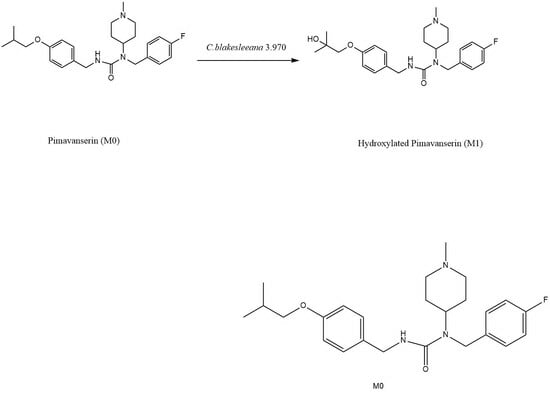
Microbial Transformation of Pimavanserin by Cunninghamella blakesleeana AS 3.970
Abstract
:1. Introduction
2. Results and Discussion
2.1. Identification of Efficient Transformation Microorganism Strains
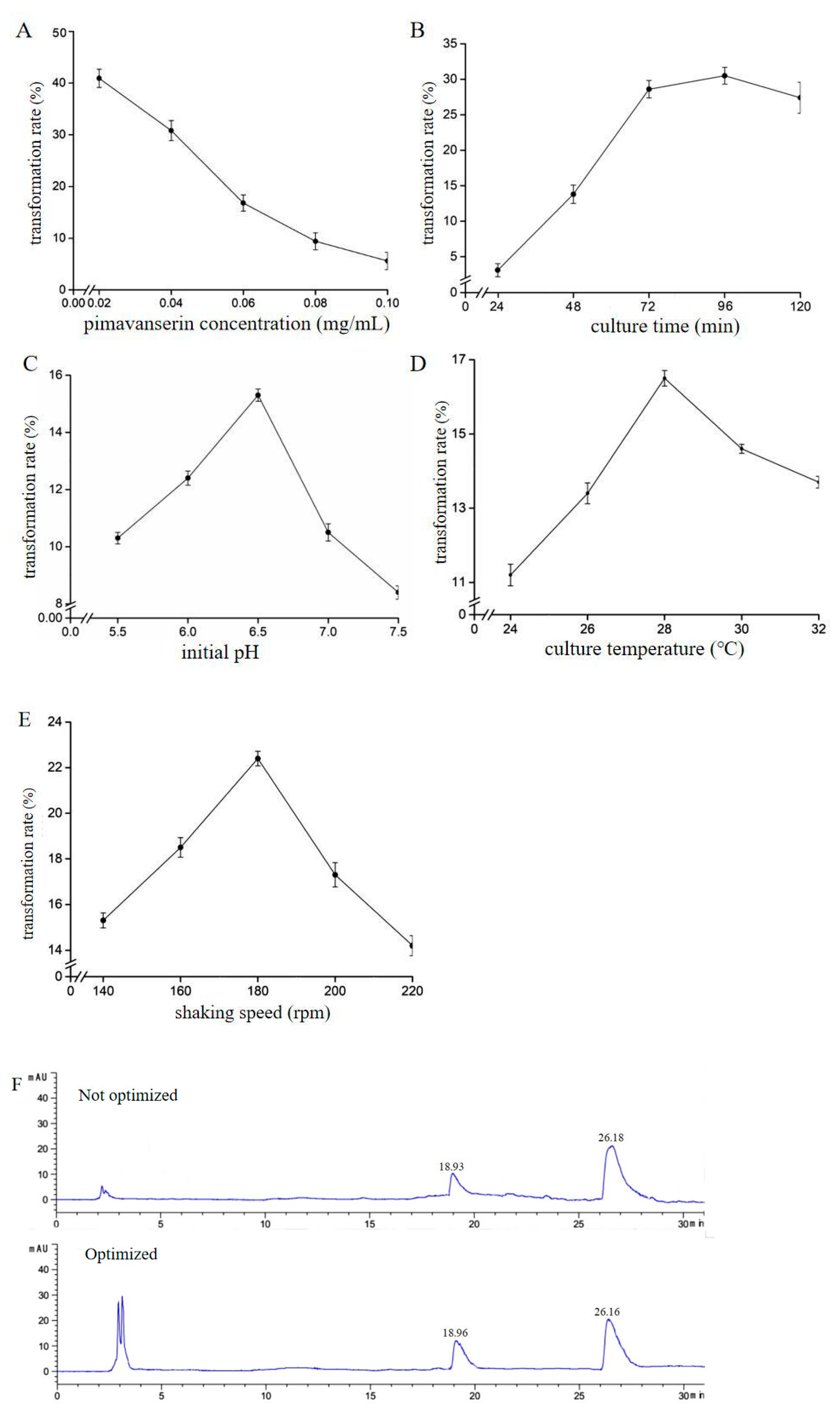
2.2. Optimization of C. blakesleeana AS 3.970 Resting Cell Biotransformation
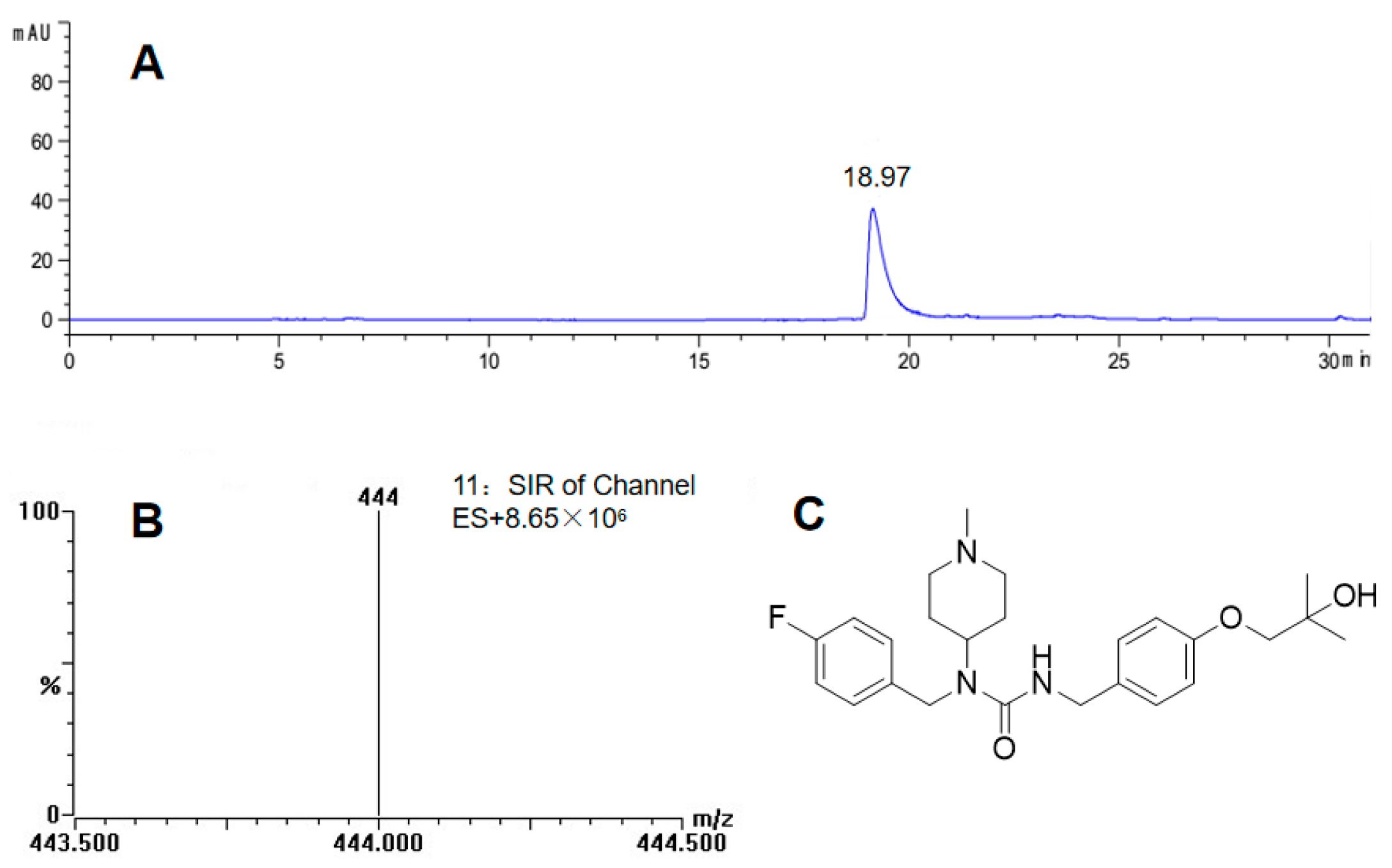
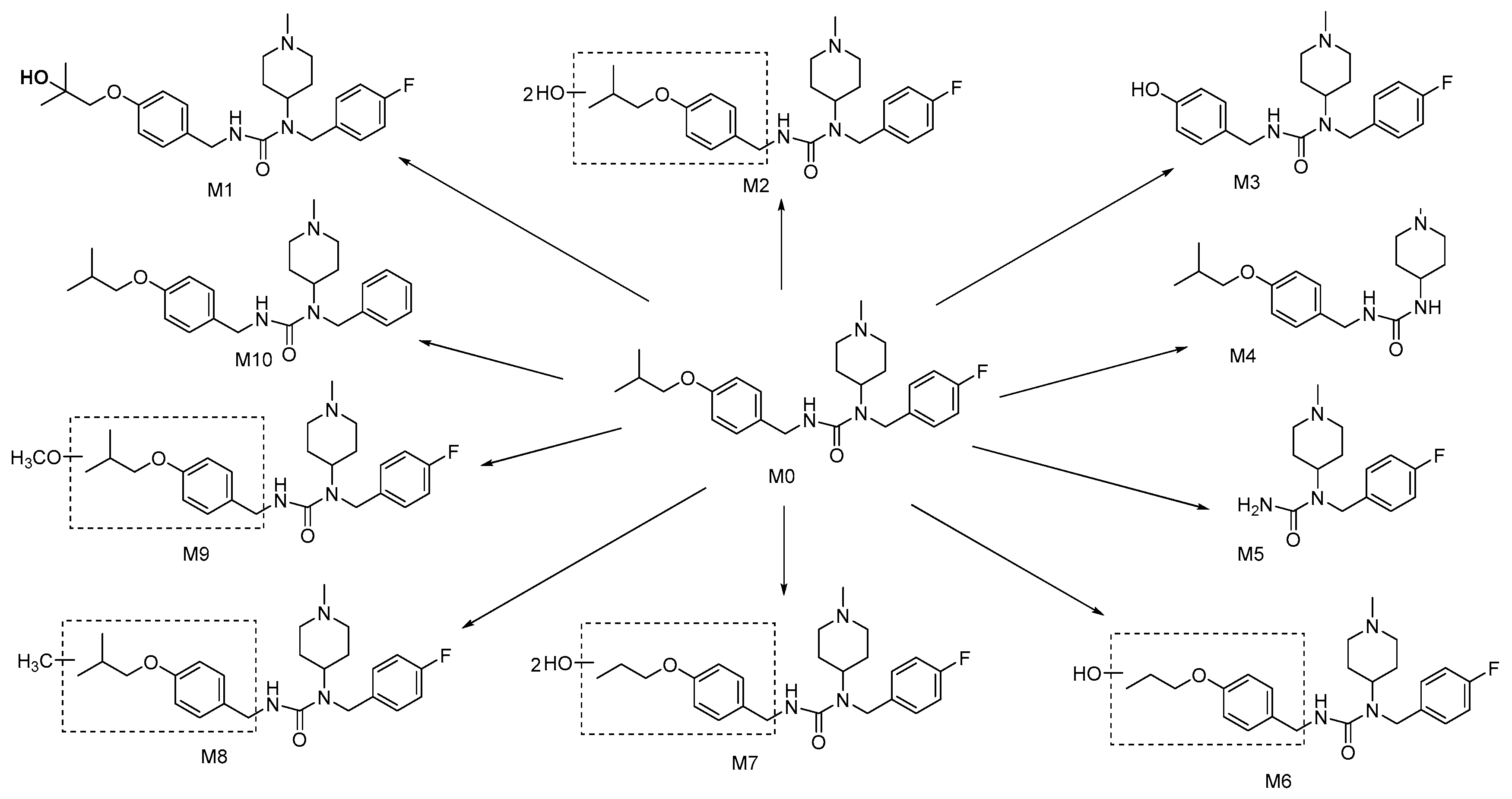
2.3. Preparation and Identification of Major Biotransformation Product M1
2.4. Analysis of Biotransformation Products by LC-MS/MS
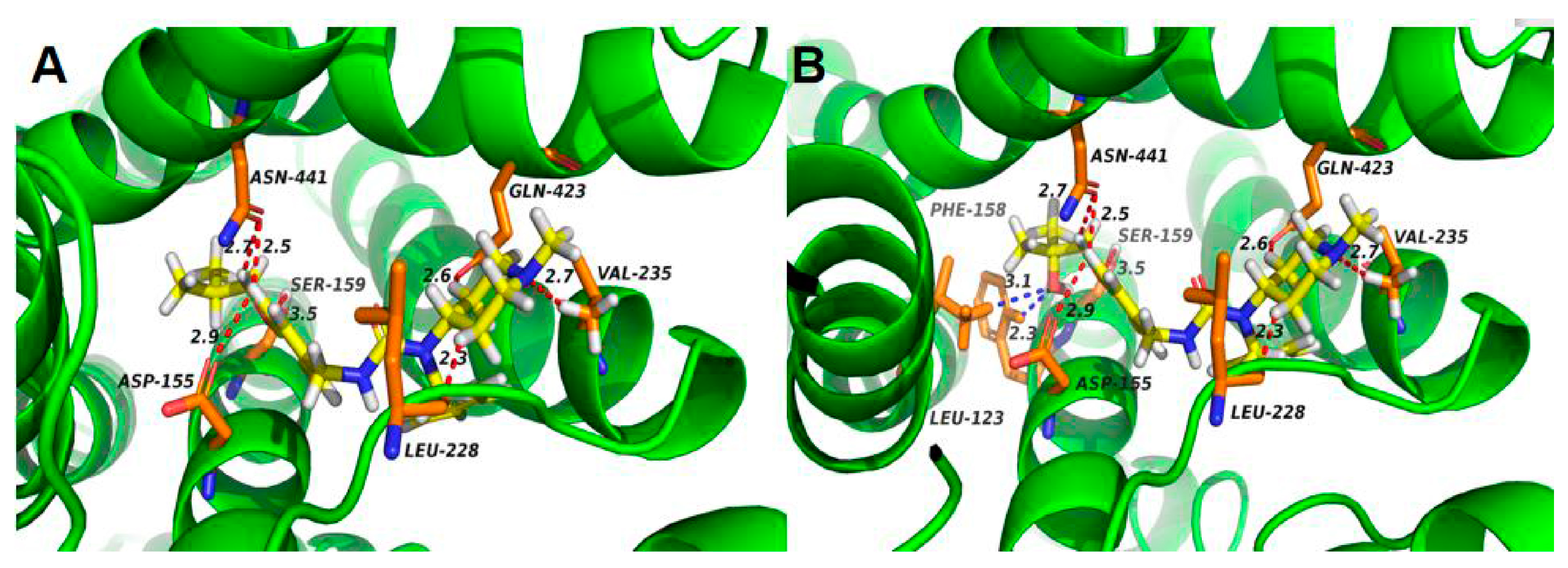
2.5. Preliminary Molecular Docking of 5-HT2A Receptor with Pimavanserin and M1
3. Material and Methods
3.1. Microorganisms and Culture Media
3.2. Substrate and Biotransformation Procedures
3.3. High Performance Liquid Chromatography (HPLC)
3.4. Optimization of the Microbial Transformation Process
3.5. Preparation and Identification of Major Biotransformation Products
3.6. Molecular Docking
4. Conclusions and Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Nasrallah, H.A.; Fedora, R.; Morton, R. Successful treatment of clozapine-nonresponsive refractory hallucinations and delusions with pimavanserin, a serotonin 5HT-2A receptor inverse agonist. Schizophr. Res. 2019, 208, 217–220. [Google Scholar] [CrossRef] [PubMed]
- Markham, A. Pimavanserin: First global approval. Drugs 2016, 76, 1053–1057. [Google Scholar] [CrossRef] [PubMed]
- Vanover, K.E.; Weiner, D.M.; Makhay, M.; Veinbergs, I.; Gardell, L.R.; Lameh, J.; Del Tredici, A.L.; Piu, F.; Schiffer, H.H.; Ott, T.R.; et al. Pharmacological and behavioral profile of N-(4-fluorophenylmethyl)-N-(1-methylpiperidin-4-yl)-N′-(4-(2-methylpropyloxy)phen ylmethyl) carbamide (2R,3R)-dihydroxybutanedioate (2:1) (ACP-103), a novel 5-hydroxytryptamine(2A) receptor inverse agonist. J. Pharmacol. Exp. Ther. 2006, 317, 910–918. [Google Scholar] [CrossRef] [PubMed]
- Yousuf, M.; Jamil, W.; Mammadova, K. Microbial bioconversion: A regio-specific method for novel drug design and toxicological study of metabolites. Curr. Pharm. Biotechnol. 2019, 20, 1156–1162. [Google Scholar] [CrossRef] [PubMed]
- Asha, S.; Vidyavathi, M. Cunninghamella—A microbial model for drug metabolism studies—A review. Biotechnol. Adv. 2009, 27, 16–29. [Google Scholar] [CrossRef] [PubMed]
- Murphy, C.D. Drug metabolism in microorganisms. Biotechnol. Lett. 2015, 37, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.T.T.; Choi, Y.J.; Lee, H.B. Isolation and characterization of three unrecorded zygomycete fungi in Korea: Cunninghamella bertholletiae, Cunninghamella echinulata, and Cunninghamella elegans. Mycobiology 2017, 45, 318–326. [Google Scholar] [CrossRef] [PubMed]
- Pekala, E.; Kubowicz, P.; Lazewska, D. Cunninghamella as a microbiological model for metabolism of histamine H(3) receptor antagonist 1-[3-(4-tert-butylphenoxy)propyl]piperidine. Appl. Biochem. Biotechnol. 2012, 168, 1584–1593. [Google Scholar] [CrossRef] [PubMed]
- de Sousa, I.P.; Sousa Teixeira, M.V.; Jacometti Cardoso Furtado, N.A. An overview of biotransformation and toxicity of diterpenes. Molecules 2018, 23, 1387. [Google Scholar] [CrossRef] [PubMed]
- Dube, A.K.; Kumar, M.S. Biotransformation of bromhexine by Cunninghamella elegans, C. echinulata and C. blakesleeana. Braz. J. Microbiol. 2017, 48, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Ye, M.; Qu, G.; Guo, H.; Guo, D. Specific 12 beta-hydroxylation of cinobufagin by filamentous fungi. Appl. Environ. Microbiol. 2004, 70, 3521–3527. [Google Scholar] [CrossRef] [PubMed]
- Bielenica, A.; Kedzierska, E.; Kolinski, M.; Kmiecik, S.; Kolinski, A.; Fiorino, F.; Severino, B.; Magli, E.; Corvino, A.; Rossi, I.; et al. 5-HT2 receptor affinity, docking studies and pharmacological evaluation of a series of 1,3-disubstituted thiourea derivatives. Eur. J. Med. Chem. 2016, 116, 173–186. [Google Scholar] [CrossRef] [PubMed]
- Shyam Prasad, G.; Narasimha Rao, K.; Preethi, R.; Girisham, S.; Reddy, S.M. Biotransformation of Meloxicam by Cunninghamella blakesleeana: Significance of Carbon and Nitrogen Source. Indian J. Microbiol. 2011, 51, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Özçinar, Ö.; Tağ, Ö.; Yusufoglu, H.; Kivçak, B.; Bedir, E. Biotransformation of ruscogenins by Cunninghamella blakesleeana NRRL 1369 and neoruscogenin by endophytic fungus Neosartorya hiratsukae. Phytochemistry 2018, 152, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kudaibergenova, B.M.; Wahab, A.T.; Siddiqui, M.; Abilov, Z.A.; Choudhary, M.I. A new metabolite from Cunninghamella blakesleeana-mediated biotransformation of an oral contraceptive drug, levonorgestrel. Nat. Prod. Res. 2021, 35, 2095–2098. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Zhang, D.; Sun, P.; Zhao, Y.; Chang, X.; Ma, Y.; Yang, L. Evaluation of microbial transformation of 10-deoxoartemisinin by UPLC-ESI-Q-TOF-MSE. Molecules 2019, 24, 3874. [Google Scholar] [CrossRef] [PubMed]
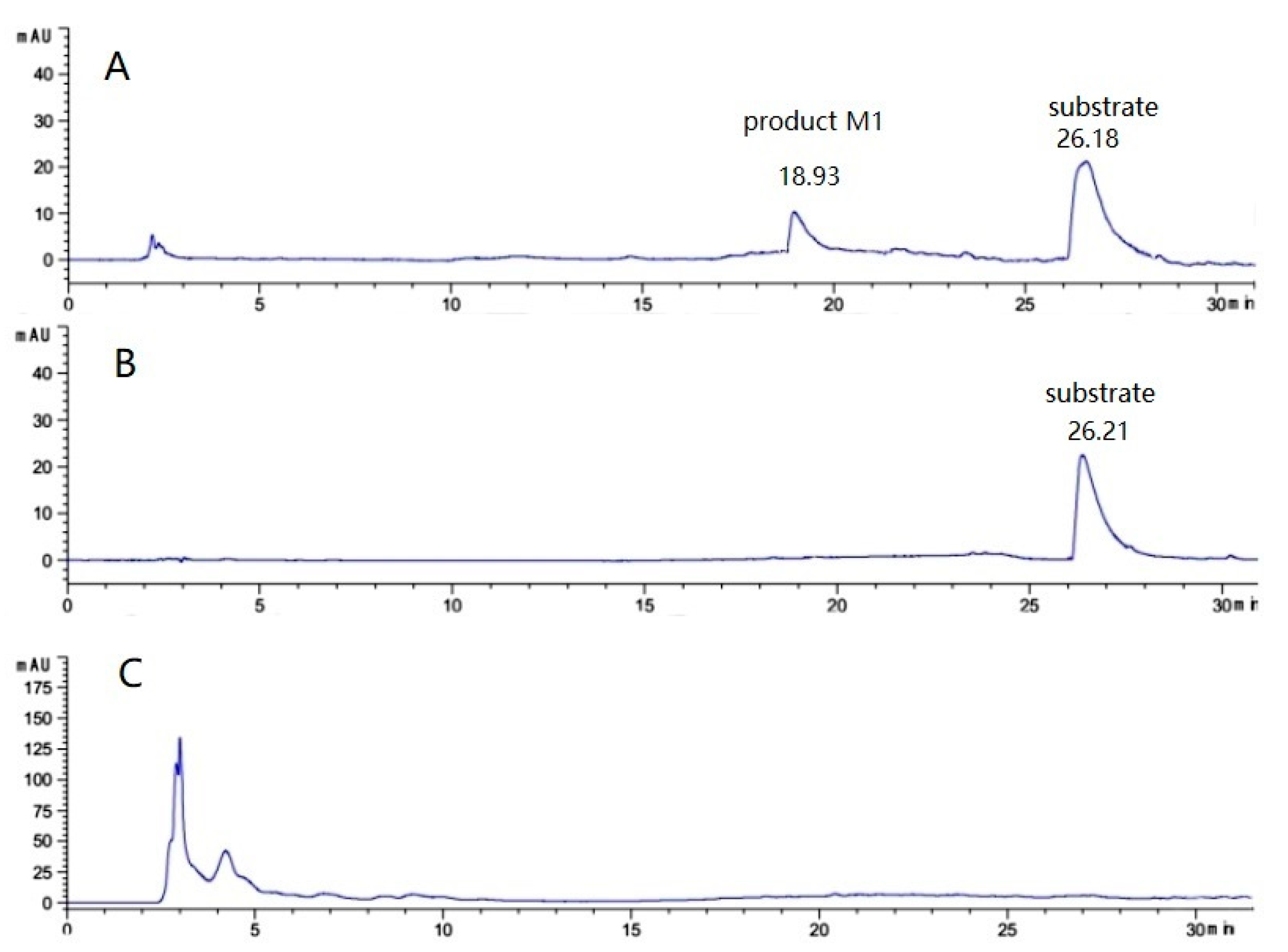
| Fungus Species | Resting Cell Transformation Rate (%) | Growth Cell Transformation Rate (%) |
|---|---|---|
| Cunninghamella blakesleeana AS 3.970 | 16.24 | 7.10 |
| Cunninghamella blakesleeana AS 3.910 | 10.46 | 4.64 |
| Cunninghamella blakesleeana AS 3.153 | 7.68 | 3.12 |
| Cunninghamella elegans AS3.156 | 7.42 | 2.44 |
| Cunninghamella elegans AS3.2028 | 4.58 | 1.12 |
| Mucor circinelloides AS 3.3421 | 2.32 | - |
| Aspergillus niger AS 3.739 | - | - |
| Eurotium cristatum AS 3.793 | - | - |
 | ||
| Position | δC | δH |
| 1 | 162.19 | - |
| 2 | 115.37 | 7.05 (1H, m) |
| 3 | 128.84 | 7.25 (1H, m) |
| 4 | 137.19 | - |
| 5 | 128.79 | 7.25 (1H, m) |
| 6 | 115.37 | 7.05 (1H, m) |
| 7 | 51.57 | 4.93 (2H, m) |
| 8 | 54.22 | 3.74 (1H, m) |
| 9 | 28.94 | 1.79 (2H, m) |
| 10 | 44.59 | 2.34 (2H, m) |
| 11 | 43.57 | 2.34 (2H, m) |
| 12 | 28.94 | 1.77 (2H, m) |
| 13 | 40.32 | 2.33 (3H, m) |
| 14 | 160.59 | - |
| 15 | 44.59 | 4.72 (2H, m) |
| 16 | 128.61 | - |
| 17 | 133.48 | 7.24 (1H, m) |
| 18 | 114.61 | 6.63–6.95 (1H, d) |
| 19 | 157.94 | - |
| 20 | 115.52 | 6.63–6.95 (1H, d) |
| 21 | 113.48 | 7.25 (1H, m) |
| 22 | 76.64 | 4.08 (2H, m) |
| 23 | 69.11 | 1.71–1.77 (2H, d) |
| 24 | 27.11 | 1.23 (3H, d) |
| 25 | 27.11 | 1.23 (3H, d) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, M.; Yu, Q.; Liu, Y.; Cai, S.; Jiang, X.; Xu, W.; Xu, W. Microbial Transformation of Pimavanserin by Cunninghamella blakesleeana AS 3.970. Catalysts 2023, 13, 1220. https://doi.org/10.3390/catal13081220
Song M, Yu Q, Liu Y, Cai S, Jiang X, Xu W, Xu W. Microbial Transformation of Pimavanserin by Cunninghamella blakesleeana AS 3.970. Catalysts. 2023; 13(8):1220. https://doi.org/10.3390/catal13081220
Chicago/Turabian StyleSong, Ming, Qi Yu, Yuqi Liu, Sulan Cai, Xuliang Jiang, Weizhuo Xu, and Wei Xu. 2023. "Microbial Transformation of Pimavanserin by Cunninghamella blakesleeana AS 3.970" Catalysts 13, no. 8: 1220. https://doi.org/10.3390/catal13081220
APA StyleSong, M., Yu, Q., Liu, Y., Cai, S., Jiang, X., Xu, W., & Xu, W. (2023). Microbial Transformation of Pimavanserin by Cunninghamella blakesleeana AS 3.970. Catalysts, 13(8), 1220. https://doi.org/10.3390/catal13081220