Preparation of Proline-Modified UIO−66 Nanomaterials and Investigation of Their Potential in Lipase Immobilization
Abstract
1. Introduction
2. Results and Discussion
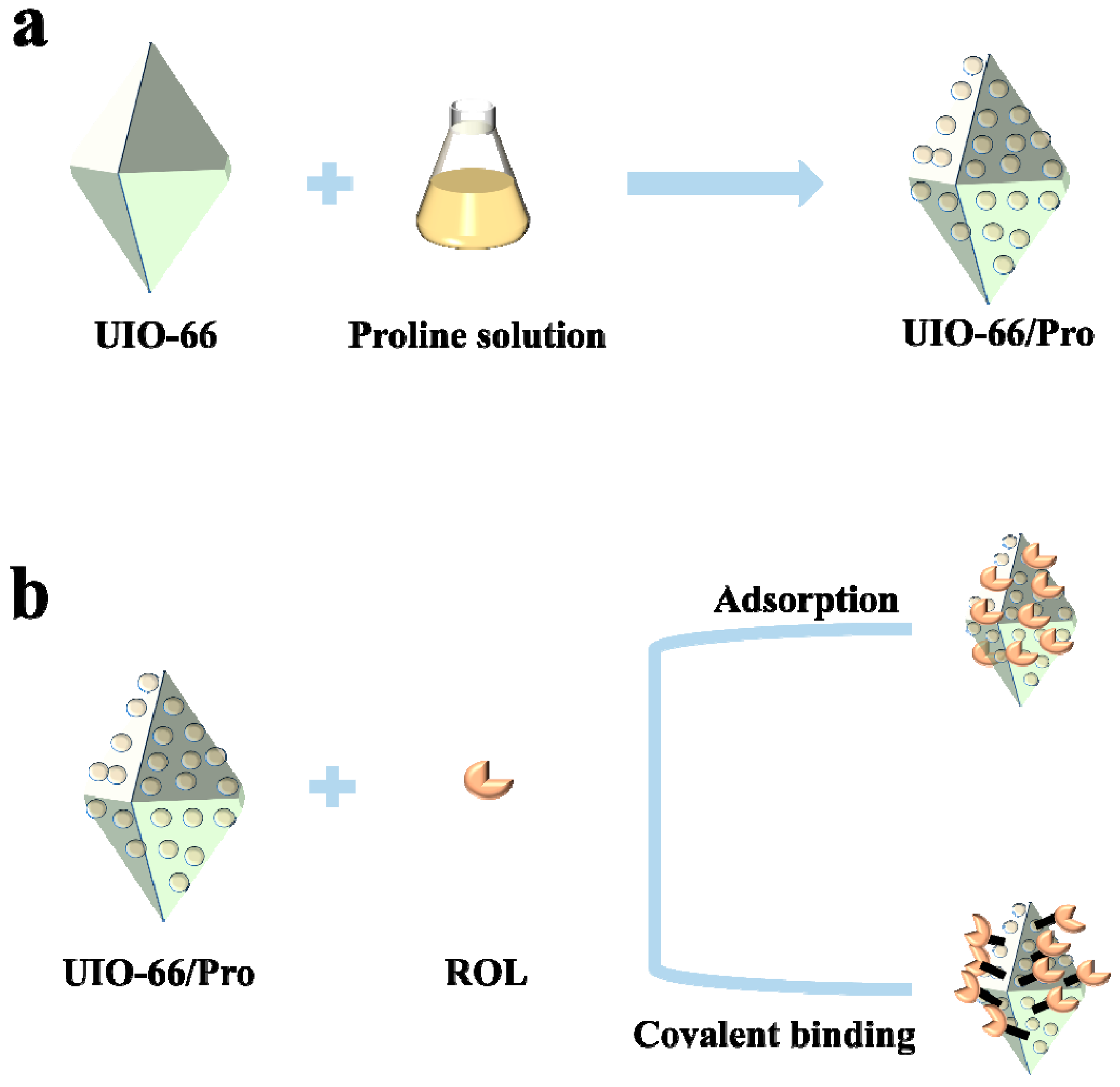
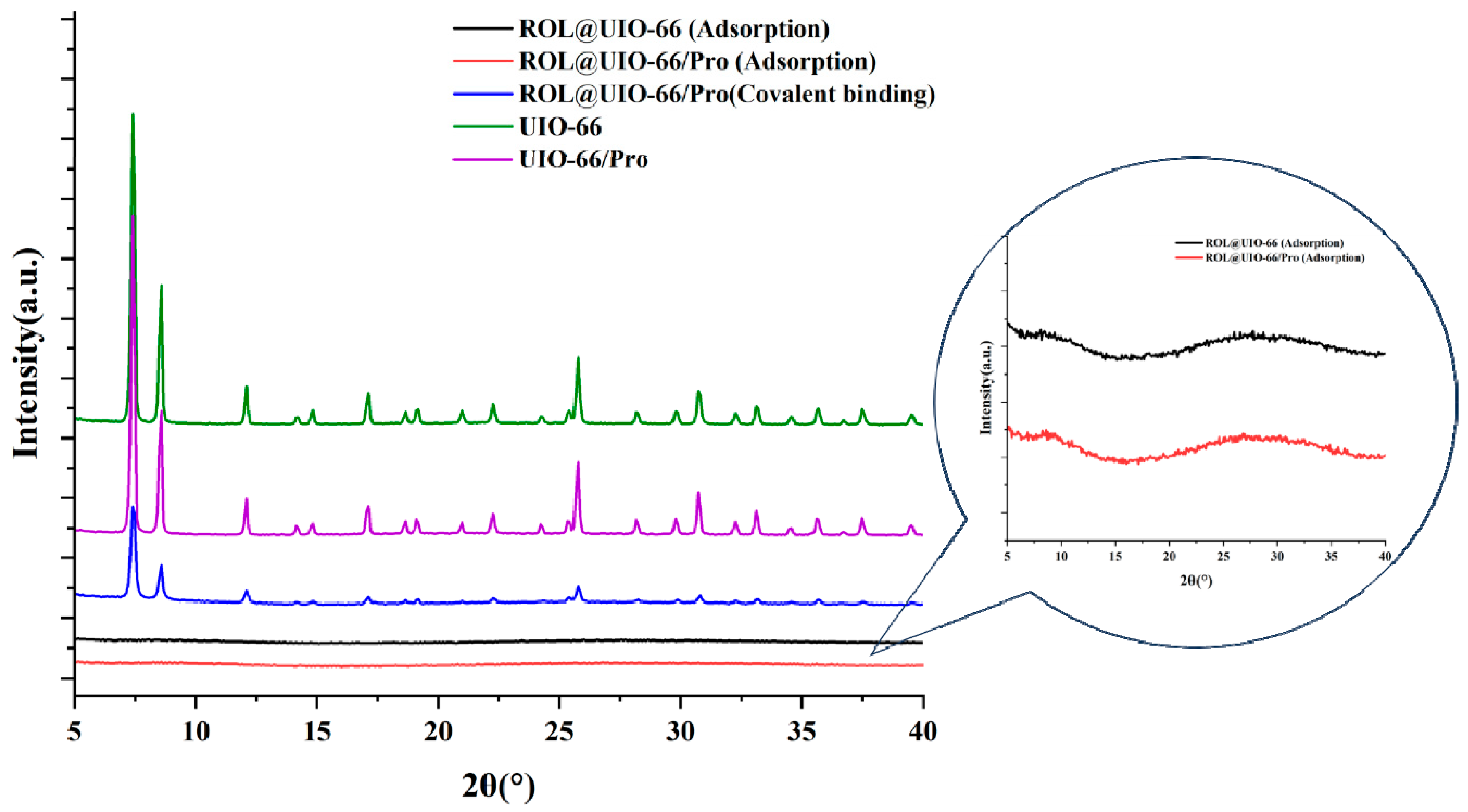
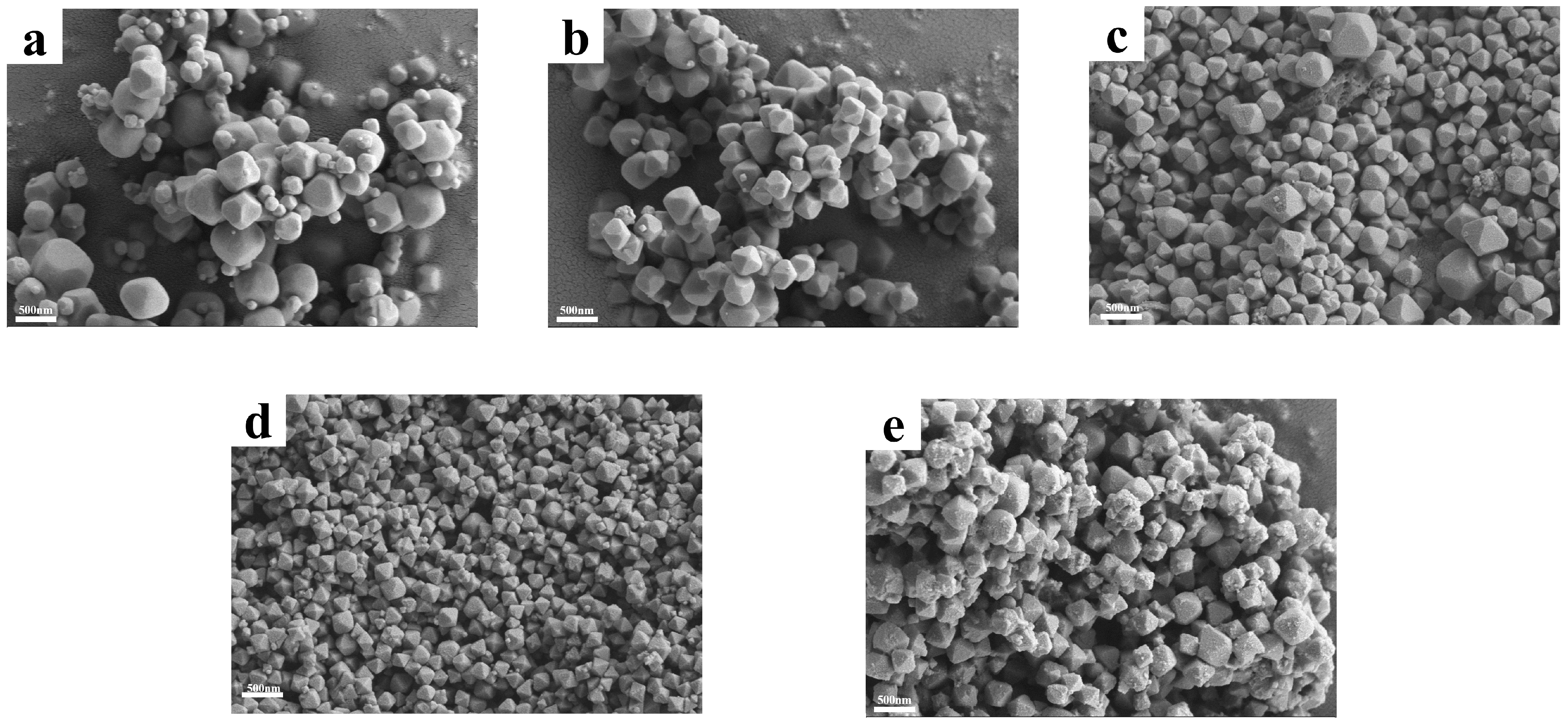
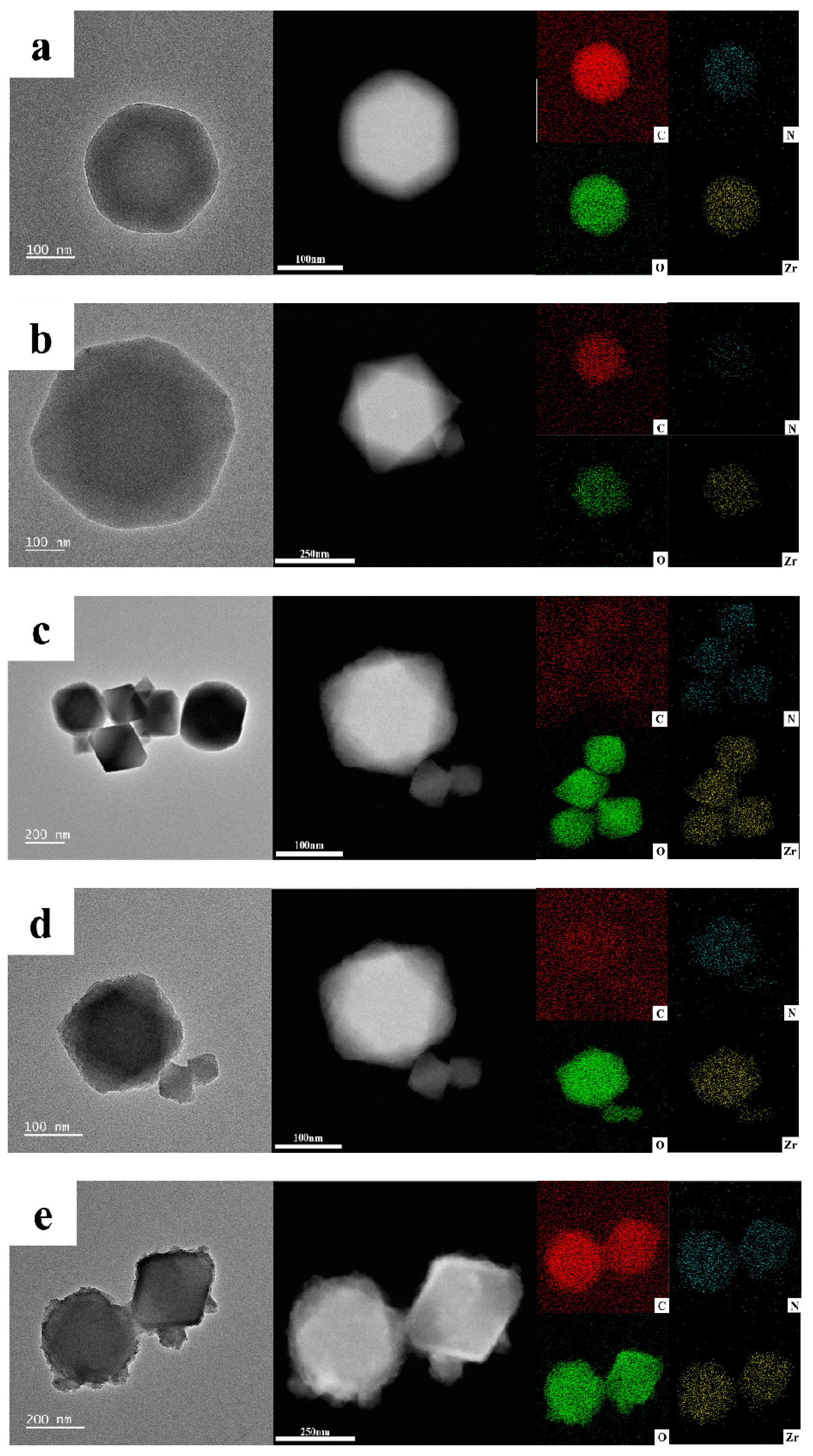
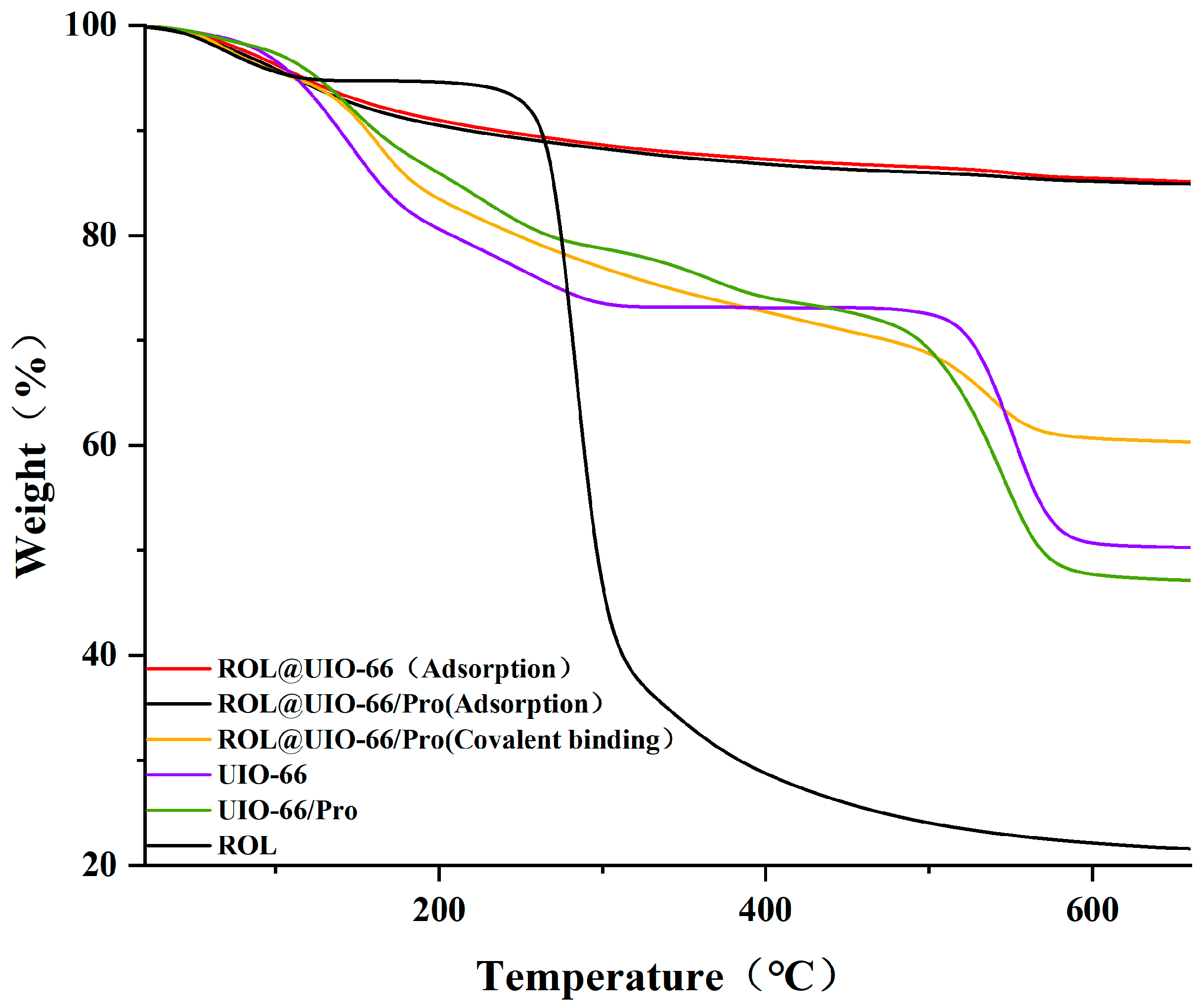
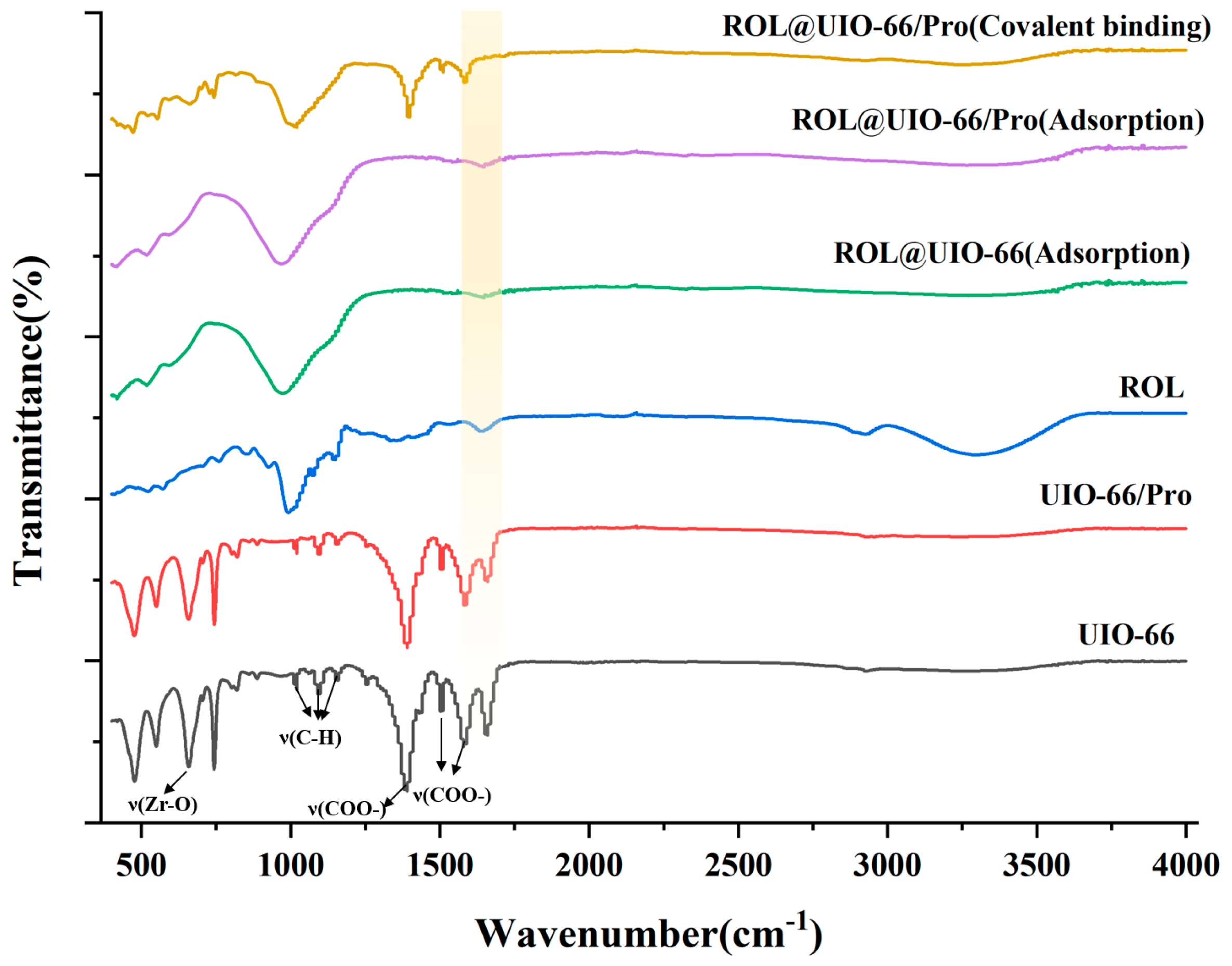
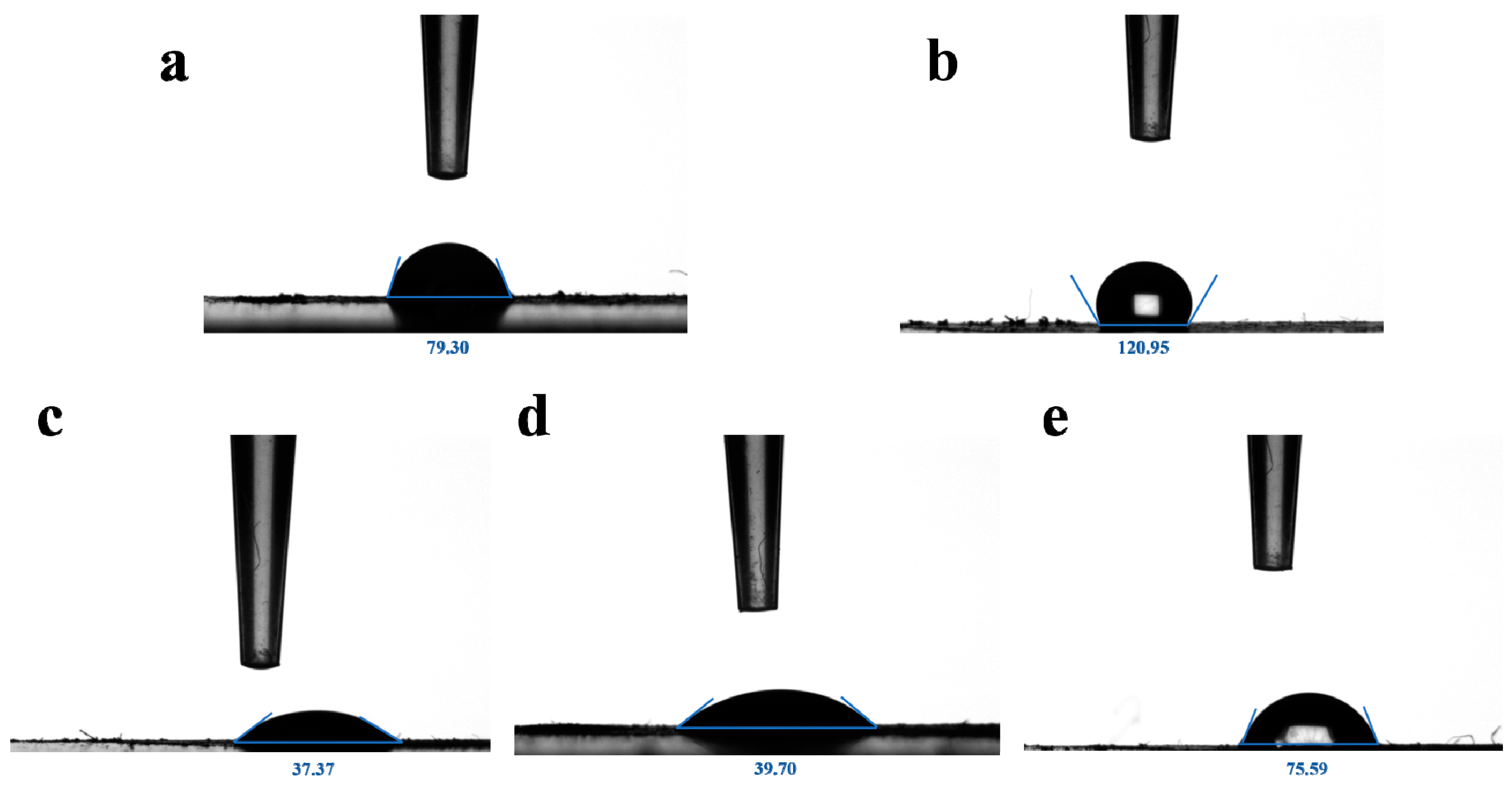
2.1. Synthesis and Characterization of UIO−66, UIO−66/Pro, ROL@ UIO−66, and ROL@ UIO−66/Pro
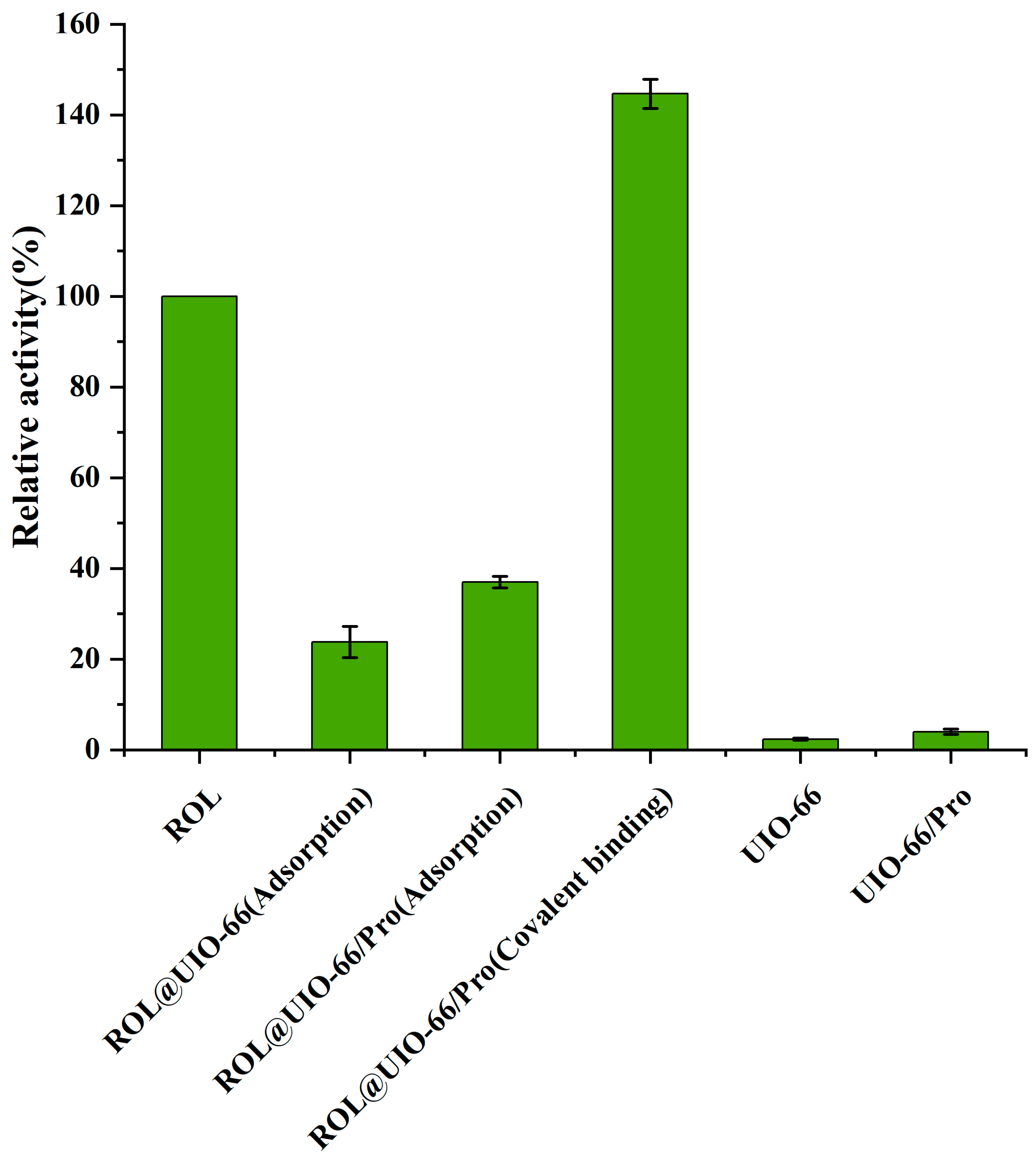
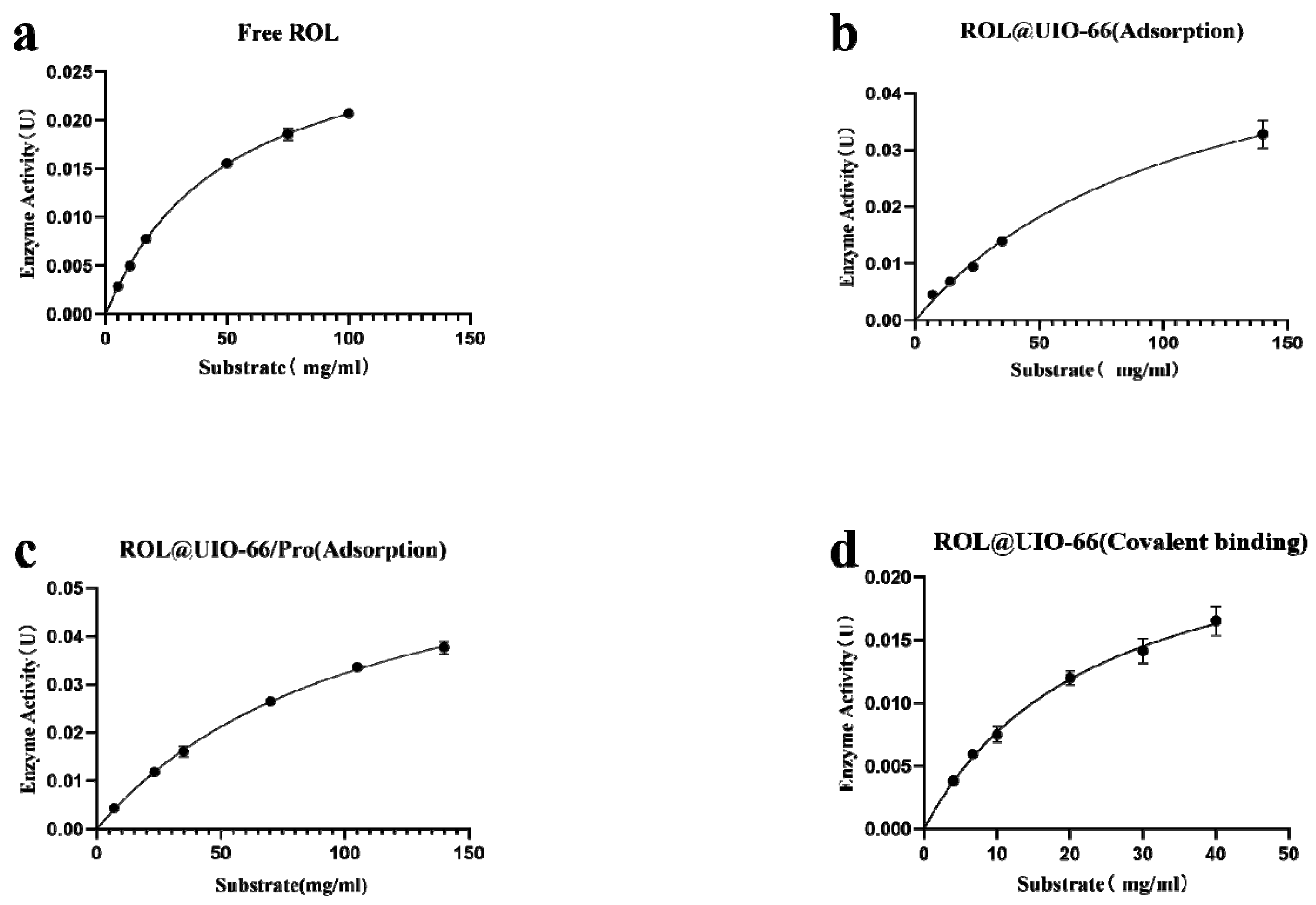
2.2. Catalytic Performance of ROL@UIO−66 and ROL@UIO−66/Pro
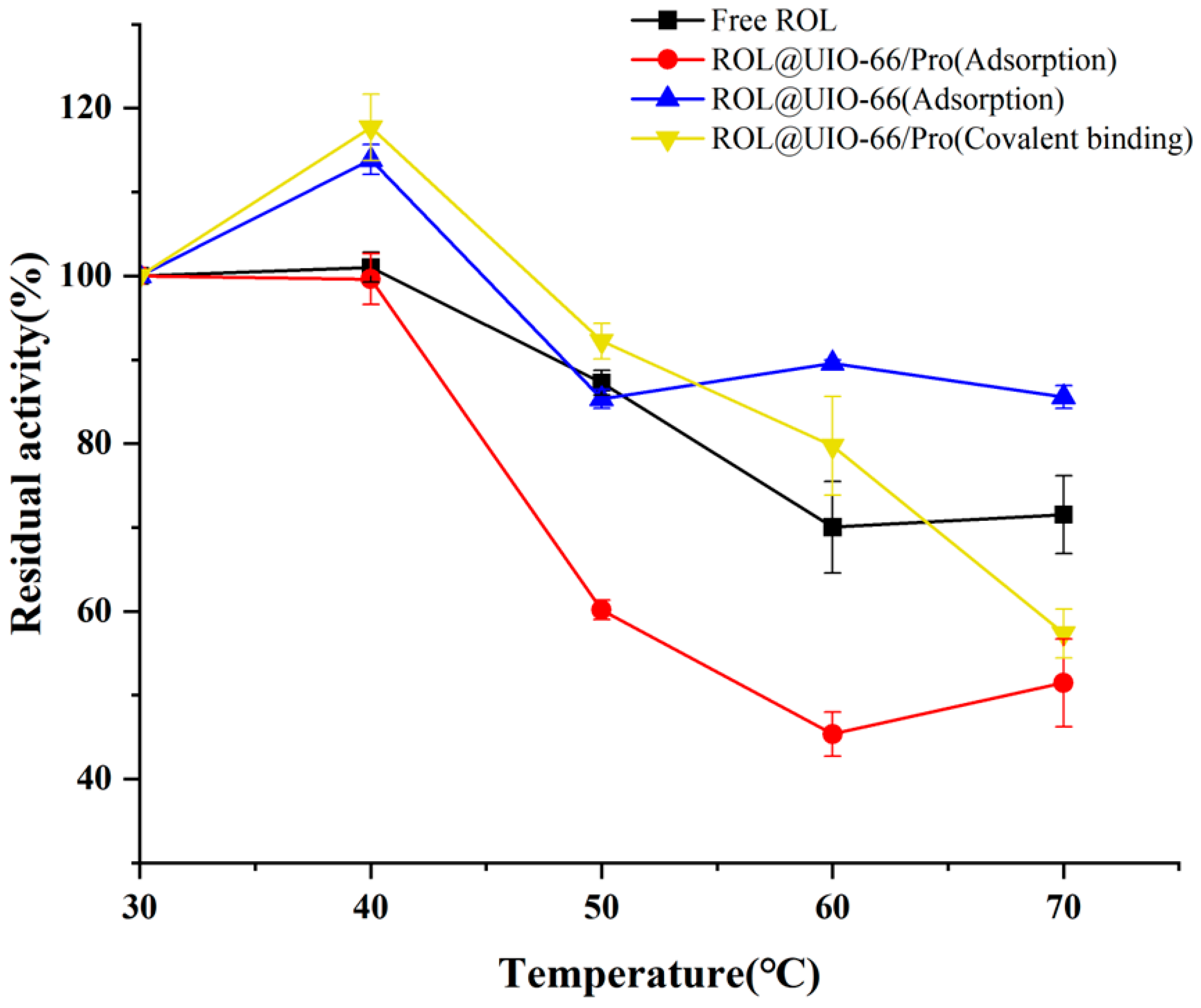
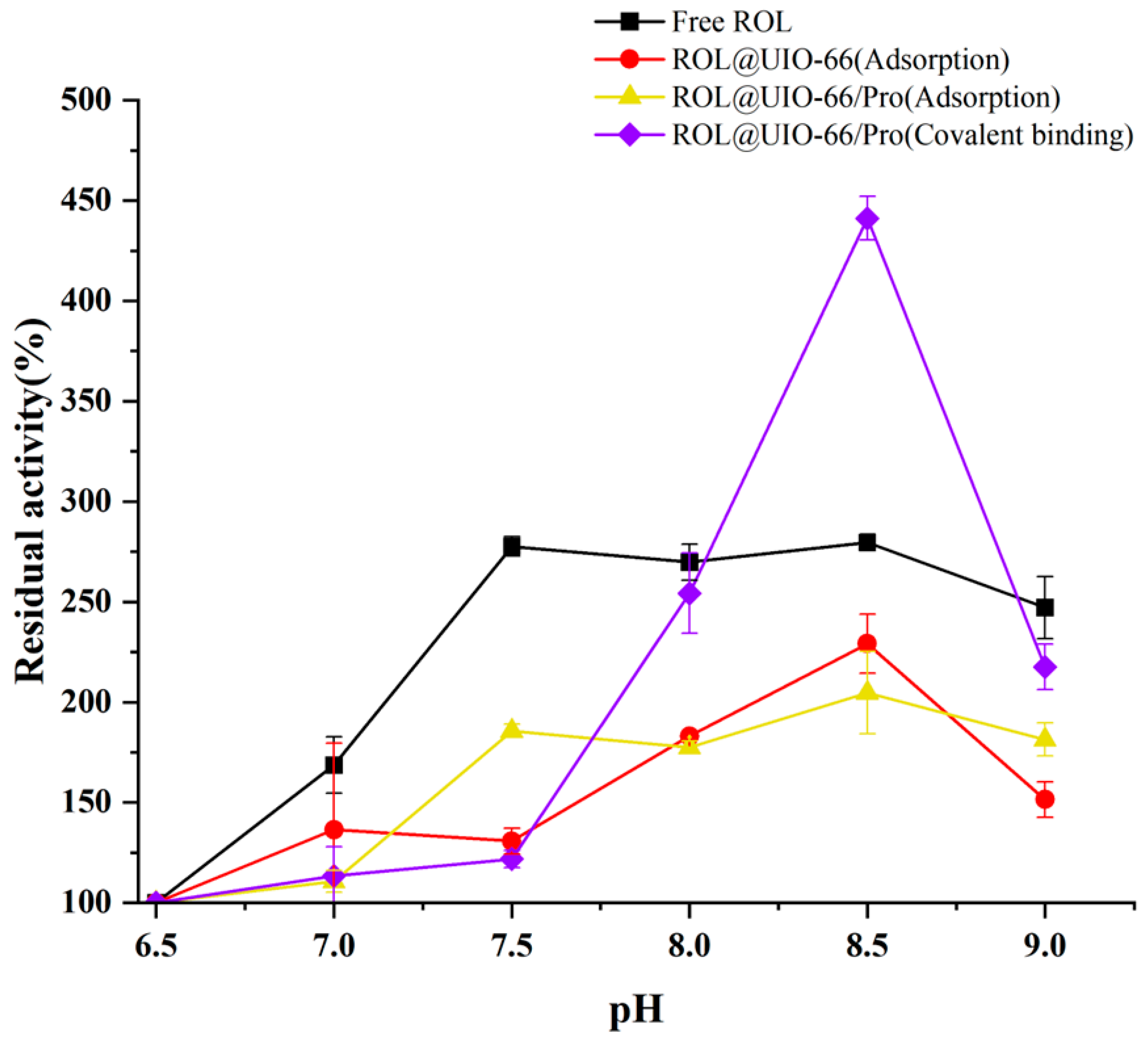
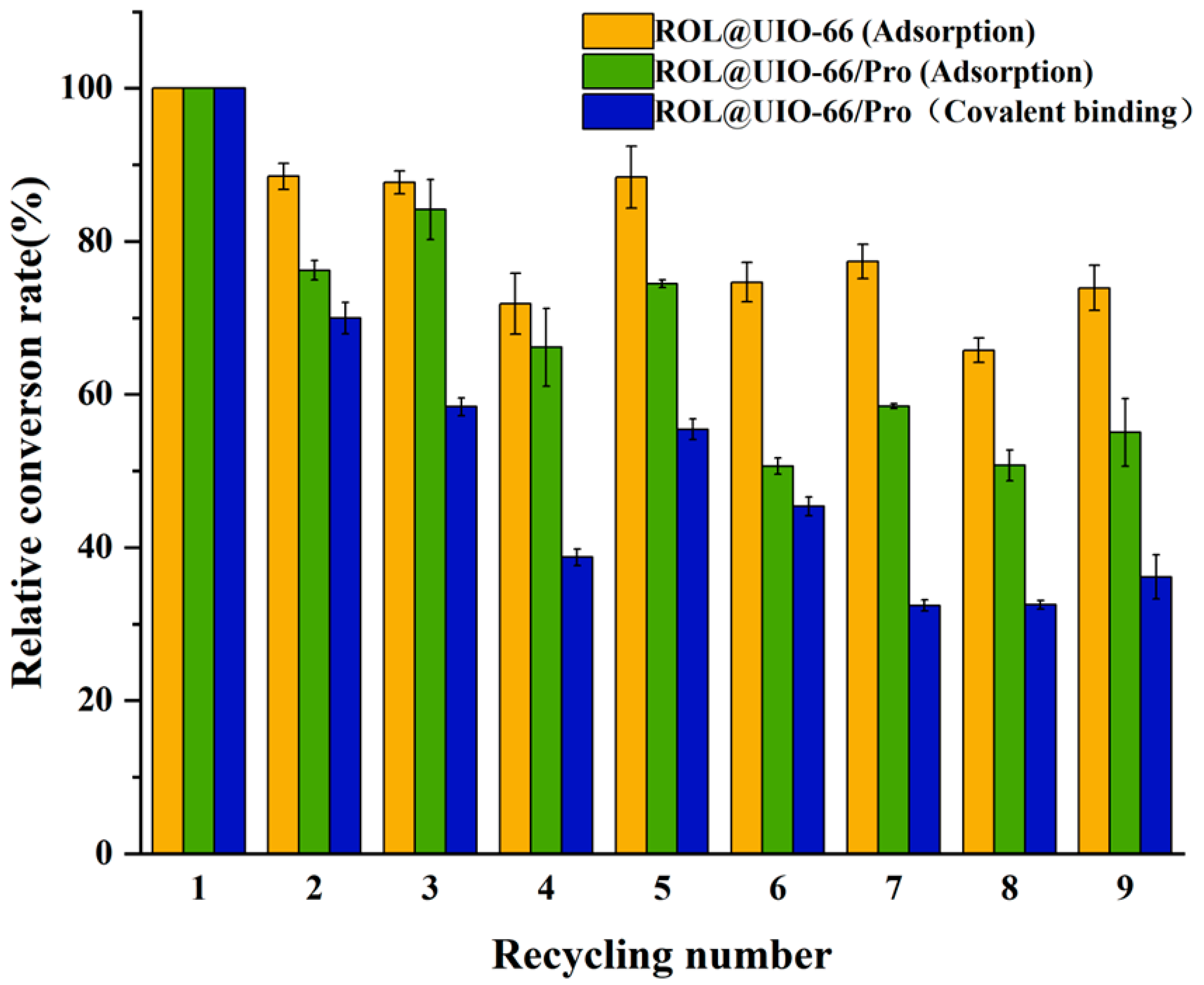
2.3. The stability of ROL@UIO−66 and ROL@UIO−66/Pro
3. Materials and Methods
3.1. Materials
3.2. Synthesis of UIO−66 and UIO−66/Pro
3.3. Immobilization of ROL
3.3.1. Adsorption
3.3.2. Covalent Binding
3.4. Lipasae Activity Assay and Stability
3.5. Kinetics Analysis
3.6. Characterization
3.7. Stability Measurements
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Patil, P.J.; Kamble, S.A.; Dhanavade, M.J.; Liang, X.; Zhang, C.; Li, X. Molecular Modeling Insights into Metal-Organic Frameworks (MOFs) as a Potential Matrix for Immobilization of Lipase: An In Silico Study. Biology 2023, 12, 1051. [Google Scholar] [CrossRef]
- Patil, P.J.; Dong, X.; Usman, M.; Bhambore, N.R.; Shah, H.; Zhang, C.; Li, X. Recent trends in metal-organic frameworks mediated lipase immobilization: A state-of-the-art review. Food Saf. Health 2023, 1, 139–169. [Google Scholar] [CrossRef]
- Li, C.; Zhang, G.; Liu, N.; Liu, L. Preparation and Properties of Rhizopus oryzae Lipase Immobilized Using an Adsorption-Crosslinking Method. Int. J. Food Prop. 2016, 19, 1776–1785. [Google Scholar] [CrossRef]
- Yu, X.W.; Xu, Y.; Xiao, R. Lipases from the genus Rhizopus: Characteristics, expression, protein engineering and application. Prog. Lipid Res. 2016, 64, 57–68. [Google Scholar] [CrossRef]
- Ken Ugo, A.; Vivian Amara, A.; Cn, I.; Kenechuwku, U. Microbial Lipases: A Prospect for Biotechnological Industrial Catalysis for Green Products: A Review. Ferment. Technol. 2017, 6, 2. [Google Scholar] [CrossRef]
- Thomas, S.M.; DiCosimo, R.; Nagarajan, A. Biocatalysis: Applications and potentials for the chemical industry. Trends Biotechnol. 2002, 20, 238–242. [Google Scholar] [CrossRef]
- López, E.N.; Medina, A.R.; Moreno, P.A.G.; Cerdán, L.E.; Grima, E.M. Extraction of microalgal lipids and the influence of polar lipids on biodiesel production by lipase-catalyzed transesterification. Bioresour. Technol. 2016, 216, 904–913. [Google Scholar] [CrossRef]
- Nematian, T.; Salehi, Z.; Shakeri, A. Conversion of bio-oil extracted from Chlorella vulgaris micro algae to biodiesel via modified superparamagnetic nano-biocatalyst. Renew. Energy 2020, 146, 1796–1804. [Google Scholar] [CrossRef]
- Fredrick, E.; Moens, K.; Heyman, B.; Fischer, S.; Van der Meeren, P.; Dewettinck, K. Monoacylglycerols in dairy recombined cream: I. The effect on milk fat crystallization. Food Res. Int. 2013, 51, 892–898. [Google Scholar] [CrossRef]
- Itabaiana, I.; Gonçalves, K.M.; Cordeiro, Y.M.L.; Zoumpanioti, M.; Leal, I.C.R.; Miranda, L.S.M.; de Souza, R.; Xenakis, A. Kinetics and mechanism of lipase catalyzed monoacylglycerols synthesis. J. Mol. Catal. B Enzym. 2013, 96, 34–39. [Google Scholar] [CrossRef]
- Feltes, M.M.C.; de Oliveira, D.; Block, J.M.; Ninow, J.L. The Production, Benefits, and Applications of Monoacylglycerols and Diacylglycerols of Nutritional Interest. Food Bioprocess Technol. 2013, 6, 17–35. [Google Scholar] [CrossRef]
- López-Fernández, J.; Benaiges, M.D.; Valero, F. Rhizopus oryzae Lipase, a Promising Industrial Enzyme: Biochemical Characteristics, Production and Biocatalytic Applications. Catalysts 2020, 10, 1277. [Google Scholar] [CrossRef]
- Marzieh, A.; Masoud, B.; Amir, R.; Abdol-Khalegh, B. Covalent immobilization of Candida rugosa lipase on a novel functionalized Fe3O4@SiO2 dip-coated nanocomposite membrane. Food Bioprod. Process. 2016, 100, 351–360. [Google Scholar]
- An, N.; Zhou, C.H.; Zhuang, X.Y.; Tong, D.S.; Yu, W.H. Immobilization of enzymes on clay minerals for biocatalysts and biosensors. Appl. Clay Sci. 2015, 114, 283–296. [Google Scholar] [CrossRef]
- Barbosa, O.; Torres, R.; Ortiz, C.; Berenguer-Murcia, A.; Rodrigues, R.C.; Fernandez-Lafuente, R. Heterofunctional Supports in Enzyme Immobilization: From Traditional Immobilization Protocols to Opportunities in Tuning Enzyme Properties. Biomacromolecules 2013, 14, 2433–2462. [Google Scholar] [CrossRef] [PubMed]
- Mendes, A.A.; Soares, C.M.F.; Tardioli, P.W. Recent advances and future prospects for biolubricant base stocks production using lipases as environmentally friendly catalysts: A mini-review. World J. Microbiol. Biotechnol. 2022, 39, 25. [Google Scholar] [CrossRef]
- Sun, X.D.; He, P.; Gao, Z.; Liao, Y.X.; Weng, S.X.; Zhao, Z.X.; Song, H.; Zhao, Z.X. Multi-crystalline N-doped Cu/CuxO/C foam catalyst derived from alkaline N-coordinated HKUST-1/CMC for enhanced 4-nitrophenol reduction. J. Colloid Interface Sci. 2019, 553, 1–13. [Google Scholar] [CrossRef]
- Cui, X.F.; Sun, X.D.; Liu, L.; Huang, Q.H.; Yang, H.C.; Chen, C.H.; Nie, S.X.; Zhao, Z.X.; Zhao, Z.X. In-situ fabrication of cellulose foam HKUST-1 and surface modification with polysaccharides for enhanced selective adsorption of toluene and acidic dipeptides. Chem. Eng. J. 2019, 369, 898–907. [Google Scholar] [CrossRef]
- Yong, J.; Hakobyan, K.; Xu, J.T.; Mellick, A.S.; Whitelock, J.; Liang, K. Comparison of protein quantification methods for protein encapsulation with ZIF-8 metal-organic frameworks. Biotechnol. J. 2023, 18, 2300015. [Google Scholar] [CrossRef]
- Babatunde, O.; Al-Zuhair, S. MOFs as potential matrices in cyclodextrin glycosyltransferase immobilization. Molecules 2021, 26, 680. [Google Scholar]
- Morris, W.; Volosskiy, B.; Demir, S.; Gándara, F.; McGrier, P.L.; Furukawa, H.; Cascio, D.; Stoddart, J.F.; Yaghi, O.M. Synthesis, Structure, and Metalation of Two New Highly Porous Zirconium Metal-Organic Frameworks. Inorg. Chem. 2012, 51, 6443–6445. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.L.; Demir, N.K.; Wu, Z.T.; Li, K. Highly Water-Stable Zirconium Metal Organic Framework UIO−66 Membranes Supported on Alumina Hollow Fibers for Desalination. J. Am. Chem. Soc. 2015, 137, 6999–7002. [Google Scholar] [CrossRef]
- Hu, P.; Liang, X.P.; Yaseen, M.; Sun, X.D.; Tong, Z.F.; Zhao, Z.X.; Zhao, Z.X. Preparation of highly-hydrophobic novel N-coordinated UIO−66(Zr) with dopamine via fast mechano-chemical method for (CHO-/Cl-)-VOCs competitive adsorption in humid environment. Chem. Eng. J. 2018, 332, 608–618. [Google Scholar] [CrossRef]
- Silva, C.G.; Luz, I.; Xamena, F.; Corma, A.; García, H. Water Stable Zr-Benzenedicarboxylate Metal-Organic Frameworks as Photocatalysts for Hydrogen Generation. Chem. Eur. J. 2010, 16, 11133–11138. [Google Scholar] [CrossRef]
- Kadhom, M.; Al-Furaiji, M.; Salih, S.; Al-Obaidi, M.A.; Abdullah, G.H.; Albayati, N. A review on UIO−66 applications in membrane-based water treatment processes. J. Water Process Eng. 2023, 51, 103402. [Google Scholar] [CrossRef]
- Chen, J.; Sun, B.Z.; Sun, C.R.; Zhang, P.L.; Xu, W.F.; Liu, Y.; Xiong, B.Q.; Tang, K.W. Immobilization of lipase AYS on UIO−66-NH2 metal-organic framework nanoparticles as a recyclable biocatalyst for ester hydrolysis and kinetic resolution. Sep. Purif. Technol. 2020, 251, 117398. [Google Scholar] [CrossRef]
- Hu, Y.L.; Dai, L.M.; Liu, D.H.; Du, W. Rationally designing hydrophobic UIO−66 support for the enhanced enzymatic performance of immobilized lipase. Green Chem. 2018, 20, 4500–4506. [Google Scholar] [CrossRef]
- Zhu, F.F.; Xu, W.; Li, X.; Meng, H.; Han, Y.D.; Wu, J.B.; Xu, J.L.; Xu, Y.; Zhang, X. Lipase immobilization on UIO−66/poly(vinylidene fluoride) hybrid membranes and active catalysis in the vegetable oil hydrolysis. N. J. Chem. 2020, 44, 14379–14388. [Google Scholar] [CrossRef]
- Feng, Y.M.; Xu, Y.; Liu, S.C.; Wu, D.; Su, Z.Q.; Chen, G.; Liu, J.H.; Li, G.L. Recent advances in enzyme immobilization based on novel porous framework materials and its applications in biosensing. Coord. Chem. Rev. 2022, 459, 214414. [Google Scholar] [CrossRef]
- Bolivar, J.M.; Woodley, J.M.; Fernandez-Lafuente, R. Is enzyme immobilization a mature discipline? Some critical considerations to capitalize on the benefits of immobilization. Chem. Soc. Rev. 2022, 51, 6251–6290. [Google Scholar] [CrossRef]
- Guo, X.Y.; Liu, D.H.; Han, T.T.; Huang, H.L.; Yang, Q.Y.; Zhong, C.L. Preparation of Thin Film Nanocomposite Membranes with Surface Modified MOF for High Flux Organic Solvent Nanofiltration. AIChE J. 2017, 63, 1303–1312. [Google Scholar] [CrossRef]
- Zhou, Z.M.; Wu, C.; Zhang, B.Q. ZIF-67 Membranes Synthesized on α-Al2O3-Plate-Supported Cobalt Nanosheets with Amine Modification for Enhanced H2/CO2 Permselectivity. Ind. Eng. Chem. Res. 2020, 59, 3182–3188. [Google Scholar] [CrossRef]
- Rani, R.; Deep, A.; Mizaikoff, B.; Singh, S. Enhanced hydrothermal stability of Cu MOF by post synthetic modification with amino acids. Vacuum 2019, 164, 449–457. [Google Scholar] [CrossRef]
- Cheng, Y.F.; Lai, O.M.; Tan, C.P.; Panpipat, W.; Cheong, L.Z.; Shen, C. Proline-Modified UIO−66 as Nanocarriers to Enhance Candida rugosa Lipase Catalytic Activity and Stability for Electrochemical Detection of Nitrofen. ACS Appl. Mater. Interfaces 2021, 13, 4146–4155. [Google Scholar] [CrossRef] [PubMed]
- Zhan, X.Q.; Tsai, F.C.; Xie, L.; Zhang, K.D.; Liu, H.L.; Ma, N.; Shi, D.; Jiang, T. Ligands-Coordinated Zr-Based MOF for Wastewater Treatment. Nanomaterials 2018, 8, 655. [Google Scholar] [CrossRef]
- Feng, Y.X.; Du, Y.J.; Kuang, G.L.; Zhong, L.; Hu, H.T.; Jia, S.R.; Cui, J.D. Hierarchical micro- and mesoporous ZIF-8 with core-shell superstructures using colloidal metal sulfates as soft templates for enzyme immobilization. J. Colloid Interface Sci. 2022, 610, 709–718. [Google Scholar] [CrossRef]
- Tatay, S.; Martínez-Giménez, S.; Rubio-Gaspar, A.; Gómez-Oliveira, E.; Castells-Gil, J.; Dong, Z.; Mayoral, Á.; Almora-Barrios, N.M.; Padial, N.; Martí-Gastaldo, C. Synthetic control of correlated disorder in UIO−66 frameworks. Nat. Commun. 2023, 14, 6962. [Google Scholar] [CrossRef]
- Sumadiyasa, M.; Manuaba, I.B.S. Determining crystallite size using scherrer formula, williamson-hull plot, and particle size with SEM. Bul. Fis. 2018, 19, 28–35. [Google Scholar] [CrossRef]
- Du, X.D.; Yi, X.H.; Wang, P.; Zheng, W.W.; Deng, J.G.; Wang, C.C. Robust photocatalytic reduction of Cr(VI) on UIO−66-NH2(Zr/Hf) metal-organic framework membrane under sunlight irradiation. Chem. Eng. J. 2019, 356, 393–399. [Google Scholar] [CrossRef]
- Jin, L.J.; Liu, H.; Xu, A.H.; Wu, Y.H.; Lu, J.K.; Liu, J.X.; Xie, S.Z.; Yao, Y.C.; Dong, L.H.; Zhang, M.; et al. Defective UIO−66-NH2 (Zr/Ce) catalyzes the synthesis of propylene carbonate under mild conditions. Microporous Mesoporous Mater. 2021, 317, 110997. [Google Scholar] [CrossRef]
- Mohammadi, N.S.; Khiabani, M.S.; Ghanbarzadeh, B.; Mokarram, R.R. Improvement of lipase biochemical properties via a two-step immobilization method: Adsorption onto silicon dioxide nanoparticles and entrapment in a polyvinyl alcohol/alginate hydrogel. J. Biotechnol. 2020, 323, 189–202. [Google Scholar] [CrossRef]
- Wu, X.L.; Yue, H.; Zhang, Y.Y.; Gao, X.Y.; Li, X.Y.; Wang, L.C.; Cao, Y.F.; Hou, M.; An, H.X.; Zhang, L.; et al. Packaging and delivering enzymes by amorphous metal-organic frameworks. Nat. Commun. 2019, 10, 5165. [Google Scholar] [CrossRef]
- Park, J.; Lee, M.; Feng, D.W.; Huang, Z.H.; Hinckley, A.C.; Yakoyenko, A.; Zou, X.D.; Cui, Y.; Bao, Z.A. Stabilization of Hexaaminobenzene in a 2D Conductive Metal-Organic Framework for High Power Sodium Storage. J. Am. Chem. Soc. 2018, 140, 10315–10323. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.C.; Luo, Y.W.; Zhang, Z.J.; Ding, W.W.; Liu, D.X.; Wang, J.C.; Guo, L.; Wen, M. Effects of functional groups of -NH2 and -NO2 on water adsorption ability of Zr-based MOFs (UIO−66). Chem. Phys. 2021, 543, 111093. [Google Scholar] [CrossRef]
- Cao, S.L.; Yue, D.M.; Li, X.H.; Smith, T.J.; Li, N.; Zong, M.H.; Wu, H.; Ma, Y.Z.; Lou, W.Y. Novel Nano-/Micro-Biocatalyst: Soybean Epoxide Hydrolase Immobilized on UIO−66-NH2 MOF for Efficient Biosynthesis of Enantiopure (R)-1, 2-Octanediol in Deep Eutectic Solvents. ACS Sustain. Chem. Eng. 2016, 4, 3586–3595. [Google Scholar] [CrossRef]
- Nezhad, M.K.; Aghaei, H. Tosylated cloisite as a new heterofunctional carrier for covalent immobilization of lipase and its utilization for production of biodiesel from waste frying oil. Renew. Energy 2021, 164, 876–888. [Google Scholar] [CrossRef]
- Zhu, C.Y.; Li, F.L.; Zhang, Y.W.; Gupta, R.K.; Patel, S.K.S.; Lee, J.K. Recent Strategies for the Immobilization of Therapeutic Enzymes. Polymers 2022, 14, 488. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Chua, Y.S.; Krungleviciute, V.; Tyagi, M.; Chen, P.; Yildirim, T.; Zhou, W. Unusual and Highly Tunable Missing-Linker Defects in Zirconium Metal-Organic Framework UIO−66 and Their Important Effects on Gas Adsorption. J. Am. Chem. Soc. 2013, 135, 10525–10532. [Google Scholar] [CrossRef] [PubMed]
- Ly, H.G.T.; Fu, G.X.; de Azambuja, F.; De Vos, D.; Parac-Vogt, T.N. Nanozymatic Activity of UIO−66 Metal-Organic Frameworks: Tuning the Nanopore Environment Enhances Hydrolytic Activity toward Peptide Bonds. ACS Appl. Nano Mater. 2020, 3, 8931–8938. [Google Scholar] [CrossRef]
- Kermasha, S.; Gill, J.K. Immobilization of enzymes and their use in biotechnological applications. In Enzymes; Elsevier: Amsterdam, The Netherlands, 2021; pp. 133–170. [Google Scholar]
- Northrop, D.B. On the meaning of Km and V/K in enzyme kinetics. J. Chem. Educ. 1998, 75, 1153. [Google Scholar] [CrossRef]
- Sun, C.; Wu, S.; Wu, Y.; Sun, B.; Zhang, P.; Tang, K. Lipase AK from Pseudomonas fluorescens immobilized on metal organic frameworks for efficient biosynthesis of enantiopure (S)− 1-(4-bromophenyl) ethanol. Process Biochem. 2023, 124, 132–139. [Google Scholar] [CrossRef]
- Banta, S.; Wheeldon, I. Theory-based development of performance metrics for comparing multireactant enzymes. ACS Catal. 2019, 10, 1123–1132. [Google Scholar] [CrossRef]
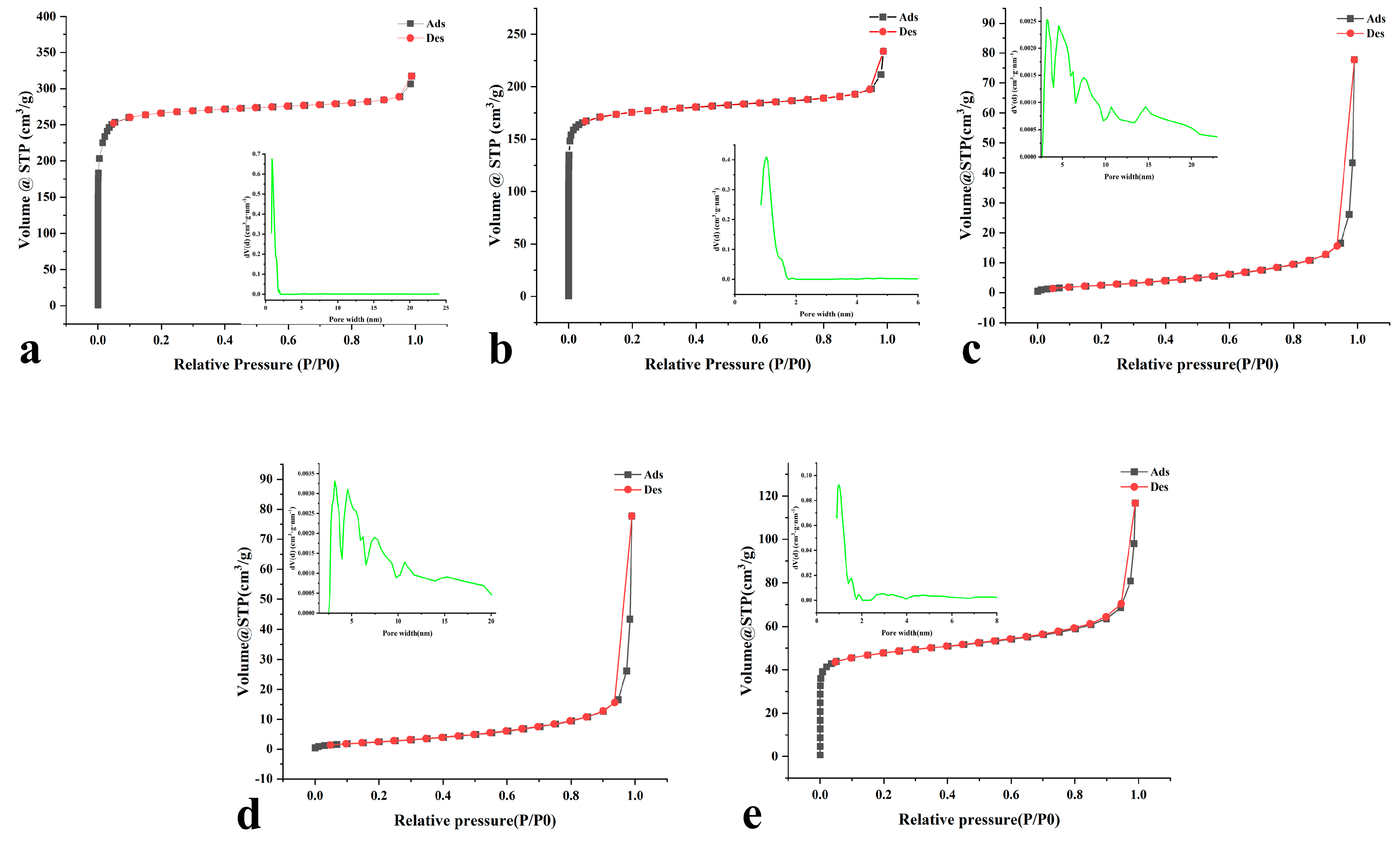
| Biocatalysts | Surface Area/(m2·g−1) | Pore Volume/(cc/g) | Pore Width (nm) | |||
|---|---|---|---|---|---|---|
| BET | BJH | DFT | BJH | HK | DFT | |
| UIO−66 | 817.944 | 0.081 | 0.399 | 3.396 | 0.593 | 0.899 |
| UIO−66/Pro | 513.717 | 0.092 | 0.274 | 3.396 | 0.603 | 1.029 |
| ROL@UIO−66(Adsorption) | 5.313 | 0.073 | 0.020 | 3.388 | 0.368 | 3.169 |
| ROL@UIO−66/Pro (Adsorption) | 6.762 | 0.121 | 0.023 | 3.388 | 0.368 | 3.169 |
| ROL@UIO−66/Pro (Covalent binding) | 160.202 | 0.111 | 0.099 | 3.799 | 0.608 | 0.984 |
| Biocatalysts | Km (mM) | Vmax (mM/min) | Vmax/Km | kcat (1/s) | kcat/Km (L·mg−1·s−1) |
|---|---|---|---|---|---|
| Free ROL | 50.653 ± 2.463 | 0.0312 ± 0.0007 | 0.000616 ± 0.00002 | 0.303 ± 0.00685 | 0.00598 ± 0.000226 |
| ROL@UIO−66(Adsorption) | 131.193 ± 36.188 | 0.05966 ± 0.0109 | 0.000463 ± 0.00005 | 0.0634 ± 0.0116 | 0.000493 ± 0.0000504 |
| ROL@UIO−66/Pro (Adsorption) | 121.367 ± 16.511 | 0.0713 ± 0.00540 | 0.000591 ± 0.00004 | 0.138 ± 0.0104 | 0.00114 ± 0.0000751 |
| ROL@UIO−66/Pro (Covalent binding) | 24.033 ± 5.758 | 0.0265 ± 0.00363 | 0.00112 ± 0.000121 | 0.0835 ± 0.0114 | 0.00354 ± 0.000380 |
| ROL@UIO−66 (Adsorption) | ROL@UIO−66/Pro (Adsorption) | ROL@UIO−66/Pro (Covalent Binding) | |
|---|---|---|---|
| Enzyme activity(U) | 0.00270 | 0.00275 | 0.00431 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, X.; Zhang, C.; Patil, P.J.; Li, W.; Li, X. Preparation of Proline-Modified UIO−66 Nanomaterials and Investigation of Their Potential in Lipase Immobilization. Catalysts 2024, 14, 180. https://doi.org/10.3390/catal14030180
Dong X, Zhang C, Patil PJ, Li W, Li X. Preparation of Proline-Modified UIO−66 Nanomaterials and Investigation of Their Potential in Lipase Immobilization. Catalysts. 2024; 14(3):180. https://doi.org/10.3390/catal14030180
Chicago/Turabian StyleDong, Xiaoxiao, Chengnan Zhang, Prasanna J. Patil, Weiwei Li, and Xiuting Li. 2024. "Preparation of Proline-Modified UIO−66 Nanomaterials and Investigation of Their Potential in Lipase Immobilization" Catalysts 14, no. 3: 180. https://doi.org/10.3390/catal14030180
APA StyleDong, X., Zhang, C., Patil, P. J., Li, W., & Li, X. (2024). Preparation of Proline-Modified UIO−66 Nanomaterials and Investigation of Their Potential in Lipase Immobilization. Catalysts, 14(3), 180. https://doi.org/10.3390/catal14030180







