025, 91, 103244. [Google Scholar] [CrossRef]
- Schwander, T.; von Borzyskowski, L.S.; Burgener, S.; Cortina, N.S.; Erb, T.J. A synthetic pathway for the fixation of carbon dioxide in vitro. Science 2016, 354, 900–904. [Google Scholar] [CrossRef]
- Cai, T.; Sun, H.B.; Qiao, J.; Zhu, L.L.; Zhang, F.; Zhang, J.; Tang, Z.J.; Wei, X.L.; Yang, J.G.; Yuan, Q.Q.; et al. Cell-free chemoenzymatic starch synthesis from carbon dioxide. Science 2021, 373, 1523–1527. [Google Scholar] [CrossRef]
- Bierbaumer, S.; Nattermann, M.; Schulz, L.; Zschoche, R.; Erb, T.J.; Winkler, C.K.; Tinzl, M.; Glueck, S.M. Enzymatic Conversion of CO2: From Natural to Artificial Utilization. Chem. Rev. 2023, 123, 5702–5754. [Google Scholar] [CrossRef]
- Chen, H.; Huang, Y.; Sha, C.; Moradian, J.M.; Yong, Y.C.; Fang, Z. Enzymatic carbon dioxide to formate: Mechanisms, challenges and opportunities. Renew. Sust. Energ. Rev. 2023, 178, 113271. [Google Scholar] [CrossRef]
- Pu, X.; Han, Y.J. Promotion of Carbon Dioxide Biofixation through Metabolic and Enzyme Engineering. Catalysts 2022, 12, 399. [Google Scholar] [CrossRef]
- Chen, J.; Wang, S.R.; Zhou, J.R.; Xu, J.X.; Wu, B.; Gao, Z.; He, B.F. Phenolic Acid Decarboxylase for Carbon Dioxide Fixation: Mining, Biochemical Characterization, and Regioselective Enzymatic β-carboxylation of -hydroxystyrene Derivatives. Catalysts 2025, 15, 210. [Google Scholar] [CrossRef]
- Wu, N.; Zhang, J.H.; Chen, Y.R.; Xu, Q.; Song, P.; Li, Y.F.; Li, K.; Liu, H. Recent advances in microbial production of L-malic acid. Appl. Microbiol. Biot. 2022, 106, 7973–7992. [Google Scholar] [CrossRef] [PubMed]
- Are, K.R.A.; Ohshima, S.; Koike, Y.; Asanuma, Y.; Kashikura, S.; Tamura, M.; Matsuda, T. Enzymatic direct carboxylation under supercritical CO2. Biochem. Eng. J. 2021, 171, 108004. [Google Scholar] [CrossRef]
- Zhang, L.C.; Jin, H.Z.; Jiao, D.N.; Lin, W.Q.; Liu, J.; Liu, H. High-yield bioconversion of glycerol to L-malic acid in Aspergillus niger via systematic engineering of glycerol catabolism and minimal glucose supplementation. Chem. Eng. J. 2025, 515, 163687. [Google Scholar] [CrossRef]
- Hu, G.P.; Zhou, J.; Chen, X.L.; Qian, Y.Y.; Gao, C.; Guo, L.; Xu, P.; Chen, W.; Chen, J.; Li, Y.; et al. Engineering synergetic CO2-fixing pathways for malate production. Metab. Eng. 2018, 47, 496–504. [Google Scholar] [CrossRef]
- Liu, X.T.; Zhao, G.; Sun, S.J.; Fan, C.L.; Feng, X.J.; Xiong, P. Biosynthetic Pathway and Metabolic Engineering of Succinic Acid. Front. Bioeng. Biotech. 2022, 10, 843887. [Google Scholar] [CrossRef] [PubMed]
- Katagiri, T.; Amao, Y. Trivalent metal ions promote the malic enzyme-catalyzed building of carbon-carbon bonds from CO2 and pyruvate. New J. Chem. 2020, 44, 17208–17214. [Google Scholar] [CrossRef]
- Ding, Q.; Ye, C. Recent advances in producing food additive L-malate: Chassis, substrate, pathway, fermentation regulation and application. Microb. Biotechnol. 2023, 16, 709–725. [Google Scholar] [CrossRef]
- Zelle, R.M.; de Hulster, E.; van Winden, W.A.; de Waard, P.; Dijkema, C.; Winkler, A.A.; Geertman, J.M.A.; van Dijken, J.P.; Pronk, J.T.; van Maris, A.J.A. Malic acid production by Engineering of pyruvate carboxylation, oxaloacetate reduction, and malate export. Appl. Environ. Microb. 2008, 74, 2766–2777. [Google Scholar] [CrossRef]
- Morimoto, Y.; Honda, K.; Ye, X.T.; Okano, K.; Ohtake, H. Directed evolution of thermotolerant malic enzyme for improved malate production. J. Biosci. Bioeng. 2014, 117, 147–152. [Google Scholar] [CrossRef]
- Dong, X.X.; Chen, X.L.; Qian, Y.Y.; Wang, Y.C.; Wang, L.; Qiao, W.H.; Liu, L.M. Metabolic engineering of W3110 to produce L-malate. Biotechnol. Bioeng. 2017, 114, 656–664. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, S.F.; Fu, S.Y.; Ren, X.D.; Zhou, Y.C.; Xue, Y.P.; Zheng, Y.G. Design of a Distal Site Saturation Test-Iterative Parallel Mutagenesis for Engineering Hydroxysteroid Dehydrogenase. J. Agric. Food Chem. 2025, 73, 9701–9713. [Google Scholar] [CrossRef] [PubMed]
- Amatto, I.V.D.; da Rosa-Garzon, N.G.; Simoes, F.A.D.; Santiago, F.; Leite, N.P.D.; Martins, J.R.; Cabral, H. Enzyme engineering and its industrial applications. Biotechnol. Appl. Biochem. 2022, 69, 389–409. [Google Scholar] [CrossRef]
- Jang, W.D.; Kim, G.B.; Kim, Y.; Lee, S.Y. Applications of artificial intelligence to enzyme and pathway design for metabolic engineering. Curr. Opin. Biotechnol. 2022, 73, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Buller, R.; Lutz, S.; Kazlauskas, R.J.; Snajdrova, R.; Moore, J.C.; Bornscheuer, U.T. From nature to industry: Harnessing enzymes for biocatalysis. Science 2023, 382, adh8615. [Google Scholar] [CrossRef]
- Liu, Y.X.; Guo, X.J.; Liu, W.J.; Wang, J.T.; Zhao, Z.K. Structural Insights into Malic Enzyme Variants Favoring an Unnatural Redox Cofactor. Chembiochem 2021, 22, 1765–17684. [Google Scholar] [CrossRef]
- Valanciute, A.; Nygaard, L.; Zschach, H.; Jepsen, M.M.; Lindorff-Larsen, K.; Stein, A. Accurate protein stability predictions from homology models. Comput. Struct. Biotechnol. J. 2023, 21, 66–73. [Google Scholar] [CrossRef]
- Feng, T.; Huang, H.X.; Xu, Y.; Che, X.Y.; Wang, M.D.; Tao, X.Y.; Feng, Y.B.; Xue, S. Multisite synergistic evolution of oleate hydratase via DCCM and the orthogonal location of distal sites. Int. J. Biol. Macromol. 2025, 312, 144001. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.P.; Fan, Y.; Jiang, X.S.; Li, X.L.; Li, S.; Feng, Y.B.; Xue, S. Efficient synthesis of L-malic acid by malic enzyme biocatalysis with CO2 fixation. Bioresour. Technol. 2024, 403, 130843. [Google Scholar] [CrossRef]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef] [PubMed]
- Madeira, F.; Madhusoodanan, N.; Lee, J.H.; Eusebi, A.; Niewielska, A.; Tivey, A.R.N.; Lopez, R.; Butcher, S. The EMBL-EBI Job Dispatcher sequence analysis tools framework in 2024. Nucleic Acids Res. 2024, 52, W521–W525. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Noh, M.H.; Park, M.; Kim, I.; Ahn, H.; Ye, D.Y.; Jung, G.Y.; Kim, S. Enzyme activity engineering based on sequence co-evolution analysis. Metab. Eng. 2022, 74, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Van Durme, J.; Delgado, J.; Stricher, F.; Serrano, L.; Schymkowitz, J.; Rousseau, F. A graphical interface for the FoldX forcefield. Bioinformatics 2011, 27, 1711–1712. [Google Scholar] [CrossRef]
- Kagami, L.; Wilter, A.; Diaz, A.; Vranken, W. The ACPYPE web server for small-molecule MD topology generation. Bioinformatics 2023, 39, btad350. [Google Scholar] [CrossRef] [PubMed]
| kcat (min−1) | Km (μM−1) | kcat/Km (mM−1·min−1) | |
|---|---|---|---|
| WT [28] | 17.7 ± 0.6 | 221 ± 31 | 80 ± 12 |
| D97E | 16.0 ± 0.5 | 121 ± 17 | 132 ± 19 |
| A464S | 16.3 ± 0.6 | 132 ± 23 | 123 ± 22 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, J.; Wang, M.; Feng, T.; Li, X.; Feng, Y.; Xue, S. Engineering Malic Enzyme CO2 Fixation Activity via a Structure–Sequence–SCANNER (3S) Co-Evolution Strategy. Catalysts 2025, 15, 789. https://doi.org/10.3390/catal15080789
Shi J, Wang M, Feng T, Li X, Feng Y, Xue S. Engineering Malic Enzyme CO2 Fixation Activity via a Structure–Sequence–SCANNER (3S) Co-Evolution Strategy. Catalysts. 2025; 15(8):789. https://doi.org/10.3390/catal15080789
Chicago/Turabian StyleShi, Jianping, Mingdong Wang, Ting Feng, Xianglong Li, Yanbin Feng, and Song Xue. 2025. "Engineering Malic Enzyme CO2 Fixation Activity via a Structure–Sequence–SCANNER (3S) Co-Evolution Strategy" Catalysts 15, no. 8: 789. https://doi.org/10.3390/catal15080789
APA StyleShi, J., Wang, M., Feng, T., Li, X., Feng, Y., & Xue, S. (2025). Engineering Malic Enzyme CO2 Fixation Activity via a Structure–Sequence–SCANNER (3S) Co-Evolution Strategy. Catalysts, 15(8), 789. https://doi.org/10.3390/catal15080789








