Diverse YqeK Diadenosine Tetraphosphate Hydrolases Control Biofilm Formation in an Iron-Dependent Manner
Abstract
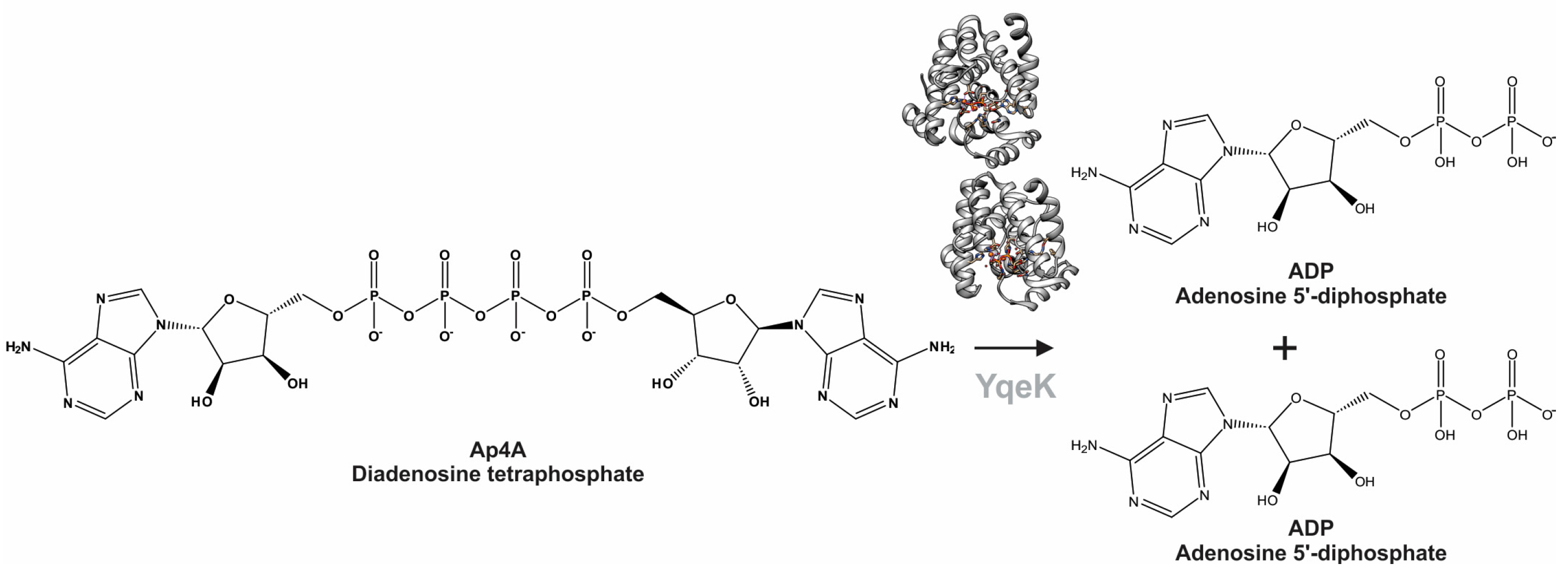
1. Introduction
2. Results
2.1. Occurrence and Evolution of YqeK Hydrolases
2.2. LB Bh and Ca YqeK Harbor Redox and Hydrolytically Active Homo- and Mixed-Metal Fe-Based Cofactors
2.3. The Diiron Cofactors of Bh and Ca YqeK
2.4. The Active Forms of the Diiron Bh and Ca YqeK
2.5. Bh YqeK Accesses Its Reduced Cofactor Forms at Higher Reduction Potentials than Ca YqeK
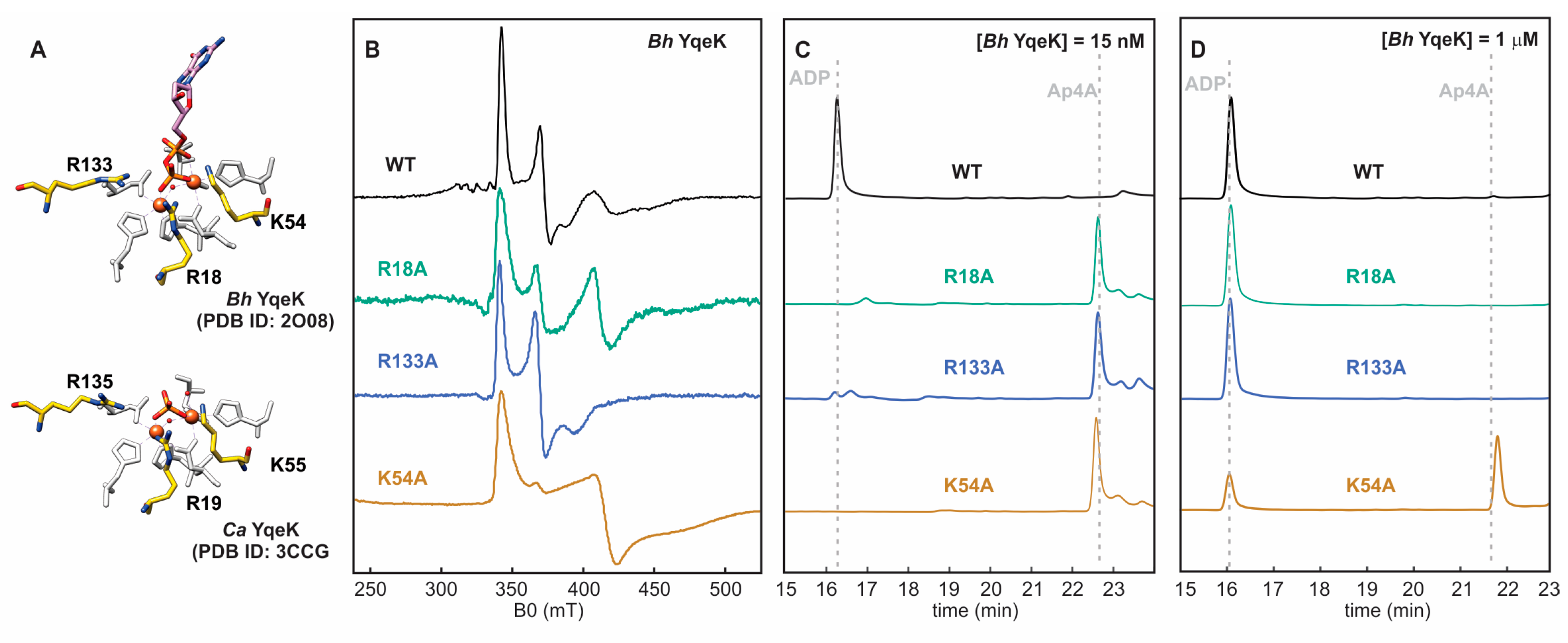
2.6. Positively Charged Residues Tune Ap4A Hydrolysis
3. Discussion
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aravind, L.; Koonin, E.V. The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem. Sci. 1998, 23, 469–472. [Google Scholar] [CrossRef] [PubMed]
- Langton, M.; Sun, S.; Ueda, C.; Markey, M.; Chen, J.; Paddy, I.; Jiang, P.; Chin, N.; Milne, A.; Pandelia, M.-E. The HD-Domain Metalloprotein Superfamily: An Apparent Common Protein Scaffold with Diverse Chemistries. Catalysts 2020, 10, 1191. [Google Scholar] [CrossRef] [PubMed]
- Minazzato, G.; Gasparrini, M.; Amici, A.; Cianci, M.; Mazzola, F.; Orsomando, G.; Sorci, L.; Raffaelli, N. Functional Charac-terization of COG1713 (YqeK) as a Novel Diadenosine Tetraphosphate Hydrolase Family. J. Bacteriol. 2020, 202, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, F.; McLennan, A.G.; Urbaniak, M.D.; Jones, N.J.; Copeland, N.A. Re-evaluation of Diadenosine Tetraphosphate (Ap4A) From a Stress Metabolite to Bona Fide Secondary Messenger. Front. Mol. Biosci. 2020, 7, 606807. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Zou, J.; Peng, H.; Stolle, A.-S.; Xie, R.; Zhang, H.; Peng, B.; Mekalanos, J.J.; Zheng, J. Alarmone Ap4A is elevated by aminoglycoside antibiotics and enhances their bactericidal activity. Proc. Natl. Acad. Sci. USA 2019, 116, 9578–9585. [Google Scholar] [CrossRef]
- Monds Russell, D.; Newell Peter, D.; Wagner Jeffrey, C.; Schwartzman Julia, A.; Lu, W.; Rabinowitz Joshua, D.; O’Toole George, A. Di-Adenosine Tetraphosphate (Ap4A) Metabolism Impacts Biofilm Formation by Pseudomonas fluorescens via Modulation of c-di-GMP-Dependent Pathways. J. Bacteriol. 2010, 192, 3011–3023. [Google Scholar] [CrossRef]
- Rapaport, E.; Zamecnik, P.C. Presence of diadenosine 5′,5′′′-P1, P4-tetraphosphate (Ap4A) in mamalian cells in levels varying widely with proliferative activity of the tissue: A possible positive “pleiotypic activator”. Proc. Natl. Acad. Sci. USA 1976, 73, 3984–3988. [Google Scholar] [CrossRef]
- Bochner, B.R.; Lee, P.C.; Wilson, S.W.; Cutler, C.W.; Ames, B.N. AppppA and related adenylylated nucleotides are synthesized as a consequence of oxidation stress. Cell 1984, 37, 225–232. [Google Scholar] [CrossRef]
- Ismail, T.M.; Hart, C.; McLennan, A.G. Regulation of Dinucleoside Polyphosphate Pools by the YgdP and ApaH Hydrolases Is Essential for the Ability of Salmonella enterica serovar Typhimurium to Invade Cultured Mammalian Cells. J. Biol. Chem. 2003, 278, 32602–32607. [Google Scholar] [CrossRef]
- Kimura, Y.; Tanaka, C.; Sasaki, K.; Sasaki, M. High concentrations of intracellular Ap4A and/or Ap5A in developing Myxococcus xanthus cells inhibit sporulation. Microbiology 2017, 163, 86–93. [Google Scholar] [CrossRef]
- Luciano, D.J.; Belasco, J.G. Np 4 A alarmones function in bacteria as precursors to RNA caps. Proc. Natl. Acad. Sci. USA 2020, 117, 3560–3567. [Google Scholar] [CrossRef] [PubMed]
- František Potužník, J.; Nešuta, O.; Škríba, A.; Volenikova, B.; Mititelu, M.B.; Mancini, F.; Serianni, V.; Fernandez, H.; Spustová, K.; Trylčová, J. Diadenosine tetraphosphate (Ap4A) serves as a 5′ RNA cap in mammalian cells. Angew. Chem. 2024, 136, e202314951. [Google Scholar] [CrossRef]
- Despotović, D.; Brandis, A.; Savidor, A.; Levin, Y.; Fumagalli, L.; Tawfik, D.S. Diadenosine tetraphosphate (Ap4A)—An E. coli alarmone or a damage metabolite? FEBS J. 2017, 284, 2194–2215. [Google Scholar] [CrossRef] [PubMed]
- Oka, M.; Takegawa, K.; Kimura, Y. Enzymatic characterization of a class II lysyl-tRNA synthetase, LysS, from Myxococcus xanthus. Arch. Biochem. Biophys. 2015, 579, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Wright, M.; Boonyalai, N.; Tanner, J.A.; Hindley, A.D.; Miller, A.D. The duality of LysU, a catalyst for both Ap4A and Ap3A formation. FEBS J. 2006, 273, 3534–3544. [Google Scholar] [CrossRef]
- Hansen, S.; Lewis, K.; Vulić, M. Role of Global Regulators and Nucleotide Metabolism in Antibiotic Tolerance in Escherichia coli. Antimicrob. Agents Chemother. 2008, 52, 2718–2726. [Google Scholar] [CrossRef]
- Zheng, T.; Jing, M.; Gong, T.; Yan, J.; Zeng, J.; Li, Y. Deletion of the yqeK gene leads to the accumulation of Ap4A and reduced biofilm formation in Streptococcus mutans. Mol. Oral Microbiol. 2021, 37, 9–21. [Google Scholar] [CrossRef]
- Branda, S.S.; González-Pastor, J.E.; Dervyn, E.; Ehrlich, S.D.; Losick, R.; Kolter, R. Genes Involved in Formation of Structured Multicellular Communities by Bacillus subtilis. J. Bacteriol. 2004, 186, 3970–3979. [Google Scholar] [CrossRef]
- McLennan, A.G. The Nudix hydrolase superfamily. Cell. Mol. Life Sci. 2006, 63, 123–143. [Google Scholar] [CrossRef]
- McLennan, A.G. Dinucleoside polyphosphates—Friend or foe? Pharmacol. Therap. 2000, 87, 73–89. [Google Scholar] [CrossRef]
- Guranowski, A. Specific and nonspecific enzymes involved in the catabolism of mononucleoside and dinucleoside polyphosphates. Pharmacol. Ther. 2000, 87, 117–139. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, M.; Takegawa, K.; Kimura, Y. Enzymatic characteristics of an ApaH-like phosphatase, PrpA, and a diadenosine tetraphosphate hydrolase, ApaH, from Myxococcus xanthus. FEBS Lett. 2014, 588, 3395–3402. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Ke, J.; Zheng, P.; Zhang, H.; Zhu, Z.; Niu, L. Structural and biochemical characterization of a nucleotide hydrolase from Streptococcus pneumonia. Structure 2024, 32, 197–1207.e4. [Google Scholar] [CrossRef] [PubMed]
- Plateau, P.; Fromant, M.; Brevet, A.; Gesquiere, A.; Blanquet, S. Catabolism of bis(5′-nucleosidyl) oligophosphates in Escherichia coli: Metal requirements and substrate specificity of homogeneous diadenosine-5′,5′′′-P1,P4-tetraphosphate pyrophosphohydrolase. Biochemistry 1985, 24, 914–922. [Google Scholar] [CrossRef] [PubMed]
- He, P.; Deng, C.; Liu, B.; Zeng, L.; Zhao, W.; Zhang, Y.; Jiang, X.; Guo, X.; Qin, J. Characterization of a bifunctional enzyme with (p)ppGpp-hydrolase/synthase activity in Leptospira interrogans. FEMS Microbiol. Lett. 2013, 348, 133–142. [Google Scholar] [CrossRef]
- Mechold, U.; Murphy, H.; Brown, L.; Cashel, M. Intramolecular Regulation of the Opposing (p)ppGpp Catalytic Activities of RelSeq, the Rel/Spo Enzyme from Streptococcus equisimilis. J. Bacteriol. 2002, 184, 2878–2888. [Google Scholar] [CrossRef]
- Jeon, Y.J.; Park, S.C.; Song, W.S.; Kim, O.-H.; Oh, B.-C.; Yoon, S.-I. Structural and biochemical characterization of bacterial YpgQ protein reveals a metal-dependent nucleotide pyrophosphohydrolase. J. Struct. Biol. 2016, 195, 113–122. [Google Scholar] [CrossRef]
- Sun, S.; He, Z.; Jiang, P.; Baral, R.; Pandelia, M.-E. Metal Dependence and Functional Diversity of Type I Cas3 Nucleases. Biochemistry 2022, 61, 327–338. [Google Scholar] [CrossRef]
- Sun, S.; Wang, R.; Pandelia, M.-E. Vibrio cholerae V-cGAP3 Is an HD-GYP Phosphodiesterase with a Metal Tunable Substrate Selectivity. Biochemistry 2022, 61, 1801–1809. [Google Scholar] [CrossRef]
- Sun, S.; Pandelia, M.-E. HD-[HD-GYP] Phosphodiesterases: Activities and Evolutionary Diversification within the HD-GYP Family. Biochemistry 2020, 59, 2340–2350. [Google Scholar] [CrossRef]
- Mulepati, S.; Bailey, S. Structural and Biochemical Analysis of Nuclease Domain of Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR)-associated Protein 3 (Cas3). J. Biol. Chem. 2011, 286, 31896–31903. [Google Scholar] [CrossRef] [PubMed]
- Gerber, E.; Bernard, R.; Castang, S.; Chabot, N.; Coze, F.; Dreux-Zigha, A.; Hauser, E.; Hivin, P.; Joseph, P.; Lazarelli, C.; et al. Deinococcus as new chassis for industrial biotechnology: Biology, physiology and tools. J. Appl. Microbiol. 2015, 119, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Hoiczyk, E.; Hansel, A. Cyanobacterial cell walls: News from an unusual prokaryotic envelope. J. Bacteriol. 2000, 182, 1191–1199. [Google Scholar] [CrossRef] [PubMed]
- Beck, J.L.; De Jersey, J.; Zerner, B.; Hendrich, M.P.; Debrunner, P.G. Properties of the Fe(II)-Fe(III) derivative of red kidney bean purple phosphatase. Evidence for a binuclear zinc-iron center in the native enzyme. J. Am. Chem. Soc. 1988, 110, 3317–3318. [Google Scholar] [CrossRef]
- Durmus, A.; Eicken, C.; Sift, B.H.; Kratel, A.; Kappl, R.; Hüttermann, J.; Krebs, B. The active site of purple acid phosphatase from sweet potatoes (Ipomoea batatas). Eur. J. Biochem. 1999, 260, 709–716. [Google Scholar] [CrossRef]
- Merkx, M.; Averill, B.A. The Activity of Oxidized Bovine Spleen Purple Acid Phosphatase Is Due to an Fe(III)Zn(II) ‘Impurity’. Biochemistry 1998, 37, 11223–11231. [Google Scholar] [CrossRef]
- Reiter, T.A.; Rusnak, F. Electrochemical Studies of the Mono-Fe, Fe−Zn, and Fe−Fe Metalloisoforms of Bacteriophage λ Protein Phosphatase. Biochemistry 2004, 43, 782–790. [Google Scholar] [CrossRef]
- Davis, J.C.; A Averill, B. Evidence for a spin-coupled binuclear iron unit at the active site of the purple acid phosphatase from beef spleen. Proc. Natl. Acad. Sci. USA 1982, 79, 4623–4627. [Google Scholar] [CrossRef]
- Wörsdörfer, B.; Lingaraju, M.; Yennawar, N.H.; Boal, A.K.; Krebs, C.; Bollinger, J.M.; Pandelia, M.-E. Organophosphonate-degrading PhnZ reveals an emerging family of HD domain mixed-valent diiron oxygenases. Proc. Natl. Acad. Sci. USA 2013, 110, 18874–18879. [Google Scholar] [CrossRef]
- Xing, G.; Barr, E.W.; Diao, Y.; Hoffart, L.M.; Prabhu, K.S.; Arner, R.J.; Reddy, C.C.; Krebs, C.; Bollinger, J.M., Jr. Oxygen ac-tivation by a mixed-valent, diiron(II/III) cluster in the glycol cleavage reaction catalyzed by myo-inositol oxygenase. Biochemistry 2006, 45, 5402–5412. [Google Scholar] [CrossRef]
- Bollinger, J.M.; Diao, Y.; Matthews, M.L.; Xing, G.; Krebs, C. myo-Inositol oxygenase: A radical new pathway for O2and C–H activation at a nonheme diiron cluster. Dalton Trans. 2008, 6, 905–914. [Google Scholar] [CrossRef] [PubMed]
- Lynch, J.; Juarez-Garcia, C.; Münck, E.; Que, L., Jr. Mössbauer and EPR studies of the binuclear iron center in ribonucleotide reductase from Escherichia coli: A new iron-to-protein stoichiometry. J. Biol. Chem. 1989, 264, 8091–8096. [Google Scholar] [CrossRef] [PubMed]
- Langton, M.; Appell, M.; Koob, J.; Pandelia, M.-E. Domain Fusion of Two Oxygenases Affords Organophosphonate Degradation in Pathogenic Fungi. Biochemistry 2022, 61, 956–962. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Calderone, L.A.; Pan, L.; Quist, T.; Pandelia, M.-E. The Fe and Zn cofactor dilemma. Biochim. Biophys. Acta-Proteins Proteom. 2023, 1871, 140931. [Google Scholar] [CrossRef] [PubMed]
- Olczak, M.; Morawiecka, B.; Watorek, W. Plant purple acid phosphatases—Genes, structures and biological function. Acta Biochim. Pol. 2003, 50, 1245–1256. [Google Scholar] [CrossRef] [PubMed]
- Funhoff, E.G.; Bollen, M.; Averill, B.A. The Fe(III)Zn(II) form of recombinant human purple acid phosphatase is not activated by proteolysis. J. Inorg. Biochem. 2005, 99, 521–529. [Google Scholar] [CrossRef]
- Goldstone, D.C.; Ennis-Adeniran, V.; Hedden, J.J.; Groom, H.C.T.; Rice, G.I.; Christodoulou, E.; Walker, P.A.; Kelly, G.; Haire, L.F.; Yap, M.W.; et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 2011, 480, 379–382. [Google Scholar] [CrossRef]
- Vorontsov, I.I.; Minasov, G.; Kiryukhina, O.; Brunzelle, J.S.; Shuvalova, L.; Anderson, W.F. Characterization of the deoxynu-cleotide triphosphate triphosphohydrolase (dNTPase) activity of the EF1143 protein from Enterococcus faecalis and crystal structure of the activator-substrate complex. J. Biol. Chem. 2011, 286, 33158–33166. [Google Scholar] [CrossRef]
- Bridwell-Rabb, J.; Kang, G.; Zhong, A.; Liu, H.-W.; Drennan, C.L. An HD domain phosphohydrolase active site tailored for oxetanocin-A biosynthesis. Proc. Natl. Acad. Sci. USA 2016, 113, 13750–13755. [Google Scholar] [CrossRef]
- Davidson, J.F.; Whyte, B.; Bissinger, P.H.; Schiestl, R.H. Oxidative stress is involved in heat-induced cell death in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1996, 93, 5116–5121. [Google Scholar] [CrossRef]
- Marcén, M.; Ruiz, V.; Serrano, M.J.; Condón, S.; Mañas, P. Oxidative stress in E. coli cells upon exposure to heat treatments. Int. J. Food Microbiol. 2017, 241, 198–205. [Google Scholar] [CrossRef] [PubMed]
- Morano, K.A.; Grant, C.M.; Moye-Rowley, W.S. The Response to Heat Shock and Oxidative Stress in Saccharomyces cerevisiae. Genetics 2012, 190, 1157–1195. [Google Scholar] [CrossRef] [PubMed]
- Kohanski, M.A.; Dwyer, D.J.; Hayete, B.; Lawrence, C.A.; Collins, J.J. A Common Mechanism of Cellular Death Induced by Bactericidal Antibiotics. Cell 2007, 130, 797–810. [Google Scholar] [CrossRef] [PubMed]
- Dwyer, D.J.; Belenky, P.A.; Yang, J.H.; MacDonald, I.C.; Martell, J.D.; Takahashi, N.; Chan, C.T.; Lobritz, M.A.; Braff, D.; Schwarz, E.G.; et al. Antibiotics induce redox-related physiological alterations as part of their lethality. Proc. Natl. Acad. Sci. USA 2014, 111, E2100–E2109. [Google Scholar] [CrossRef] [PubMed]
- Dukan, S.; Farewell, A.; Ballesteros, M.; Taddei, F.; Radman, M.; Nyström, T. Protein oxidation in response to increased transcriptional or translational errors. Proc. Natl. Acad. Sci. USA 2000, 97, 5746–5749. [Google Scholar] [CrossRef]
- Ding, H.; Hidalgo, E.; Demple, B. The Redox State of the [2Fe-2S] Clusters in SoxR Protein Regulates Its Activity as a Transcription Factor. J. Biol. Chem. 1996, 271, 33173–33175. [Google Scholar] [CrossRef]
- Traoré, D.A.K.; El Ghazouani, A.; Jacquamet, L.; Borel, F.; Ferrer, J.-L.; Lascoux, D.; Ravanat, J.-L.; Jaquinod, M.; Blondin, G.; Caux-Thang, C.; et al. Structural and functional characterization of 2-oxo-histidine in oxidized PerR protein. Nat. Chem. Biol. 2008, 5, 53–59. [Google Scholar] [CrossRef]
- Fuangthong, M.; Herbig, A.F.; Bsat, N.; Helmann, J.D. Regulation of the Bacillus subtilis fur and perR genes by PerR: Not all members of the PerR regulon are peroxide inducible. J. Bacteriol. 2002, 184, 3276–3286. [Google Scholar] [CrossRef]
- Porcheron, G.; Garénaux, A.; Proulx, J.; Sabri, M.; Dozois, C.M. Iron, copper, zinc, and manganese transport and regulation in pathogenic Enterobacteria: Correlations between strains, site of infection and the relative importance of the different metal transport systems for virulence. Front. Cell. Infect. Microbiol. 2013, 3, 90. [Google Scholar] [CrossRef]
- Payne, S.M.; Neilands, I.B. Iron and Virulence in the Family Enterobacteriaceae. CRC Crit. Rev. Microbiol. 1988, 16, 81–111. [Google Scholar] [CrossRef]
- Caza, M.; Kronstad, J.W. Shared and distinct mechanisms of iron acquisition by bacterial and fungal pathogens of humans. Front. Cell. Infect. Microbiol. 2013, 3, 80. [Google Scholar] [CrossRef] [PubMed]
- Zallot, R.; Oberg, N.; Gerlt, J.A. The EFI Web Resource for Genomic Enzymology Tools: Leveraging Protein, Genome, and Metagenome Databases to Discover Novel Enzymes and Metabolic Pathways. Biochemistry 2019, 58, 4169–4182. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Briefings Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Hoang, D.T.; Chernomor, O.; Von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef]
- Leslie Dutton, P. [23] Redox potentiometry: Determination of midpoint potentials of oxidation-reduction components of bio-logical electron-transfer systems. In Methods Enzymol; Academic Press: Cambridge, MA, USA, 1978; Volume 54, pp. 411–435. [Google Scholar]
- Schenk, G.; Boutchard, C.L.; Carrington, L.E.; Noble, C.J.; Moubaraki, B.; Murray, K.S.; de Jersey, J.; Hanson, G.R.; Hamilton, S. A Purple Acid Phosphatase from Sweet Potato Contains an Antiferromagnetically Coupled Binuclear Fe-Mn Center. J. Biol. Chem. 2001, 276, 19084–19088. [Google Scholar] [CrossRef]






| Metal (mol/mol Protein Monomer) | ||||||
|---|---|---|---|---|---|---|
| Fe | Co | Mn | Ni | Zn | ||
| Bh YqeK | LB | 0.58 ± 0.15 | 0.01 ± 0.01 | 0.06 ± 0.01 | 0.04 ± 0.02 | 0.40 ± 0.17 |
| M9-Fe | 1.31 ± 0.32 | 0.01 ± 0.01 | 0.03 ± 0.04 | 0.05 ± 0.05 | 0.10 ± 0.08 | |
| Ca YqeK | LB | 0.49 ± 0.23 | 0.03 ± 0.04 | 0.05 ± 0.02 | 0.05 ± 0.02 | 0.13 ± 0.03 |
| M9-Fe | 1.5 ± 0.06 | 0.01 ± 0.01 | 0.01 ± 0.01 | 0.06 ± 0.04 | 0.07 ± 0.06 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ueda, C.; Chin, N.; Yang, Q.; Pan, L.; Ponniah, R.; Pandelia, M.-E. Diverse YqeK Diadenosine Tetraphosphate Hydrolases Control Biofilm Formation in an Iron-Dependent Manner. Catalysts 2024, 14, 652. https://doi.org/10.3390/catal14090652
Ueda C, Chin N, Yang Q, Pan L, Ponniah R, Pandelia M-E. Diverse YqeK Diadenosine Tetraphosphate Hydrolases Control Biofilm Formation in an Iron-Dependent Manner. Catalysts. 2024; 14(9):652. https://doi.org/10.3390/catal14090652
Chicago/Turabian StyleUeda, Chie, Natalie Chin, Qianyi Yang, Luying Pan, Rheann Ponniah, and Maria-Eirini Pandelia. 2024. "Diverse YqeK Diadenosine Tetraphosphate Hydrolases Control Biofilm Formation in an Iron-Dependent Manner" Catalysts 14, no. 9: 652. https://doi.org/10.3390/catal14090652
APA StyleUeda, C., Chin, N., Yang, Q., Pan, L., Ponniah, R., & Pandelia, M.-E. (2024). Diverse YqeK Diadenosine Tetraphosphate Hydrolases Control Biofilm Formation in an Iron-Dependent Manner. Catalysts, 14(9), 652. https://doi.org/10.3390/catal14090652








