Inhibition Mechanism of Lecithin-Dependent Hemolysin from Vibrio parahaemolyticus by Flavonoids: An Enzyme Kinetic and Structural Approach
Abstract
1. Introduction
2. Results and Discussion
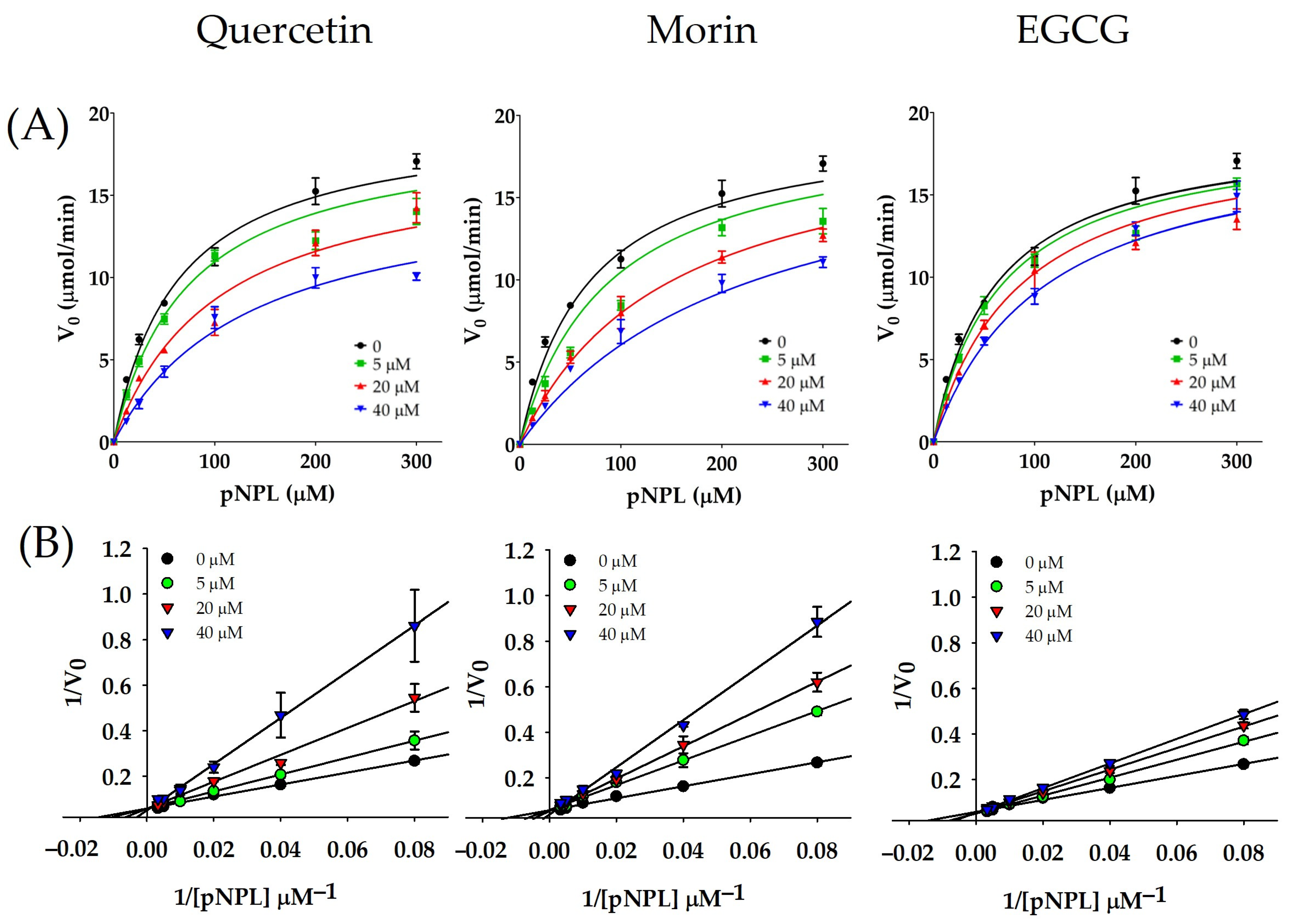
2.1. Inhibition Kinetics of vpLDH by Flavonoids
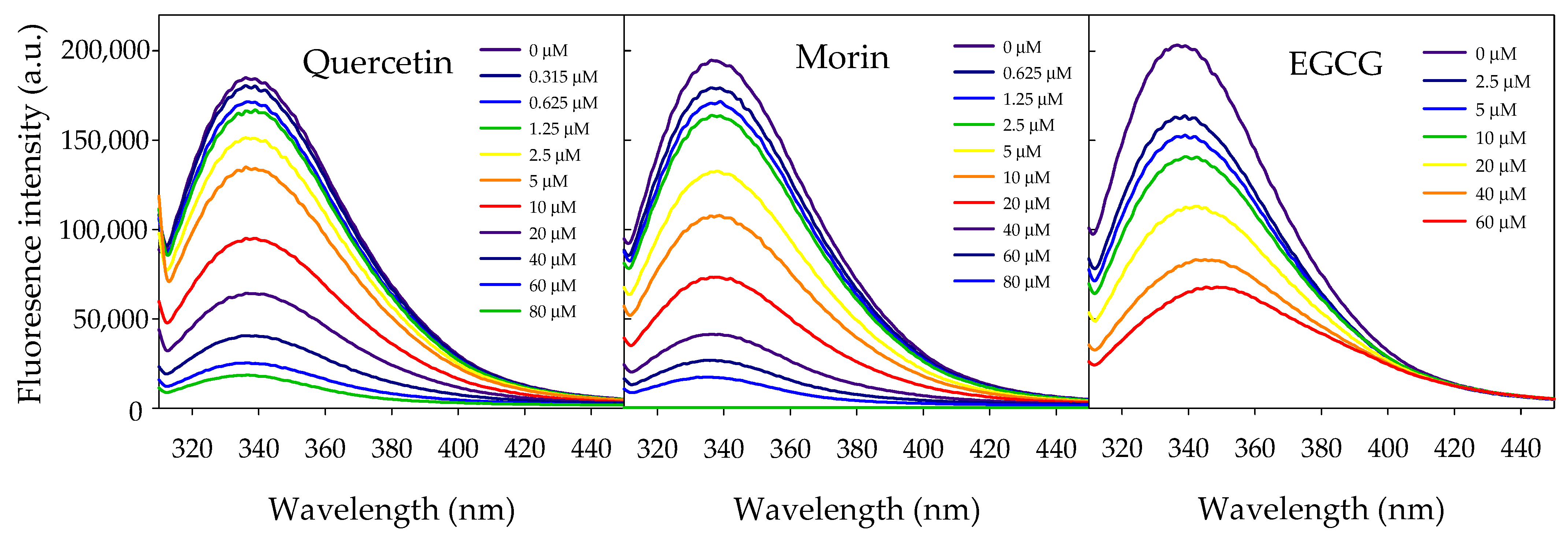
2.2. vpLDH–Flavonoid Interaction by Quenching Fluorescence
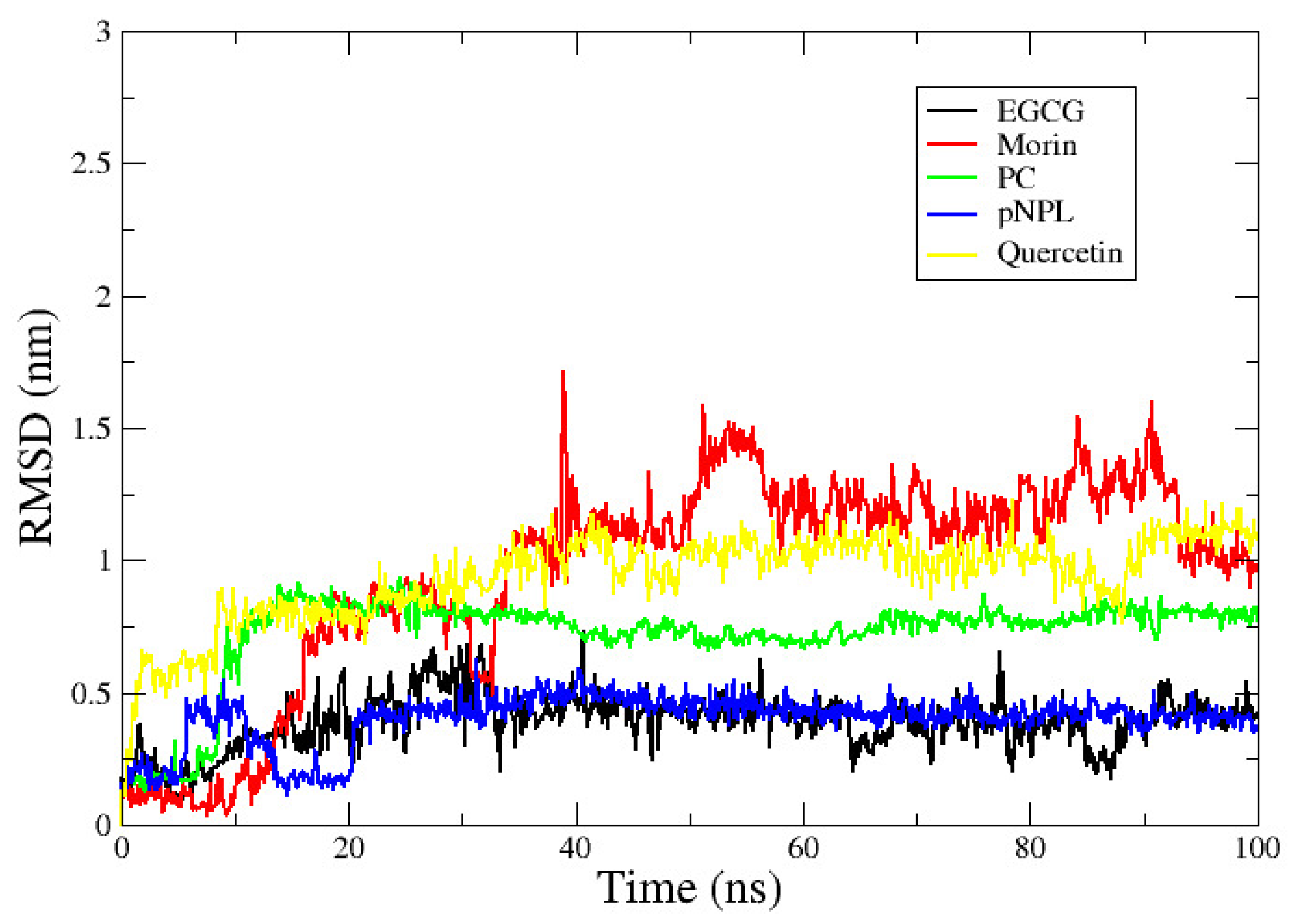
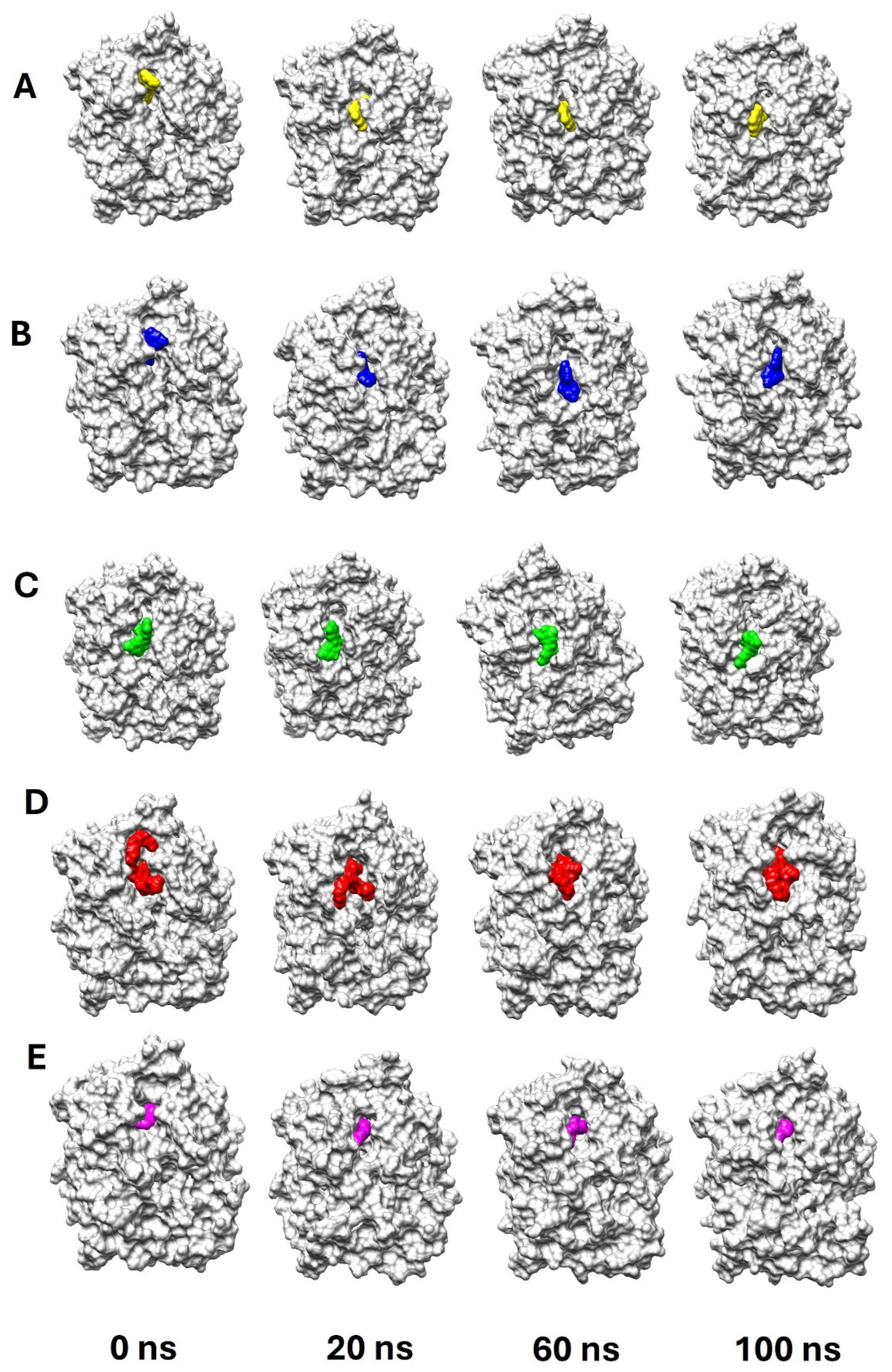
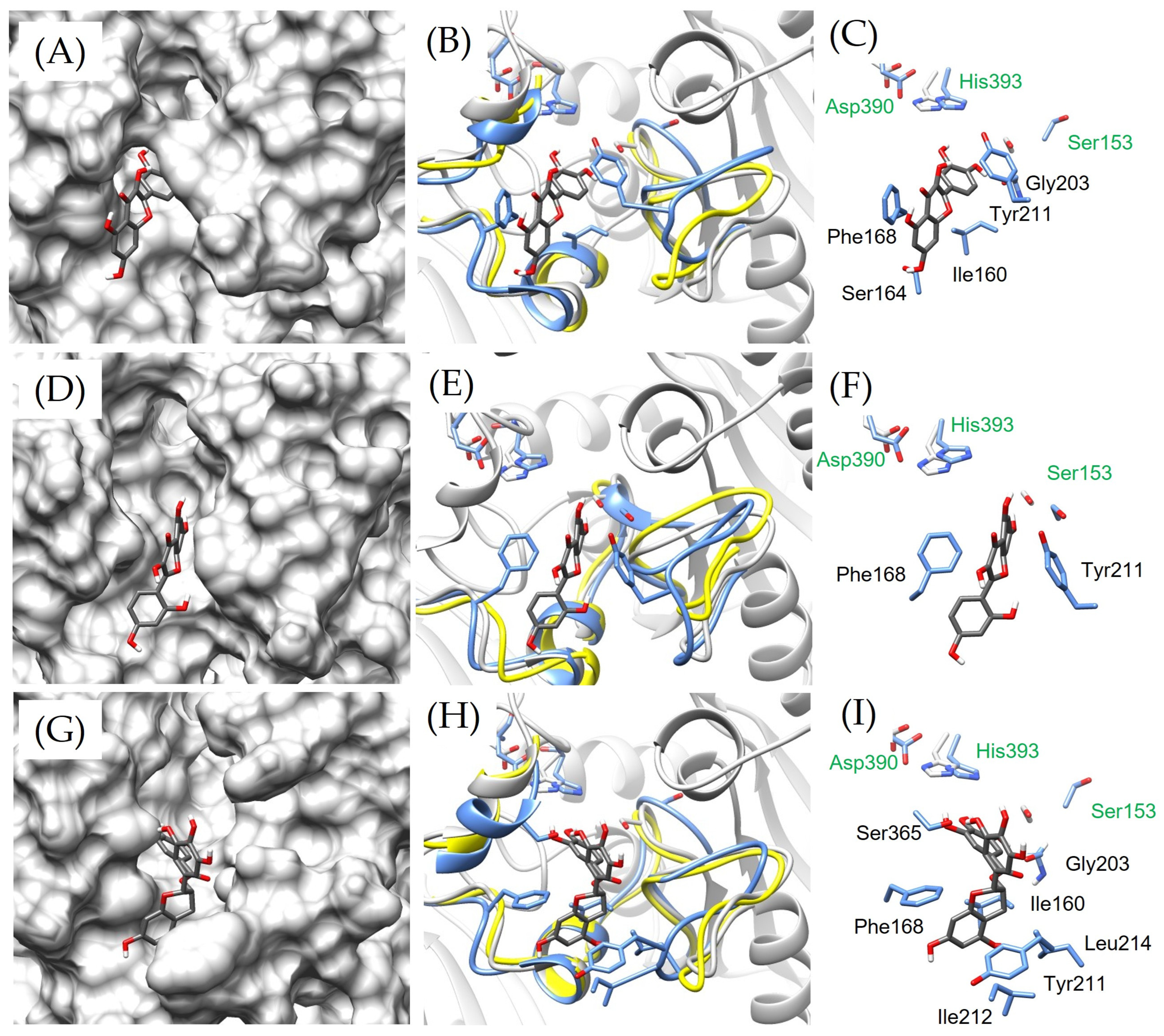
2.3. vpLDH–Flavonoid Structural–Binding Mode by MD Simulations
3. Materials and Methods
3.1. vpLDH Protein Overexpression and Purification
3.2. Enzymatic Activity
3.3. vpLDH Inhibition Kinetics
3.4. Determination of Binding Constants Between Flavonoids and vpLDH by Intrinsic Tryptophan Fluorescence
3.5. Molecular Dynamics Simulation and Surface Area
3.6. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Namadi, P.; Deng, Z. Optimum environmental conditions controlling prevalence of Vibrio parahaemolyticus in marine environment. Mar. Environ. Res. 2023, 183, 105828. [Google Scholar] [CrossRef]
- Yang, F.; Xu, L.; Huang, W.; Li, F. Highly lethal Vibrio parahaemolyticus strains cause acute mortality in Penaeus vannamei post-larvae. Aquaculture 2022, 548, 737605. [Google Scholar] [CrossRef]
- Han, J.E.; Tang, K.F.J.; Lightner, D.V. Genotyping of virulence plasmid from Vibrio parahaemolyticus isolates causing acute hepatopancreatic necrosis disease in shrimp. Dis. Aquat. Org. 2015, 115, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Han, J.E.; Tang, K.F.J.; Tran, L.H.; Lightner, D.V. Photorhabdus insect-related (Pir) toxin-like genes in a plasmid of Vibrio parahaemolyticus, the causative agent of acute hepatopancreatic necrosis disease (AHPND) of shrimp. Dis. Aquat. Org. 2015, 113, 33–40. [Google Scholar] [CrossRef]
- de Souza Valente, C.; Wan, A.H.L. Vibrio and major commercially important vibriosis diseases in decapod crustaceans. J. Invertebr. Pathol. 2021, 181, 107527. [Google Scholar] [CrossRef]
- Fang, G.-Y.; Liu, X.-Q.; Mu, X.-J.; Huang, B.-W.; Jiang, Y.-J. Distinct increase in antimicrobial resistance genes among Vibrio parahaemolyticus in recent decades worldwide. Chemosphere 2023, 340, 139905. [Google Scholar] [CrossRef]
- Wang, D.; Flint, S.H.; Palmer, J.S.; Gagic, D.; Fletcher, G.C.; On, S.L.W. Global expansion of Vibrio parahaemolyticus threatens the seafood industry: Perspective on controlling its biofilm formation. LWT 2022, 158, 113182. [Google Scholar] [CrossRef]
- Balta, I.; Linton, M.; Pinkerton, L.; Kelly, C.; Stef, L.; Pet, I.; Stef, D.; Criste, A.; Gundogdu, O.; Corcionivoschi, N. The effect of natural antimicrobials against Campylobacter spp. and its similarities to Salmonella spp, Listeria spp., Escherichia coli, Vibrio spp., Clostridium spp. and Staphylococcus spp. Food Control 2021, 121, 107745. [Google Scholar] [CrossRef]
- Li, A.; Shi, C.; Qian, S.; Wang, Z.; Zhao, S.; Liu, Y.; Xue, Z. Evaluation of antibiotic combination of Litsea cubeba essential oil on Vibrio parahaemolyticus inhibition mechanism and anti-biofilm ability. Microb. Pathog. 2022, 168, 105574. [Google Scholar] [CrossRef]
- Yu, H.; Pei, J.; Qiu, W.; Mei, J.; Xie, J. The Antimicrobial Effect of Melissa officinalis L. Essential Oil on Vibrio parahaemolyticus: Insights Based on the Cell Membrane and External Structure. Front. Microbiol. 2022, 13, 812792. [Google Scholar] [CrossRef]
- Dehbanipour, R.; Ghalavand, Z. Anti-virulence therapeutic strategies against bacterial infections: Recent advances. Germs 2022, 12, 262–275. [Google Scholar] [CrossRef]
- Brannon, J.R.; Hadjifrangiskou, M. The arsenal of pathogens and antivirulence therapeutic strategies for disarming them. Drug Des. Dev. Ther. 2016, 10, 1795–1806. [Google Scholar] [CrossRef]
- Calvert, M.B.; Jumde, V.R.; Titz, A. Pathoblockers or antivirulence drugs as a new option for the treatment of bacterial infections. Beilstein J. Org. Chem. 2018, 14, 2607–2617. [Google Scholar] [CrossRef]
- Vazquez-Armenta, F.J.; Aros-Corrales, M.O.; Alvarez-Ainza, M.L.; Bernal-Mercado, A.T.; Ayala-Zavala, J.F.; Ochoa-Leyva, A.; Lopez-Zavala, A.A. Antibacterial and anti-virulence potential of plant phenolic compounds against Vibrio parahaemolyticus. F1000Res 2023, 12, 1256. [Google Scholar] [CrossRef] [PubMed]
- Roy, P.K.; Park, S.-H.; Song, M.G.; Park, S.Y. Antimicrobial Efficacy of Quercetin against Vibrio parahaemolyticus Biofilm on Food Surfaces and Downregulation of Virulence Genes. Polymers 2022, 14, 3847. [Google Scholar] [CrossRef] [PubMed]
- Faleye, O.S.; Lee, J.-H.; Lee, J. Selected flavonoids exhibit antibiofilm and antibacterial effects against Vibrio by disrupting membrane integrity, virulence and metabolic activities. Biofilm 2023, 6, 100165. [Google Scholar] [CrossRef]
- Aguirre-Guzmán, G.; Sánchez-Martínez, J.G.; Pérez-Castañeda, R.; Palacios-Monzón, A.; Trujillo-Rodríguez, T.; De La Cruz-Hernández, N.I. Pathogenicity and Infection Route of Vibrio parahaemolyticus in American White Shrimp, LitoPenaeus vannamei. J. World Aquac. Soc. 2010, 41, 464–470. [Google Scholar] [CrossRef]
- Khimmakthong, U.; Sukkarun, P. The spread of Vibrio parahaemolyticus in tissues of the Pacific white shrimp LitoPenaeus vannamei analyzed by PCR and histopathology. Microb. Pathog. 2017, 113, 107–112. [Google Scholar] [CrossRef]
- Tang, K.F.; Bondad-Reantaso, M.G.; Arthur, J.R.; MacKinnon, B.; Hao, B.; Alday-Sanz, V.; Liang, Y.; Dong, X. Shrimp Acute Hepatopancreatic Necrosis Disease Strategy Manual; FAO Fisheries and Aquaculture Circular; FAO: Rome, Italy, 2020. [Google Scholar]
- Paria, P.; Kunal, S.P.; Behera, B.K.; Mohapatra, P.K.D.; Das, A.; Parida, P.K.; Das, B.K. Molecular characterization and genetic diversity study of Vibrio parahaemolyticus isolated from aquaculture farms in India. Aquaculture 2019, 509, 104–111. [Google Scholar] [CrossRef]
- Beshiru, A.; Igbinosa, E.O. Surveillance of Vibrio parahaemolyticus pathogens recovered from ready-to-eat foods. Sci. Rep. 2023, 13, 4186. [Google Scholar] [CrossRef]
- Che, J.; Hu, S.; Fang, Q.; Liu, B.; Liu, Z.; Hu, C.; Wang, L.; Li, L.; Bao, B. Construction and characterization of different hemolysin gene deletion strains in Vibrio parahaemolyticus (ΔhlyA, ΔhlyIII) and evaluation of their virulence. J. Invertebr. Pathol. 2024, 207, 108210. [Google Scholar] [CrossRef] [PubMed]
- Lian, L.; Xue, J.; Li, W.; Ren, J.; Tang, F.; Liu, Y.; Xue, F.; Dai, J. VscF in T3SS1 Helps to Translocate VPA0226 in Vibrio parahaemolyticus. Front. Cell. Infect. Microbiol. 2021, 11, 652432. [Google Scholar] [CrossRef] [PubMed]
- Ananda Raja, R.; Sridhar, R.; Balachandran, C.; Palanisammi, A.; Ramesh, S.; Nagarajan, K.; Paulpandi, S. Identification of Vibrio parahaemolyticus from farmed penaeid shrimp, Penaeus vannamei (Boone, 1931) by multiplex PCR targeting toxR and tlh genes. Indian J. Fish. 2023, 70, 162–167. [Google Scholar] [CrossRef]
- Paria, P.; Behera, B.K.; Mohapatra, P.K.D.; Parida, P.K. Virulence factor genes and comparative pathogenicity study of tdh, trh and tlh positive Vibrio parahaemolyticus strains isolated from Whiteleg shrimp, LitoPenaeus vannamei (Boone, 1931) in India. Infect. Genet. Evol. 2021, 95, 105083. [Google Scholar] [CrossRef]
- Chimalapati, S.; de Souza Santos, M.; Lafrance, A.E.; Ray, A.; Lee, W.-R.; Rivera-Cancel, G.; Vale, G.; Pawlowski, K.; Mitsche, M.A.; McDonald, J.G.; et al. Vibrio deploys type 2 secreted lipase to esterify cholesterol with host fatty acids and mediate cell egress. eLife 2020, 9, e58057. [Google Scholar] [CrossRef]
- Carter, P.; Wells, J.A. Dissecting the catalytic triad of a serine protease. Nature 1988, 332, 564–568. [Google Scholar] [CrossRef]
- Taniguchi, H.; Hirano, H.; Kubomura, S.; Higashi, K.; Mizuguchi, Y. Comparison of the nucleotide sequences of the genes for the thermostable direct hemolysin and the thermolabile hemolysin from Vibrio parahaemolyticus. Microb. Pathog. 1986, 1, 425–432. [Google Scholar] [CrossRef]
- Akoh, C.C.; Lee, G.-C.; Liaw, Y.-C.; Huang, T.-H.; Shaw, J.-F. GDSL family of serine esterases/lipases. Prog. Lipid Res. 2004, 43, 534–552. [Google Scholar] [CrossRef]
- Wan, Y.; Liu, C.; Ma, Q. Structural analysis of a Vibrio phospholipase reveals an unusual Ser–His–chloride catalytic triad. J. Biol. Chem. 2019, 294, 11391–11401. [Google Scholar] [CrossRef]
- Wang, C.; Liu, C.; Zhu, X.; Peng, Q.; Ma, Q. Catalytic site flexibility facilitates the substrate and catalytic promiscuity of Vibrio dual lipase/transferase. Nat. Commun. 2023, 14, 4795. [Google Scholar] [CrossRef]
- Vazquez-Morado, L.E.; Robles-Zepeda, R.E.; Ochoa-Leyva, A.; Arvizu-Flores, A.A.; Garibay-Escobar, A.; Castillo-Yañez, F.; Lopez-Zavala, A.A. Biochemical characterization and inhibition of thermolabile hemolysin from Vibrio parahaemolyticus by phenolic compounds. PeerJ 2021, 9, e10506. [Google Scholar] [CrossRef] [PubMed]
- Roy, P.K.; Song, M.G.; Park, S.Y. The Inhibitory Effect of Quercetin on Biofilm Formation of Listeria monocytogenes Mixed Culture and Repression of Virulence. Antioxidants 2022, 11, 1733. [Google Scholar] [CrossRef]
- Wu, Y.; Jiang, L.; Ran, W.; Zhong, K.; Zhao, Y.; Gao, H. Antimicrobial activities of natural flavonoids against foodborne pathogens and their application in food industry. Food Chem. 2024, 460, 140476. [Google Scholar] [CrossRef] [PubMed]
- Nag, D.; Dastidar, D.G.; Chakrabarti, G. Natural flavonoid morin showed anti-bacterial activity against Vibrio cholera after binding with cell division protein FtsA near ATP binding site. Biochim. Biophys. Acta (BBA)—Gen. Subj. 2021, 1865, 129931. [Google Scholar] [CrossRef] [PubMed]
- Mottaghi, S.; Abbaszadeh, H. The anticarcinogenic and anticancer effects of the dietary flavonoid, morin: Current status, challenges, and future perspectives. Phytother. Res. 2021, 35, 6843–6861. [Google Scholar] [CrossRef]
- Palei, N.N.; Mounika, G.; Mohanta, B.C.; Rajangam, J. Quercetin and Morin dual drug loaded nanostructured lipid carriers: Formulation and in vitro cytotoxicity study on MCF7 breast cancer cells. J. Dispers. Sci. Technol. 2024, 45, 2146–2154. [Google Scholar] [CrossRef]
- Chen, X.; Liu, B.; Tong, R.; Ding, S.; Wu, J.; Lei, Q.; Fang, W. Improved stability and targeted cytotoxicity of epigallocatechin-3-gallate palmitate for anticancer therapy. Langmuir 2021, 37, 969–977. [Google Scholar] [CrossRef]
- Copeland, R.A. Evaluation of Enzyme Inhibitors in Drug Discovery: A guide for Medicinal Chemists and Pharmacologists; John Wiley & Sons: Hoboken, NJ, USA, 2013. [Google Scholar]
- Jia, A.; Woo, N.Y.; Zhang, X.-H. Expression, purification, and characterization of thermolabile hemolysin (TLH) from Vibrio alginolyticus. Dis. Aquat. Org. 2010, 90, 121–127. [Google Scholar] [CrossRef]
- Sakatoku, A.; Hatano, K.; Takada, K.; Shimizu, R.; Suzuki, T.; Seki, M.; Suzuki, N.; Tanaka, D.; Nakamura, S.; Isshiki, T. Purification and Characterization of the Lecithin-Dependent Thermolabile Hemolysin Vhe1 from the Vibrio sp. Strain MA3 Associated with Mass Mortality of Pearl Oyster (Pinctada fucata). Curr. Microbiol. 2023, 80, 288. [Google Scholar] [CrossRef]
- Vazquez-Armenta, F.J.; Valdez-Olmos, U.F.; Arvizu-Flores, A.A.; Ayala-Zavala, J.F.; Ochoa-Leyva, A.; Lopez-Zavala, A.A. Metal Ions and Chemical Modification Reagents Inhibit the Enzymatic Activity of Lecithin-Dependent Hemolysin from Vibrio parahaemolyticus. Toxins 2022, 14, 609. [Google Scholar] [CrossRef]
- Pereañez, J.A.; Núñez, V.; Patiño, A.C.; Londoño, M.; Quintana, J.C. Inhibitory Effects Of Plant Phenolic Compounds On Enzymatic And Cytotoxic Activities Induced By A Snake Venom Phospholipase A2. Vitae 2011, 18, 295–304. [Google Scholar] [CrossRef]
- Wicka, M.; Wanarska, M.; Krajewska, E.; Pawlak-Szukalska, A.; Kur, J.; Cieśliński, H. Cloning, expression, and biochemical characterization of a cold-active GDSL-esterase of a Pseudomonas sp. S9 isolated from Spitsbergen island soil. Acta Biochim. Pol. 2016, 63, 117–125. [Google Scholar] [CrossRef]
- Slater, L.S.; Callis, P.R. Molecular Orbital Theory of the 1La and 1Lb States of Indole. 2. An ab Initio Study. J. Phys. Chem. 1995, 99, 8572–8581. [Google Scholar] [CrossRef]
- Ghisaidoobe, A.B.; Chung, S.J. Intrinsic tryptophan fluorescence in the detection and analysis of proteins: A focus on Förster resonance energy transfer techniques. Int. J. Mol. Sci. 2014, 15, 22518–22538. [Google Scholar] [CrossRef]
- Stachurska, K.; Antosiewicz, J.M. Contribution of Each Tryptophan to the Total Fluorescence Emitted by a Protein with Multiple Tryptophan Residues Depends on the Energy of the Excitation Radiation Quantum. ACS Omega 2024, 9, 43998–44004. [Google Scholar] [CrossRef]
- Motz, R.N.; Sun, A.C.; Lehnherr, D.; Ruccolo, S. High-Throughput Determination of Stern–Volmer Quenching Constants for Common Photocatalysts and Quenchers. ACS Org. Inorg. Au 2023, 3, 266–273. [Google Scholar] [CrossRef]
- Gayer, A.V.; Yakimov, B.P.; Budylin, G.S.; Shirshin, E.A. Evaluating the number of ligand binding sites on protein from tryptophan fluorescence quenching under typical experimental conditions. J. Biomed. Photonics Eng. 2020, 6, 020303. [Google Scholar] [CrossRef]
- Muñoz-Bacasehua, C.; Rosas-Rodríguez, J.A.; López-Zavala, A.A.; Valenzuela-Soto, E.M. Spectroscopic analysis of coenzyme binding to betaine aldehyde dehydrogenase dependent on potassium. Luminescence 2021, 36, 1733–1742. [Google Scholar] [CrossRef]
- Berg, O.G.; von Hippel, P.H. Diffusion-controlled macromolecular interactions. Annu. Rev. Biophys. Biophys. Chem. 1985, 14, 131–158. [Google Scholar] [CrossRef]
- Lau, W.Y.V.; Taylor, P.K.; Brinkman, F.S.; Lee, A.H. Pathogen-associated gene discovery workflows for novel antivirulence therapeutic development. eBioMedicine 2023, 88, 104429. [Google Scholar] [CrossRef]
- Ogawara, H. Possible drugs for the treatment of bacterial infections in the future: Anti-virulence drugs. J. Antibiot. 2021, 74, 24–41. [Google Scholar] [CrossRef]
- Al-Shabib, N.A.; Khan, J.M.; Malik, A.; Alsenaidy, M.A.; Rehman, M.T.; AlAjmi, M.F.; Alsenaidy, A.M.; Husain, F.M.; Khan, R.H. Molecular insight into binding behavior of polyphenol (rutin) with beta lactoglobulin: Spectroscopic, molecular docking and MD simulation studies. J. Mol. Liq. 2018, 269, 511–520. [Google Scholar] [CrossRef]
- Siddiqui, G.A.; Siddiqi, M.K.; Khan, R.H.; Naeem, A. Probing the binding of phenolic aldehyde vanillin with bovine serum albumin: Evidence from spectroscopic and docking approach. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 203, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Duan, H.; Song, Z.; Xu, R. Comparative study on the interaction between fibrinogen and flavonoids. J. Mol. Struct. 2022, 1262, 132963. [Google Scholar] [CrossRef]
- Li, R.; Huang, L.; Zhang, Z.; Chen, J.; Tang, H. Integrated multispectroscopic analysis and molecular docking analyses of the structure-affinity relationship and mechanism of the interaction of flavonoids with zein. Food Chem. 2022, 386, 132839. [Google Scholar] [CrossRef]
- Sandu, N.; Chilom, C.G.; Popescu, A.I. Structural and molecular aspects of flavonoids as ligands for serum transferrin. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 254, 119600. [Google Scholar] [CrossRef]
- Song, W.; Wang, L.; Zhao, Y.; Lanzi, G.; Wang, X.; Zhang, C.; Guan, J.; Wang, W.; Guo, X.; Meng, Y. Hibifolin, a natural sortase a inhibitor, attenuates the pathogenicity of Staphylococcus aureus and enhances the antibacterial activity of cefotaxime. Microbiol. Spectr. 2022, 10, e00950-22. [Google Scholar] [CrossRef]
- Song, W.; Wang, L.; Jin, M.; Guo, X.; Wang, X.; Guan, J.; Zhao, Y. Punicalagin, an Inhibitor of Sortase A, Is a Promising Therapeutic Drug to Combat Methicillin-Resistant Staphylococcus aureus Infections. Antimicrob. Agents Chemother. 2022, 66, e00224-22. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, X.; Liu, S.; Li, G.; Zhang, B.; Deng, X.; Niu, X. Novel inhibitor discovery and the conformational analysis of inhibitors of listeriolysin O via protein-ligand modeling. Sci. Rep. 2015, 5, 8864. [Google Scholar] [CrossRef]
- Moreno-Córdova, E.N.; Arvizu-Flores, A.A.; Valenzuela-Soto, E.M.; García-Orozco, K.D.; Wall-Medrano, A.; Alvarez-Parrilla, E.; Ayala-Zavala, J.F.; Domínguez-Avila, J.A.; González-Aguilar, G.A. Gallotannins are uncompetitive inhibitors of pancreatic lipase activity. Biophys. Chem. 2020, 264, 106409. [Google Scholar] [CrossRef]
- Billowria, K.; Ali, R.; Rangra, N.K.; Kumar, R.; Chawla, P.A. Bioactive Flavonoids: A Comprehensive Review on Pharmacokinetics and Analytical Aspects. Crit. Rev. Anal. Chem. 2024, 54, 1002–1016. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.; Rickles, L.F.; Halpin, W.A. The crystal and molecular structure of quercetin: A biologically active and naturally occurring flavonoid. Bioorganic Chem. 1986, 14, 55–69. [Google Scholar] [CrossRef]
- Cody, V.; Luft, J.R. Conformational analysis of flavonoids: Crystal and molecular structures of morin hydrate and myricetin (1:2) triphenylphosphine oxide complex. J. Mol. Struct. 1994, 317, 89–97. [Google Scholar] [CrossRef]
- Mehmood, S.; Maqsood, M.; Mahtab, N.; Khan, M.I.; Sahar, A.; Zaib, S.; Gul, S. Epigallocatechin gallate: Phytochemistry, bioavailability, utilization challenges, and strategies. J. Food Biochem. 2022, 46, e14189. [Google Scholar] [CrossRef] [PubMed]
- Valdés-Tresanco, M.S.; Valdés-Tresanco, M.E.; Valiente, P.A.; Moreno, E. gmx_MMPBSA: A New Tool to Perform End-State Free Energy Calculations with GROMACS. J. Chem. Theory Comput. 2021, 17, 6281–6291. [Google Scholar] [CrossRef]
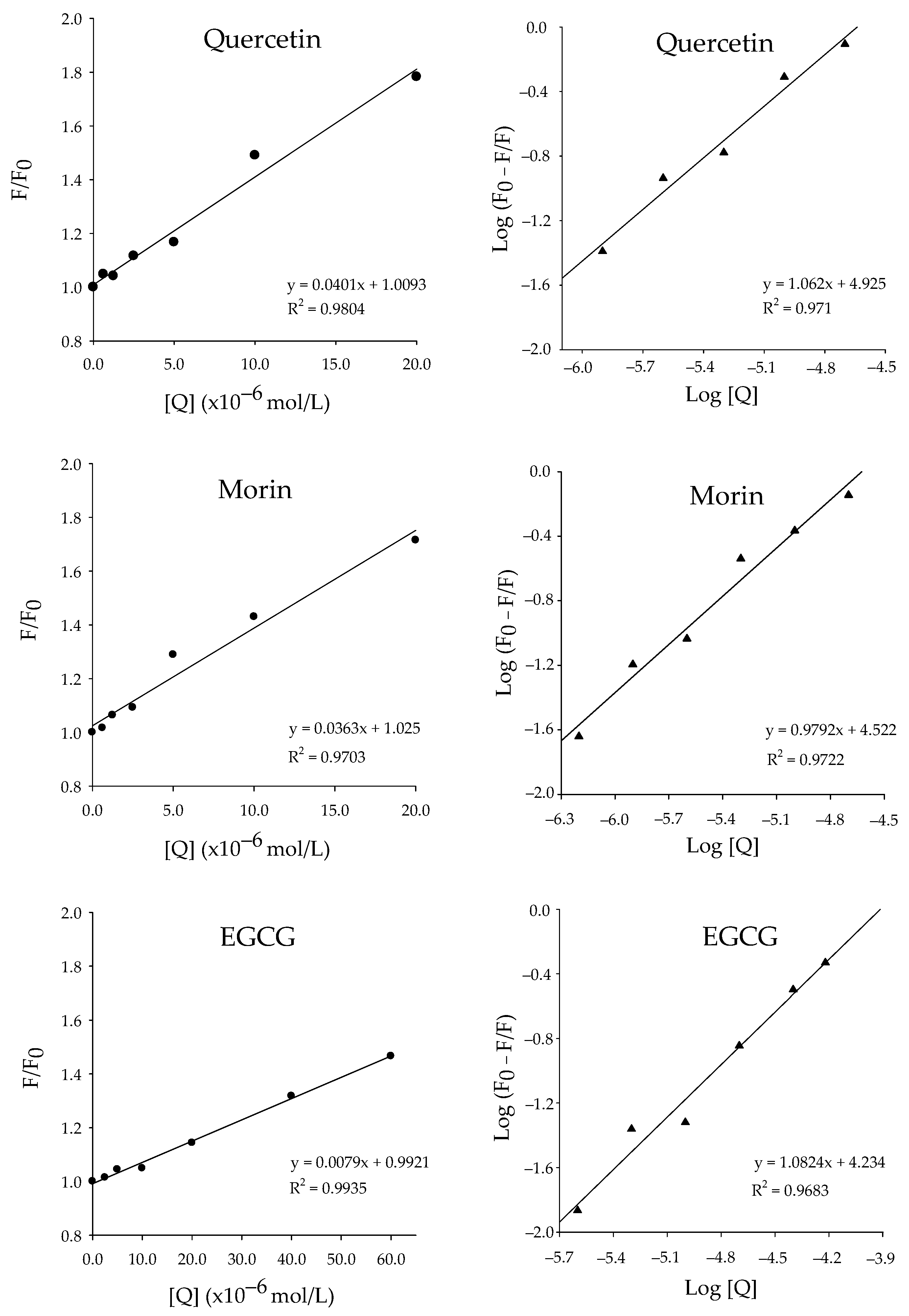
| Flavonoids | Ksv (104 mol−1) | Kq (1012 mol−1 s−1) | Kd (μM) | n |
|---|---|---|---|---|
| Quercetin | 4.01 ± 0.05 | 4.01 ± 0.05 | 7.91 ± 0.55 a | 1.06 |
| Morin | 3.63 ± 0.07 | 3.63 ± 0.07 | 25.05 ± 1.63 b | 0.98 |
| EGCG | 0.79 ± 0.02 | 0.79 ± 0.02 | 44.69 ± 2.88 c | 1.08 |
| Residue | EGCG | Morin | PC | pNPL | Quercetin |
|---|---|---|---|---|---|
| Asp152 | 1.10 ± 0.58 | 0.88 ± 0.32 | |||
| Ser153 | −0.16 ± 0.27 | −1.02 ± 0.61 | −0.60 ± 0.35 | −0.16 ± 0.52 | |
| Leu154 | −1.27 ± 0.41 | −0.43 ± 0.24 | |||
| Ile160 | −1.28 ± 0.62 | −0.91 ± 0.38 | |||
| Ser164 | −0.61 ± 0.40 | ||||
| Phe168 | −2.05 ± 0.56 | −1.61 ± 0.88 | −2.11 ± 0.59 | −0.86 ± 0.33 | −3.06 ± 1.02 |
| Phe179 | −0.55 ± 0.29 | −0.29 ± 0.37 | −0.84 ± 0.43 | −0.53 ± 0.35 | |
| Trp185 | −0.43 ± 0.32 | ||||
| Gly203 | −0.99 ± 0.66 | −2.12 ± 0.75 | |||
| Gly204 | −0.55 ± 0.31 | −1.76 ± 0.63 | |||
| Tyr211 | −2.16 ± 0.91 | −0.64 ± 0.50 | |||
| Leu214 | −0.45 ± 0.36 | ||||
| Leu247 | −1.05 ± 0.32 | −1.05 ± 0.30 | |||
| Asn248 | −0.11 ± 0.24 | −1.32 ± 1.29 | −0.91 ± 0.38 | −0.10 ± 0.44 | |
| Met251 | −0.98 ± 0.38 | −1.06 ± 0.39 | |||
| Asn252 | −0.22 ± 0.57 | −0.44 ± 0.96 | −1.02 ± 0.95 | −0.83 ± 0.40 | |
| Tyr253 | −0.19 ± 0.44 | 0.33 ± 0.34 | |||
| Met282 | −0.44 ± 0.23 | ||||
| Pro285 | −0.92 ± 0.36 | −0.34 ± 0.27 | |||
| Ala287 | −0.80 ± 0.25 | −0.76 ± 0.25 | |||
| Ala290 | −0.45 ± 0.20 | −0.28 ± 0.18 | |||
| Gln292 | −0.10 ± 0.31 | −0.26 ± 0.39 | −0.19 ± 0.22 | ||
| Thr334 | −0.66 ± 0.23 | ||||
| Phe338 | −0.37 ± 0.31 | −0.14 ± 0.18 | |||
| Ser363 | −0.18 ± 0.52 | ||||
| Ser364 | −0.14 ± 0.31 | −0.14 ± 0.22 | |||
| Ser365 | −1.27 ± 0.80 | −0.69 ± 0.56 | −0.46 ± 0.47 | −0.74 ± 0.33 | −1.29 ± 0.39 |
| Tyr368 | −0.66 ± 0.30 | −0.62 ± 0.81 | −1.50 ± 0.43 | −1.53 ± 0.54 | −0.45 ± 0.34 |
| Met369 | −0.34 ± 0.38 | ||||
| Phe388 | −0.16 ± 0.15 | −0.17 ± 0.13 | |||
| Val391 | −0.25 ± 0.17 | ||||
| Thr392 | −0.31 ± 0.77 | −1.59 ± 0.37 | −1.52 ± 0.40 | −0.10 ± 0.19 | |
| His393 | −0.35 ± 0.86 | −0.24 ± 0.57 | 0.30 ± 0.41 | ||
| Pro394 | −0.69 ± 0.31 | −0.34 ± 0.23 | |||
| Thr398 | −0.78 ± 0.22 | −0.15 ± 0.13 | |||
| His399 | 0.63 ± 0.31 | 0.22 ± 0.08 | |||
| Val402 | −0.72 ± 0.26 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vazquez-Armenta, F.J.; Alvarez-Armenta, A.; Sugich-Miranda, R.; Ayala-Zavala, F.; Morales-Ortega, A.; Arvizu-Flores, A.A.; Lopez-Zavala, A.A. Inhibition Mechanism of Lecithin-Dependent Hemolysin from Vibrio parahaemolyticus by Flavonoids: An Enzyme Kinetic and Structural Approach. Catalysts 2025, 15, 257. https://doi.org/10.3390/catal15030257
Vazquez-Armenta FJ, Alvarez-Armenta A, Sugich-Miranda R, Ayala-Zavala F, Morales-Ortega A, Arvizu-Flores AA, Lopez-Zavala AA. Inhibition Mechanism of Lecithin-Dependent Hemolysin from Vibrio parahaemolyticus by Flavonoids: An Enzyme Kinetic and Structural Approach. Catalysts. 2025; 15(3):257. https://doi.org/10.3390/catal15030257
Chicago/Turabian StyleVazquez-Armenta, Francisco J., Andres Alvarez-Armenta, Rocio Sugich-Miranda, Fernando Ayala-Zavala, Adriana Morales-Ortega, Aldo A. Arvizu-Flores, and Alonso A. Lopez-Zavala. 2025. "Inhibition Mechanism of Lecithin-Dependent Hemolysin from Vibrio parahaemolyticus by Flavonoids: An Enzyme Kinetic and Structural Approach" Catalysts 15, no. 3: 257. https://doi.org/10.3390/catal15030257
APA StyleVazquez-Armenta, F. J., Alvarez-Armenta, A., Sugich-Miranda, R., Ayala-Zavala, F., Morales-Ortega, A., Arvizu-Flores, A. A., & Lopez-Zavala, A. A. (2025). Inhibition Mechanism of Lecithin-Dependent Hemolysin from Vibrio parahaemolyticus by Flavonoids: An Enzyme Kinetic and Structural Approach. Catalysts, 15(3), 257. https://doi.org/10.3390/catal15030257







