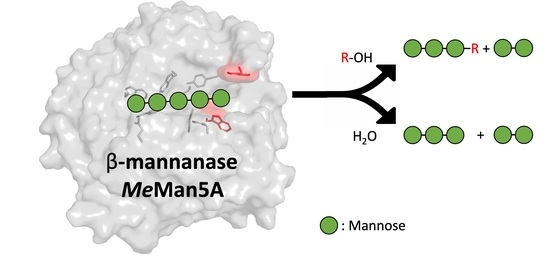
Flexibility and Function of Distal Substrate-Binding Tryptophans in the Blue Mussel β-Mannanase MeMan5A and Their Role in Hydrolysis and Transglycosylation
Abstract
:1. Introduction
2. Results
2.1. Basic Enzyme Characterization
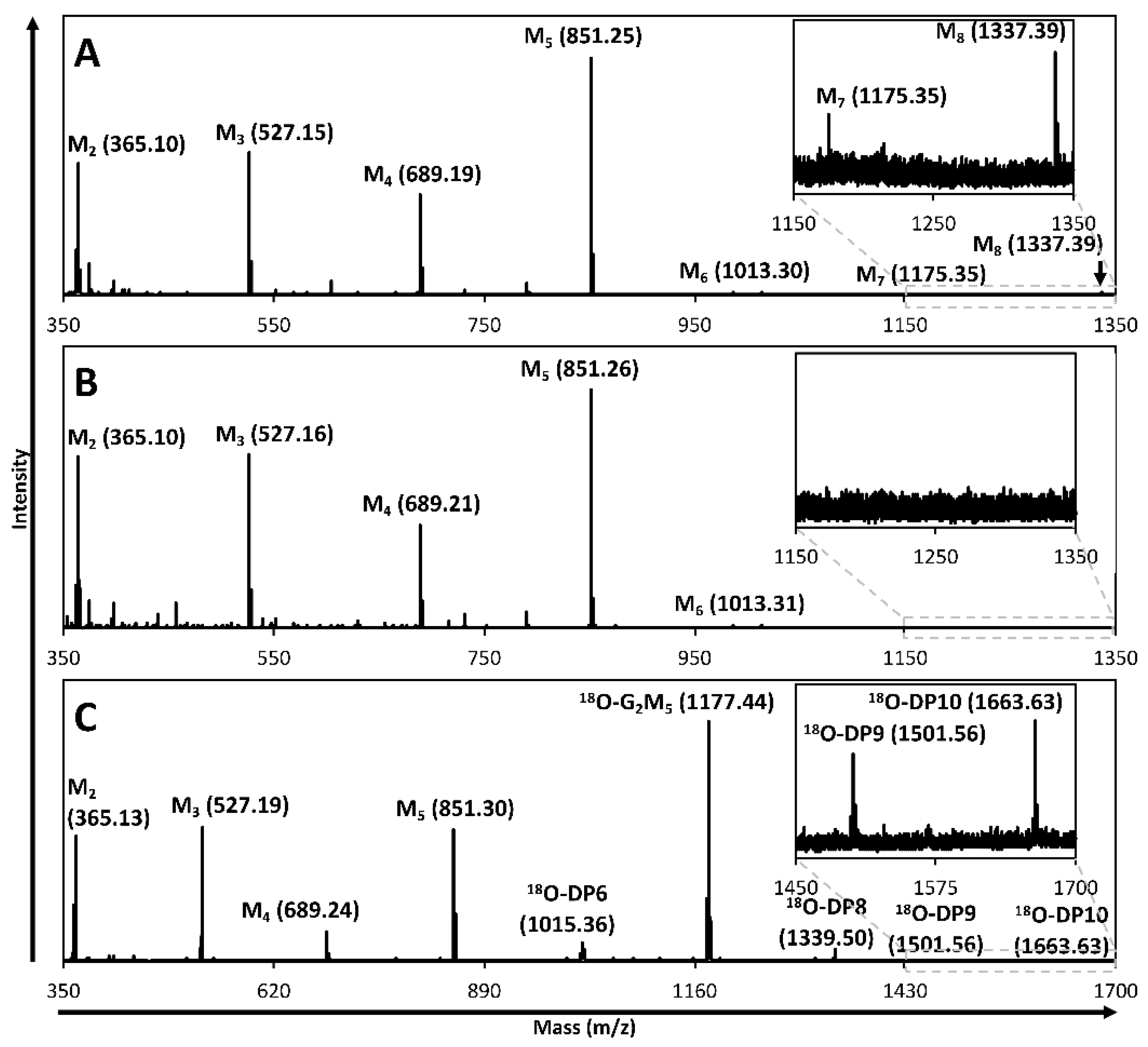
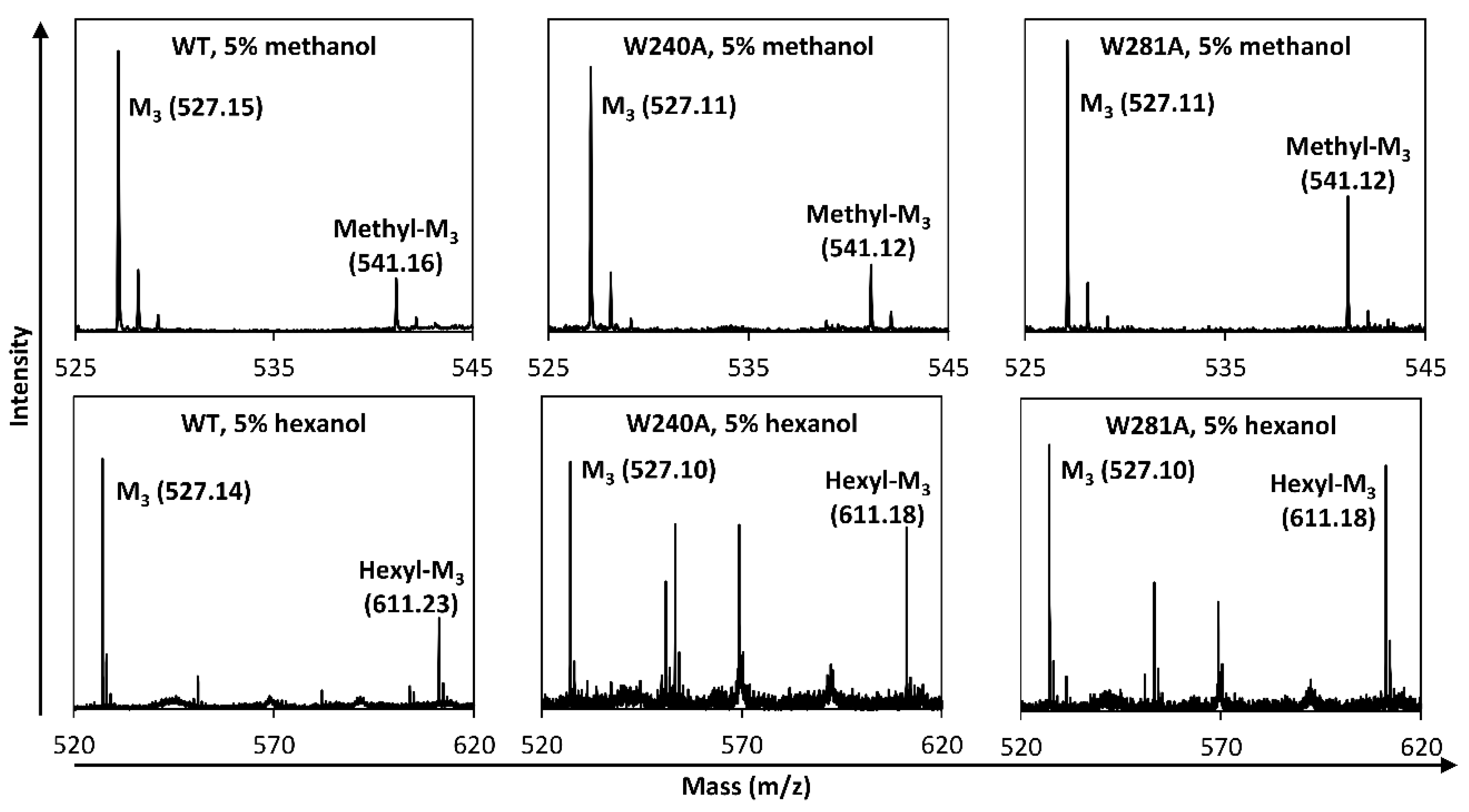
2.2. Hydrolysis and Transglycosylation Product Formation
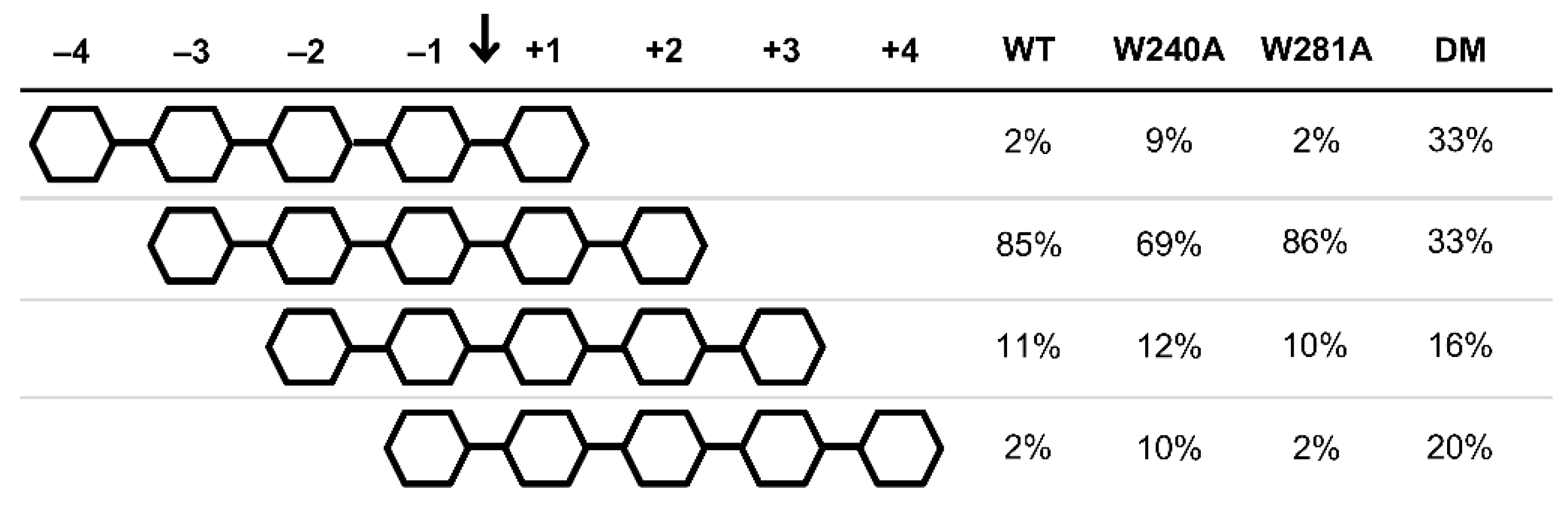
2.3. Productive Binding Mode Preferences
2.4. Enzyme Kinetics
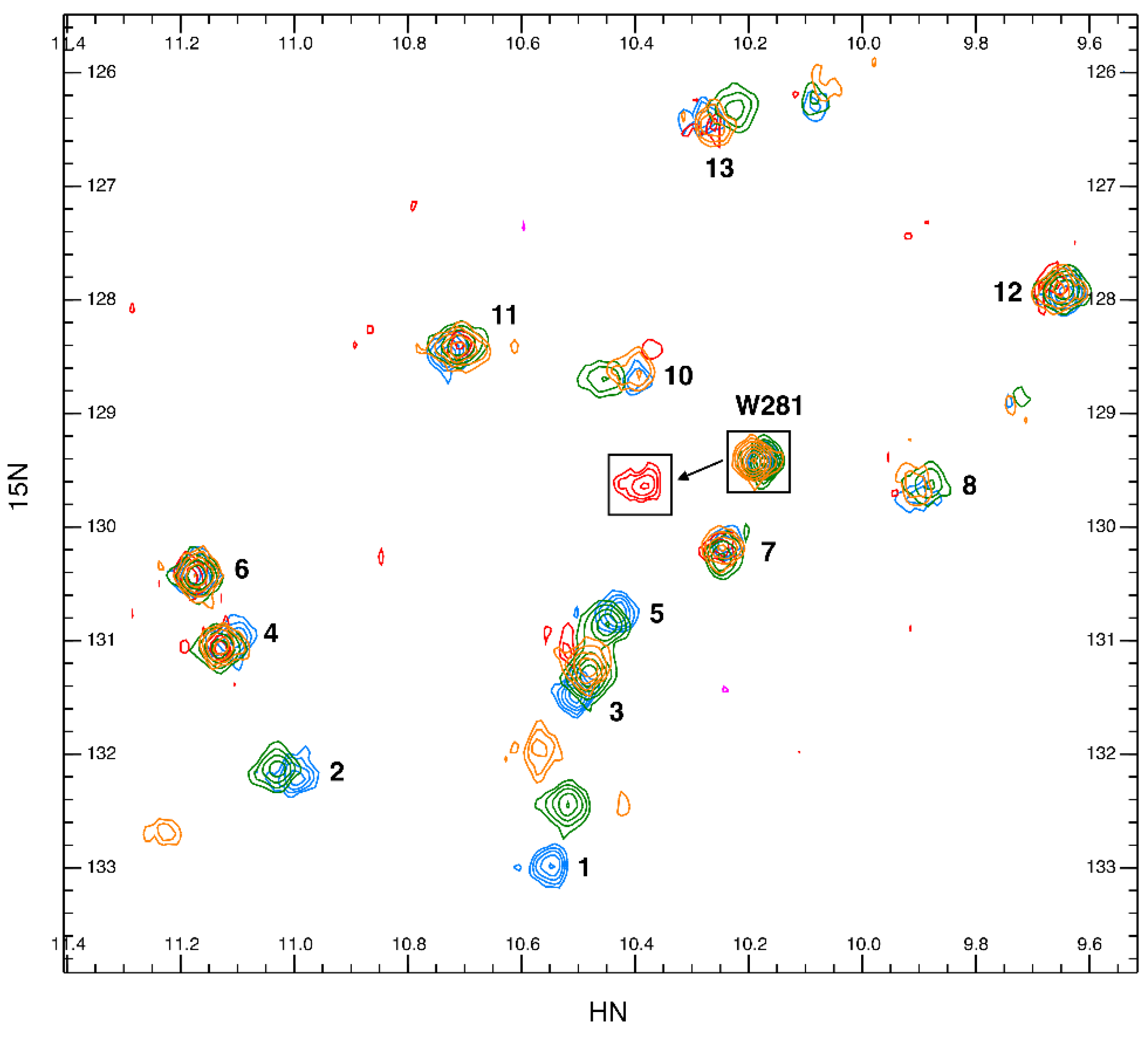
2.5. Tryptophan Flexibility and Glycan Interactions
2.6. Glycan Interaction and Flexibility Analyzed by Molecular Dynamics Simulation
3. Discussion
4. Materials and Methods
4.1. Carbohydrates and Media
4.2. Construction of Enzyme Variants
4.3. Transformation of Pichia Pastoris and β-Mannanase Assay
4.4. Expression in Pichia Pastoris
4.5. Enzyme Purification
4.6. Basic Enzyme Characterization
4.7. Enzyme Kinetics
4.8. Product Formation and Productive Binding Mode Preferences
4.9. Transglycosylation with Saccharides and Alcohols as Acceptors
4.10. NMR Spectroscopy
4.11. Molecular Dynamics Simulation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Dawood, A.; Ma, K. Applications of microbial β-mannanases. Front. Bioeng. Biotechnol. 2020, 8, 598630. [Google Scholar] [CrossRef] [PubMed]
- Henrissat, B. A classification of glycoside hydrolases based on amino acid sequence similarities. Biochem. J. 1991, 280, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Lombard, V.; Golaconde Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed]
- Aspeborg, H.; Coutinho, P.; Wang, Y.; Brumer, H.; Henrissat, B. Evolution, substrate specificity and subfamily classification of glycoside hydrolase family 5 (GH5). BMC Evol. Biol. 2012, 12, 186. [Google Scholar] [CrossRef]
- Rye, C.S.; Withers, S.G. Glycosidase mechanisms. Curr. Opin. Chem. Biol. 2000, 4, 573–580. [Google Scholar] [CrossRef]
- Bissaro, B.; Monsan, P.; Faure, R.; O’Donohue, M.J. Glycosynthesis in a waterworld: New insight into the molecular basis of transglycosylation in retaining glycoside hydrolases. Biochem. J. 2015, 467, 17–35. [Google Scholar] [CrossRef]
- Rosengren, A.; Hägglund, P.; Anderson, L.; Pavon-Orozco, P.; Peterson-Wulff, R.; Nerinckx, W.; Stålbrand, H. The role of subsite +2 of the Trichoderma reesei β-mannanase TrMan5A in hydrolysis and transglycosylation. Biocatal. Biotransform. 2012, 30, 338–352. [Google Scholar] [CrossRef]
- Rosengren, A.; Reddy, S.K.; Sjöberg, J.S.; Aurelius, O.; Logan, D.T.; Kolenová, K.; Stålbrand, H. An Aspergillus nidulans β-mannanase with high transglycosylation capacity revealed through comparative studies within glycosidase family 5. Appl. Microbiol. Biotechnol. 2014, 98, 10091–10104. [Google Scholar] [CrossRef]
- Butler, S.J.; Birgersson, S.; Wiemann, M.; Arcos-Hernandez, M.; Stålbrand, H. Transglycosylation by β-mannanase TrMan5A variants and enzyme synergy for synthesis of allyl glycosides from galactomannan. Process Biochem. 2022, 112, 154–166. [Google Scholar] [CrossRef]
- Dilokpimol, A.; Nakai, H.; Gotfredsen, C.H.; Baumann, M.J.; Nakai, N.; Abou Hachem, M.; Svensson, B. Recombinant production and characterisation of two related GH5 endo-β-1,4-mannanases from Aspergillus nidulans FGSC A4 showing distinctly different transglycosylation capacity. Biochim. Biophys. Acta 2011, 1814, 1720–1729. [Google Scholar] [CrossRef]
- Couturier, M.; Roussel, A.; Rosengren, A.; Leone, P.; Stålbrand, H.; Berrin, J.G. Structural and biochemical analyses of glycoside hydrolase families 5 and 26 β-(1,4)-mannanases from Podospora anserina reveal differences upon manno-oligosaccharide catalysis. J. Biol. Chem. 2013, 288, 14624–14635. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, P.K.; Kapoor, M. Production, properties, and applications of endo-β-mannanases. Biotechnol. Adv. 2017, 35, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Lundqvist, J.; Teleman, A.; Junel, L.; Zacchi, G.; Dahlman, O.; Tjerneld, F.; Stålbrand, H. Isolation and characterization of galactoglucomannan from spruce (Picea abies). Carbohydr. Polym. 2002, 48, 29–39. [Google Scholar] [CrossRef]
- Scheller, H.V.; Ulvskov, P. Hemicelluloses. Annu. Rev. Plant Biol. 2010, 61, 263–289. [Google Scholar] [CrossRef] [PubMed]
- Voiniciuc, C. Modern mannan: A hemicellulose’s journey. New Phytol. 2022, 234, 1175–1184. [Google Scholar] [CrossRef]
- Agger, J.W.; Zeuner, B. Bio-based surfactants: Enzymatic functionalization and production from renewable resources. Curr. Opin. Biotechnol. 2022, 78, 102842. [Google Scholar] [CrossRef]
- Arcos-Hernandez, M.; Naidjonoka, P.; Butler, S.J.; Nylander, T.; Stålbrand, H.; Jannasch, P. Thermoresponsive Glycopolymers Based on Enzymatically Synthesized Oligo-β-Mannosyl Ethyl Methacrylates and N-Isopropylacrylamide. Biomacromolecules 2021, 22, 2338–2351. [Google Scholar] [CrossRef]
- Morrill, J.; Månberger, A.; Rosengren, A.; Naidjonoka, P.; von Freiesleben, P.; Krogh, K.B.R.M.; Bergquist, K.E.; Nylander, T.; Nordberg Karlsson, E.; Adlercreutz, P.; et al. β-Mannanase-catalyzed synthesis of alkyl mannooligosides. Appl. Microbiol. Biotechnol. 2018, 102, 5149–5163. [Google Scholar] [CrossRef]
- Wang, Y.; Vilaplana, F.; Brumer, H.; Aspeborg, H. Enzymatic characterization of a glycoside hydrolase family 5 subfamily 7 (GH5_7) mannanase from Arabidopsis thaliana. Planta 2014, 239, 653–665. [Google Scholar] [CrossRef]
- Brusa, C.; Belloy, N.; Gérard, D.; Muzard, M.; Dauchez, M.; Plantier-Royon, R.; Rémond, C. Exploring the aglycone subsite of a GH11 xylanase for the synthesis of xylosides by transglycosylation reactions. J. Biotechnol. 2018, 272–273, 56–63. [Google Scholar] [CrossRef]
- Eide, K.B.; Lindbom, A.R.; Eijsink, V.G.H.; Norberg, A.L.; Sørlie, M. Analysis of productive binding modes in the human chitotriosidase. FEBS Lett. 2013, 587, 3508–3513. [Google Scholar] [CrossRef] [PubMed]
- Abdul Manas, N.H.; Md. Ilias, R.; Mahadi, N.M. Strategy in manipulating transglycosylation activity of glycosyl hydrolase for oligosaccharide production. Crit. Rev. Biotechnol. 2018, 38, 272–293. [Google Scholar] [CrossRef] [PubMed]
- Teze, D.; Zhao, J.; Wiemann, M.; Kazi, Z.G.A.; Lupo, R.; Zeuner, B.; Vuillemin, M.; Rønne, M.E.; Carlström, G.; Duus, J.O.; et al. Rational Enzyme Design without Structural Knowledge: A Sequence-Based Approach for Efficient Generation of Transglycosylases. Chem. Eur. J. 2021, 27, 10323–10334. [Google Scholar] [CrossRef] [PubMed]
- Ueda, M.; Hirano, Y.; Fukuhara, H.; Naka, Y.; Nakazawa, M.; Sakamoto, T.; Ogata, Y.; Tamada, T. Gene cloning, expression, and X-ray crystallographic analysis of a β-mannanase from Eisenia fetida. Enzym. Microb. Technol. 2018, 117, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Schröder, R.; Wegrzyn, T.F.; Sharma, N.N.; Atkinson, R.G. LeMAN4 endo-β-mannanase from ripe tomato fruit can act as a mannan transglycosylase or hydrolase. Planta 2006, 224, 1091–1102. [Google Scholar] [CrossRef]
- Xu, B.; Hägglund, P.; Stålbrand, H.; Janson, J.C. endo-β-1,4-Mannanases from blue mussel, Mytilus edulis: Purification, characterization, and mode of action. J. Biotechnol. 2002, 92, 267–277. [Google Scholar] [CrossRef]
- Xu, B.; Muñoz, I.G.; Janson, J.C.; Ståhlberg, J. Crystallization and X-ray analysis of native and selenomethionyl β-mannanase Man5A from blue mussel, Mytilus edulis, expressed in Pichia pastoris. Acta Cryst. 2002, D58, 542–545. [Google Scholar] [CrossRef]
- Xu, B.; Sellos, D.; Janson, J.C. Cloning and expression in Pichia pastoris of a blue mussel (Mytilus edulis) β-mannanase gene. Eur. J. Biochem. 2002, 269, 1753–1760. [Google Scholar] [CrossRef]
- Larsson, A.M.; Anderson, L.; Xu, B.; Muñoz, I.G.; Usón, I.; Janson, J.C.; Stålbrand, H.; Ståhlberg, J. Three-dimensional crystal structure and enzymic characterization of β-mannanase Man5A from blue mussel Mytilus edulis. J. Mol. Biol. 2006, 357, 1500–1510. [Google Scholar] [CrossRef]
- Lobo-da-Cunha, A. Structure and function of the digestive system in molluscs. Cell Tissue Res. 2019, 377, 475–503. [Google Scholar] [CrossRef]
- Mizutani, K.; Tsuchiya, S.; Toyoda, M.; Nanbu, Y.; Tominaga, K.; Yuasa, K.; Takahashi, N.; Tsuji, A.; Mikami, B. Structure of β-1,4-mannanase from the common sea hare Aplysia kurodai at 1.05 Å resolution. Acta Cryst. 2012, F68, 1164–1168. [Google Scholar] [CrossRef]
- Kim, M.K.; An, Y.J.; Song, J.M.; Jeong, C.S.; Kang, M.H.; Kwon, K.K.; Lee, Y.H.; Cha, S.S. Structure-based investigation into the functional roles of the extended loop and substrate-recognition sites in an endo-β-1,4-D-mannanase from the Antarctic springtail, Cryptopygus antarcticus. Proteins 2014, 82, 3217–3223. [Google Scholar] [CrossRef]
- Hekmat, O.; Lo Leggio, L.; Rosengren, A.; Kamarauskaite, J.; Kolenova, K.; Stålbrand, H. Rational engineering of mannosyl binding in the distal glycone subsites of Cellulomonas fimi endo-β-1,4-mannanase: Mannosyl binding promoted at subsite −2 and demoted at subsite−3. Biochemistry 2010, 49, 4884–4896. [Google Scholar] [CrossRef]
- Sabini, E.; Schubert, H.; Murshudov, G.; Wilson, K.S.; Siika-Aho, M.; Penttilä, M. The three-dimensional structure of a Trichoderma reesei β-mannanase from glycoside hydrolase family 5. Acta Cryst. 2000, D56, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Kumagai, Y.; Yamashita, K.; Tagami, T.; Uraji, M.; Wan, K.; Okuyama, M.; Yao, M.; Kimura, A.; Hatanaka, T. The loop structure of Actinomycete glycoside hydrolase family 5 mannanases governs substrate recognition. FEBS J. 2015, 282, 4001–4014. [Google Scholar] [CrossRef] [PubMed]
- Gagné, D.; Narayanan, C.; Nguyen-Thi, N.; Roux, L.D.; Bernard, D.N.; Brunzelle, J.S.; Couture, J.-F.; Agarwal, P.K.; Doucet, N. Ligand binding enhances millisecond conformational exchange in xylanase B2 from Streptomyces lividans. Biochemistry 2016, 55, 4184–4196. [Google Scholar] [CrossRef]
- Smith, C.A.; Stewart, N.K.; Toth, M.; Vakulenko, S.B. Structural insights into the mechanism of carbapenemase activity of the OXA-48 β-lactamase. Antimicrob. Agents Chemother. 2019, 63, 10. [Google Scholar] [CrossRef]
- Madhuprakash, J.; Singh, A.; Kumar, S.; Sinha, M.; Kaur, P.; Sharma, S.; Podile, A.R.; Singh, T.P. Structure of chitinase D from Serratia proteamaculans reveals the structural basis of its dual action of hydrolysis and transglycosylation. Int. J. Biochem. Mol. Biol. 2013, 4, 166–178. [Google Scholar]
- Wu, M.; Bu, L.; Vuong, T.V.; Wilson, D.B.; Crowley, M.F.; Sandgren, M.; Ståhlberg, J.; Beckham, G.T.; Hansson, H. Loop motions important to product expulsion in the Thermobifida fusca glycoside hydrolase family 6 cellobiohydrolase from structural and computational studies. J. Biol. Chem. 2013, 288, 33107–33117. [Google Scholar] [CrossRef]
- Miki, A.; Inaba, S.; Maruno, T.; Kobayashi, Y.; Oda, M. Tryptophan introduction can change β-glucan binding ability of the carbohydrate-binding module of endo-1,3-β-glucanase. Biosci. Biotechnol. Biochem. 2017, 81, 951–957. [Google Scholar] [CrossRef]
- Kiessling, L.L.; Diehl, R. CH-π Interactions in Glycan Recognition. ACS Chem. Biol. 2021, 16, 1884–1893. [Google Scholar] [CrossRef] [PubMed]
- Song, K.M.; Okuyama, M.; Nishimura, M.; Tagami, T.; Mori, H.; Kimura, A. Aromatic residue on β→α loop 1 in the catalytic domain is important to the transglycosylation specificity of glycoside hydrolase family 31 α-glucosidase. Biosci. Biotechnol. Biochem. 2013, 72, 1759–1765. [Google Scholar] [CrossRef]
- Bertani, G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 1951, 62, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Stålbrand, H.; Siika-Aho, M.; Tenkanen, M.; Viikari, L. Purification and characterization of two β-mannanases from Trichoderma reesei. J. Biotechnol. 1993, 29, 229–242. [Google Scholar] [CrossRef]
- Suganuma, T.; Matsuno, R.; Ohnishi, M.; Hiromi, K. A study of the mechanism of action of Taka-amylase A on linear oligosaccharides by product analysis and computer simulation. J. Biochem. 1978, 84, 293–316. [Google Scholar] [CrossRef]
- Matsui, I.; Ishikawa, K.; Matsui, E.; Miyairi, S.; Fukui, S.; Honda, K. Subsite structure of Saccharomycopsis α-amylase secreted from Saccharomyces cerevisiae. J. Biochem. 1991, 109, 566–569. [Google Scholar] [CrossRef]
- Hogg, D.; Pell, G.; Dupree, P.; Goubet, F.; Martin-Orué, S.M.; Armand, S.; Gilbert, H.J. The modular architecture of Cellvibrio japonicus mannanases in glycoside hydrolase families 5 and 26 points to differences in their role in mannan degradation. Biochem. J. 2003, 371, 1027–1043. [Google Scholar] [CrossRef]
- Bågenholm, V.; Reddy, S.K.; Bouraoui, H.; Morrill, J.; Kulcinskaja, E.; Bahr, C.M.; Aurelius, O.; Rogers, T.; Xiao, Y.; Logan, D.T.; et al. Galactomannan catabolism conferred by a polysaccharide utilization locus of Bacteroides ovatus: Enzyme synergy and crystal structure of a β-mannanase. J. Biol. Chem. 2017, 292, 229–243. [Google Scholar] [CrossRef] [PubMed]
- Koerdel, J.; Skelton, N.J.; Akke, M.; Palmer, A.G.; Chazin, W.J. Backbone dynamics of calcium-loaded calbindin D9k studied by two-dimensional proton-detected nitrogen-15 NMR spectroscopy. Biochemistry 1992, 31, 4856–4866. [Google Scholar] [CrossRef]
- Farrow, N.A.; Muhandiram, R.; Singer, A.U.; Pascal, S.M.; Kay, C.M.; Gish, I.G.; Shoelson, S.E.; Pawson, T.; Forman-Kay, J.D.; Kay, L.E. Backbone Dynamics of a Free and a Phosphopeptide-Complexed Src Homology 2 Domain Studied by 15N NMR Relaxation. Biochemistry 1994, 33, 5984–6003. [Google Scholar] [CrossRef]
- Carr, H.Y.; Purcell, E.M. Effects of diffusion on free precession in nuclear magnetic resonance experiments. Phys. Rev. 1954, 94, 630–638. [Google Scholar] [CrossRef]
- Meiboom, S.; Gill, D. Modified spin-echo method for measuring nuclear relaxation times. Rev. Sci. Instrum. 1958, 29, 688–691. [Google Scholar] [CrossRef]
- Loria, J.P.; Rance, M.; Palmer, A.G.R. A relaxation-compensated Carr-Purcell-Meiboom-Gill sequence for characterizing chemical exchange by NMR spectroscopy. J. Am. Chem. Soc. 1999, 121, 2331–2332. [Google Scholar] [CrossRef]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef] [PubMed]
- Vranken, W.F.; Boucher, W.; Stevens, T.J.; Fogh, R.H.; Pajon, A.; Llinas, M.; Ulrich, E.L.; Markley, J.L.; Ionides, J.; Laue, E.D. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins 2005, 59, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Ahlner, A.; Carlsson, M.; Jonsson, B.H.; Lundström, P. PINT: A software for integration of peak volumes and extraction of relaxation rates. J. Biomol. NMR 2013, 56, 191–202. [Google Scholar] [CrossRef]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef]
- Allouche, A. Software News and Updates Gabedit—A Graphical User Interface for Computational Chemistry Softwares. J. Comput. Chem. 2012, 32, 174–182. [Google Scholar] [CrossRef]

| Product | WT | W240A | W281A | W240A/W281A |
|---|---|---|---|---|
| [M1] (µM) | 7.7 ± 0.1 | 36 ± 0.6 | 8.7 ± 0.1 | 89 ± 0.9 |
| [M2] (µM) | 183 ± 1 | 153 ± 2 | 235 ± 0.4 | 82 ± 0.9 |
| [M3] (µM) | 191 ± 0.8 | 163 ± 3 | 251 ± 0.6 | 96 ± 1 |
| [M4] (µM) | 48 ± 0.4 | 64 ± 1 | 16 ± 0.9 | 89 ± 1 |
| [M4]: [M1] ratio | 6.2 | 1.8 | 1.8 | 1.0 |
| Enzyme Variant | LBG kcat (s−1) | LBG KM (g·L−1) | LBG kcat/KM (g·L−1·s−1) | M4 kcat/KM (s−1·mM−1) | M5 kcat/KM (s−1·mM−1) |
|---|---|---|---|---|---|
| WT | 332 ± 5 | 0.75 ± 0.05 | 446 ± 30 | 0.71 ± 0.05 | 36 ± 2 |
| W240A | 105 ± 3 | 1.3 ± 0.1 | 83 ± 9 | 0.18 ± 0.02 | 2.8 ± 0.5 |
| W281A | 121 ± 3 | 0.73 ± 0.1 | 165 ± 15 | 0.55 ± 0.08 | 17 ± 3 |
| W240A/W281A | 66.4 ± 18 | 3.7 ± 0.4 | 18 ± 5 | 0.23 ± 0.01 | 0.84 ± 0.03 |
| Residue | R2 (apo) | R2 (M2) | R2 (G2M5) |
|---|---|---|---|
| Trp #1 | 21.2 ± 1.55 | 21.3 ± 1.65 | n.d. a |
| Trp #2 | 25.7 ± 1.73 | 23.3 ± 1.21 | n.d. a |
| Trp #3 | 19.7 ± 0.52 | 21.8 ± 1.01 | n.d. a |
| Trp #4 | 24.0 ± 2.79 | 26.4 ± 1.75 | 29.1 ± 4.81 |
| Trp #5 | 20.3 ± 1.01 | 21.8 ± 0.94 | n.d. a |
| Trp #6 | 22.1 ± 1.08 | 25.4 ± 1.83 | 27.2 ± 2.12 |
| Trp #7 | 23.5 ± 2.06 | 23.3 ± 1.44 | 22.0 ± 3.02 |
| Trp #8 | 31.2 ± 4.04 | 28.1 ± 3.34 | n.d. a |
| Trp #9 (W281) | 12.0 ± 0.31 | 12.4 ± 0.27 | 18.2 ± 1.82 |
| Trp #10 | 31.6 ± 7.59 | 31.1 ± 5.17 | 19.1 ± 7.92 |
| Trp #11 | 22.5 ± 1.11 | 24.1 ± 1.86 | 23.3 ± 4.16 |
| Trp #12 | 20.8 ± 1.40 | 21.1 ± 0.73 | 19.6 ± 2.35 |
| Trp #13 | 30.9 ± 7.79 | 27.2 ± 4.38 | 24.3 ± 3.21 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Birgersson, S.; Morrill, J.; Stenström, O.; Wiemann, M.; Weininger, U.; Söderhjelm, P.; Akke, M.; Stålbrand, H. Flexibility and Function of Distal Substrate-Binding Tryptophans in the Blue Mussel β-Mannanase MeMan5A and Their Role in Hydrolysis and Transglycosylation. Catalysts 2023, 13, 1281. https://doi.org/10.3390/catal13091281
Birgersson S, Morrill J, Stenström O, Wiemann M, Weininger U, Söderhjelm P, Akke M, Stålbrand H. Flexibility and Function of Distal Substrate-Binding Tryptophans in the Blue Mussel β-Mannanase MeMan5A and Their Role in Hydrolysis and Transglycosylation. Catalysts. 2023; 13(9):1281. https://doi.org/10.3390/catal13091281
Chicago/Turabian StyleBirgersson, Simon, Johan Morrill, Olof Stenström, Mathias Wiemann, Ulrich Weininger, Pär Söderhjelm, Mikael Akke, and Henrik Stålbrand. 2023. "Flexibility and Function of Distal Substrate-Binding Tryptophans in the Blue Mussel β-Mannanase MeMan5A and Their Role in Hydrolysis and Transglycosylation" Catalysts 13, no. 9: 1281. https://doi.org/10.3390/catal13091281
APA StyleBirgersson, S., Morrill, J., Stenström, O., Wiemann, M., Weininger, U., Söderhjelm, P., Akke, M., & Stålbrand, H. (2023). Flexibility and Function of Distal Substrate-Binding Tryptophans in the Blue Mussel β-Mannanase MeMan5A and Their Role in Hydrolysis and Transglycosylation. Catalysts, 13(9), 1281. https://doi.org/10.3390/catal13091281