SOEing PCR/Docking Optimization of Protein A-G/scFv-Fc-Bioconjugated Au Nanoparticles for Interaction with Meningitidis Bacterial Antigen
Abstract
1. Introduction
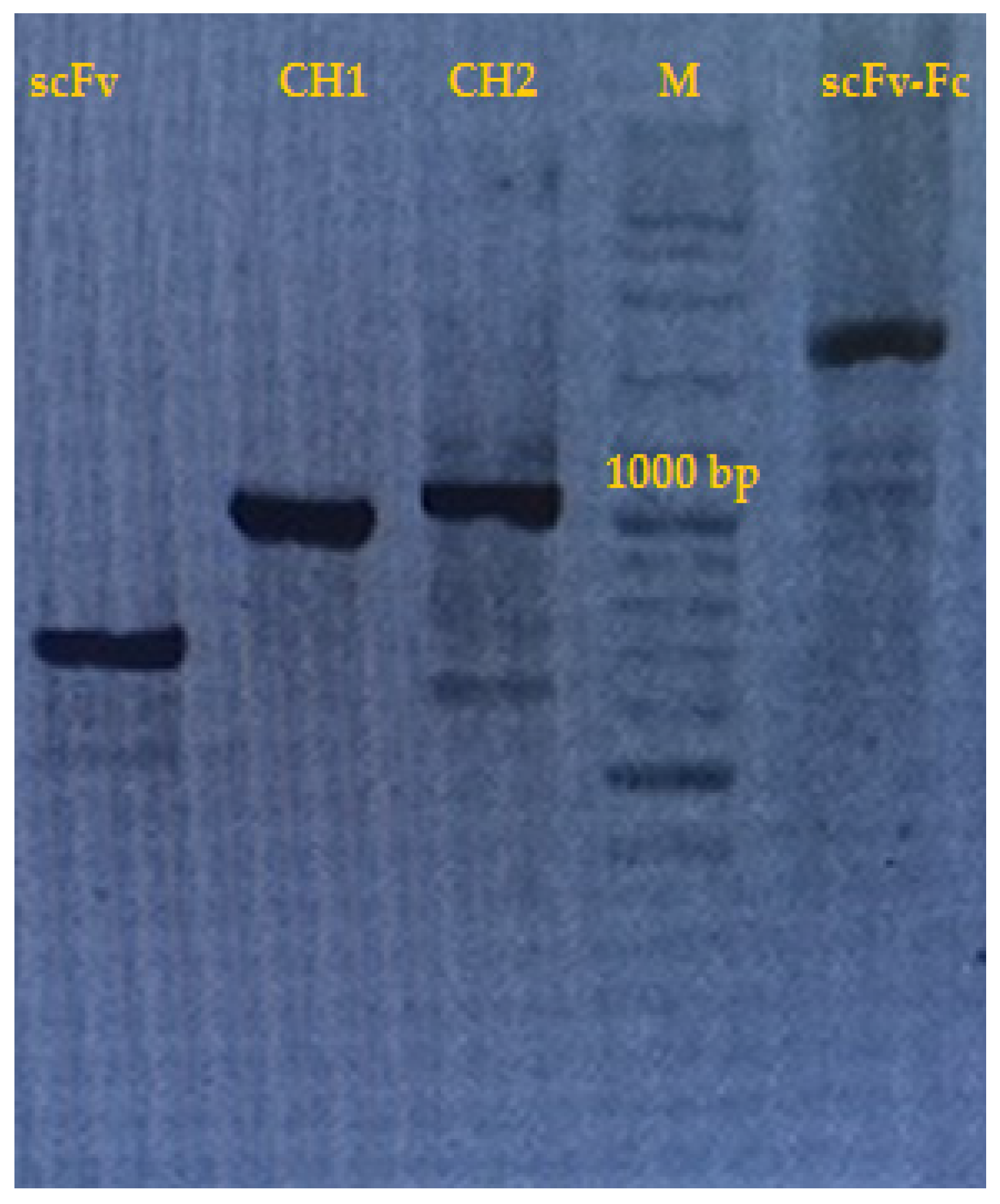
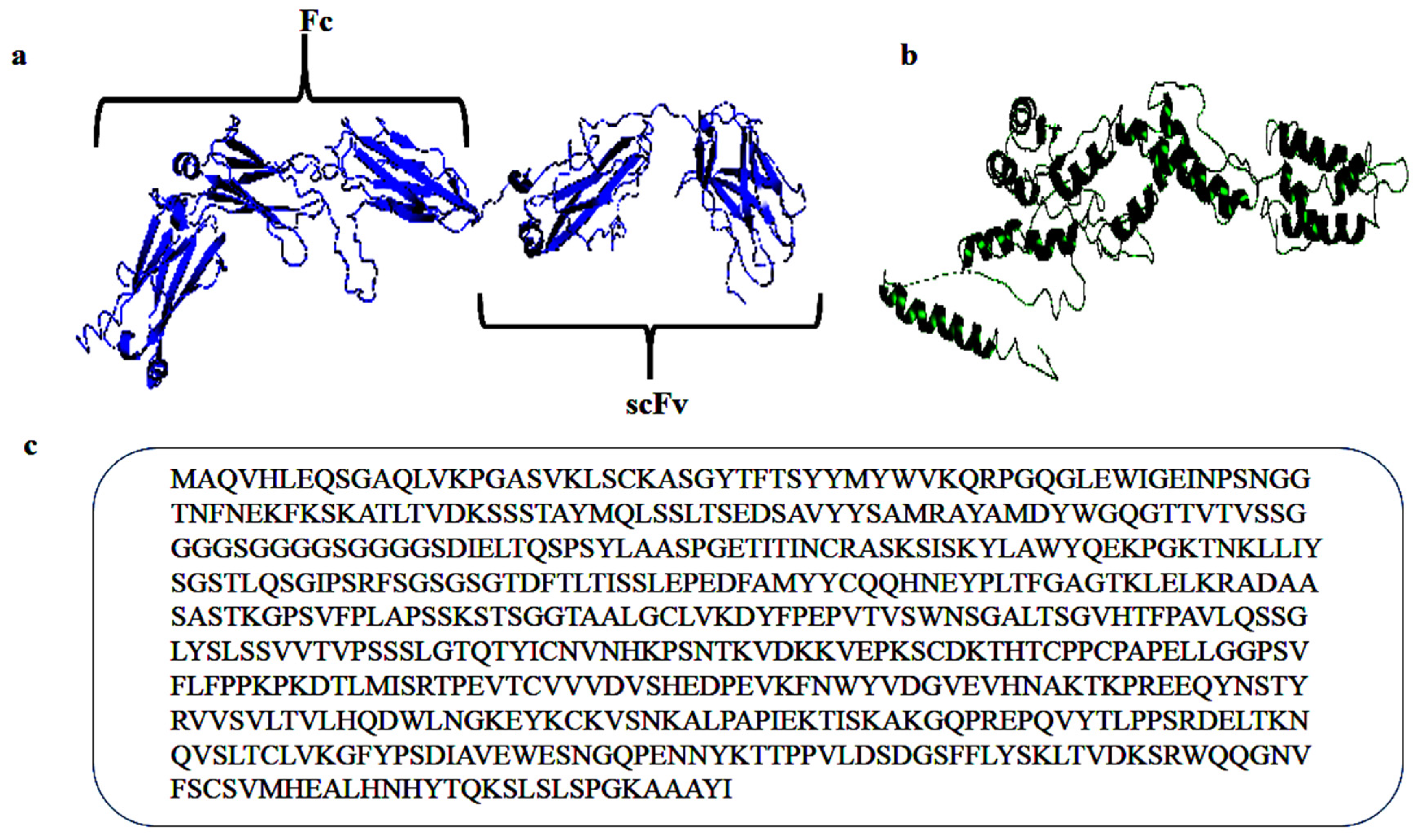
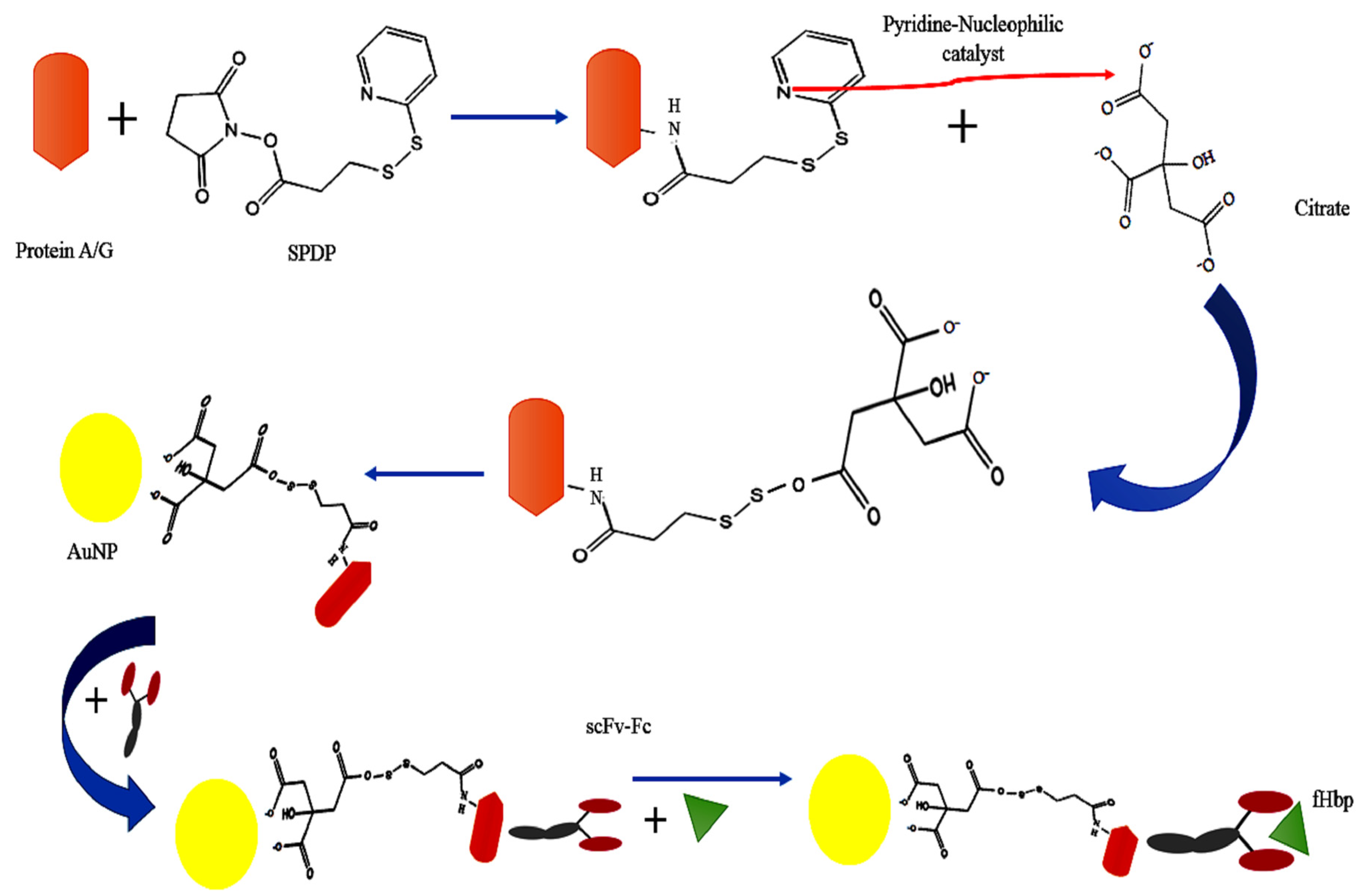
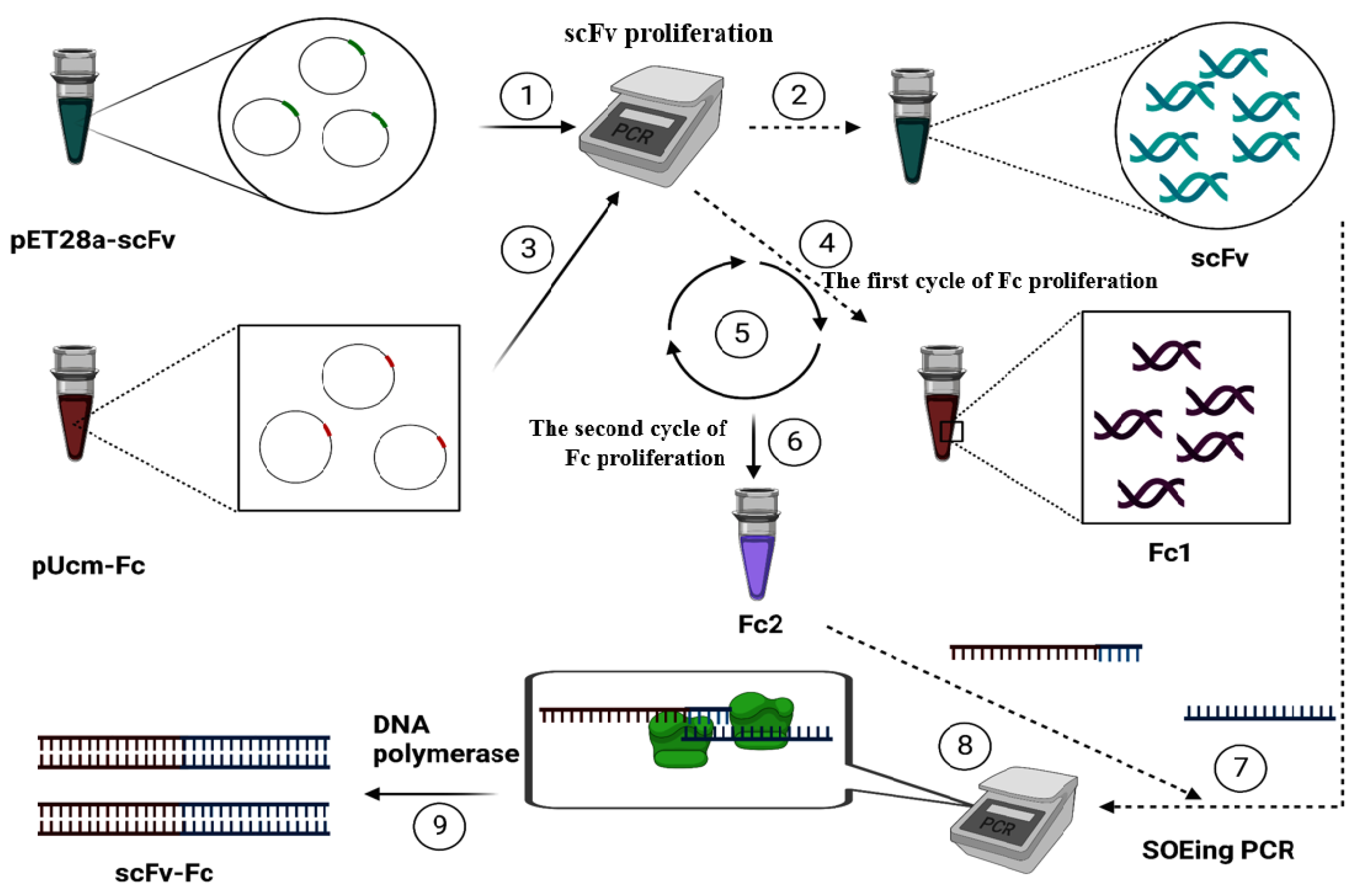
2. Results and Discussion
3. Materials and Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jazayeri, M.H.; Amani, H.; Pourfatollah, A.A.; Pazoki-Toroudi, H.; Sedighimoghaddam, B. Various methods of gold nanoparticles (GNPs) conjugation to antibodies. Sens. Bio-Sens. Res. 2016, 9, 17–22. [Google Scholar] [CrossRef]
- Yao, J.; Wang, Y.; Dai, Y.; Liu, C.C. Bioconjugated, single-use biosensor for the detection of biomarkers of prostate cancer. ACS Omega 2018, 3, 6411–6418. [Google Scholar] [CrossRef]
- Okyem, S.; Awotunde, O.; Ogunlusi, T.; Riley, M.B.; Driskell, J.D. High-Affinity Points Of Interaction on Antibody Allow Synthesis of Stable and Highly Functional Antibody–Gold Nanoparticle Conjugates. Bioconjug. Chem. 2021, 32, 1753–1762. [Google Scholar] [CrossRef]
- Conroy, P.J.; Hearty, S.; Leonard, P.; O’Kennedy, R.J. (Eds.) Antibody production, design and use for biosensor-based applications. In Seminars in Cell & Developmental Biology; Elsevier: Amsterdam, The Netherlands, 2009. [Google Scholar]
- Assali, A.; Razzazan, S.; Akhavan, O.; Mottaghitalab, F.; Adeli, M.; Atyabi, F. The bio-interface between functionalized Au NR@ GO nanoplatforms with protein corona and their impact on delivery and release system. Coll. Surf. B Biointerfaces 2019, 173, 891–898. [Google Scholar] [CrossRef]
- Raval, K.; Ganatra, T. Basics, types and applications of molecular docking: A review. IP Int. J. Compr. Adv. Pharmacol. 2022, 7, 12–16. [Google Scholar] [CrossRef]
- Bartuzi, D.; Kaczor, A.A.; Targowska-Duda, K.M.; Matosiuk, D. Recent advances and applications of molecular docking to G protein-coupled receptors. Molecules 2017, 22, 340. [Google Scholar] [CrossRef] [PubMed]
- Assali, A.; Akhavan, O.; Adeli, M.; Razzazan, S.; Dinarvand, R.; Zanganeh, S.; Soleimani, M.; Dinarvand, M.; Atyabi, F. Multifunctional core-shell nanoplatforms (gold@ graphene oxide) with mediated NIR thermal therapy to promote miRNA delivery. Nanomed. Nanotechnol. Biol. Med. 2018, 14, 1891–1903. [Google Scholar] [CrossRef] [PubMed]
- Rad, M.; Ebrahimipour, G.; Bandehpour, M.; Akhavan, O.; Yarian, F. Enzymatic Formation of Recombinant Antibody-Conjugated Gold Nanoparticles in the Presence of Citrate Groups and Bacteria. Catalysts 2022, 12, 1048. [Google Scholar] [CrossRef]
- Mirica, A.-C.; Stan, D.; Chelcea, I.-C.; Mihailescu, C.M.; Ofiteru, A.; Bocancia-Mateescu, L.-A. Latest Trends in Lateral Flow Immunoassay (LFIA) Detection Labels and Conjugation Process. Front. Bioeng. Biotechnol. 2022, 10, 922772. [Google Scholar] [CrossRef]
- Tripathi, K.; Driskell, J.D. Quantifying bound and active antibodies conjugated to gold nanoparticles: A comprehensive and robust approach to evaluate immobilization chemistry. ACS Omega 2018, 3, 8253–8259. [Google Scholar] [CrossRef]
- Gao, S.; Rojas-Vega, F.; Rocha-Martin, J.; Guisán, J.M. Oriented immobilization of antibodies through different surface regions containing amino groups: Selective immobilization through the bottom of the Fc region. Int. J. Biol. Macromol. 2021, 177, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Rispens, T.; Vidarsson, G. Human IgG Subclasses. In Antibody Fc; Elsevier: Amsterdam, The Netherlands, 2014; pp. 159–177. [Google Scholar]
- Zeng, X.; Shen, Z.; Mernaugh, R. Recombinant antibodies and their use in biosensors. Anal. Bioanal. Chem. 2012, 402, 3027–3038. [Google Scholar] [CrossRef]
- Xu, X.; Zhang, R.; Chen, X. Application of a single-chain fragment variable (scFv) antibody for the confirmatory diagnosis of hydatid disease in non-endemic areas. Electron. J. Biotechnol. 2017, 29, 57–62. [Google Scholar] [CrossRef]
- Ahangarzadeh, S.; Bandehpour, M.; Kazemi, B. Selection of single-chain variable fragments specific for Mycobacterium tuberculosis ESAT-6 antigen using ribosome display. Iran. J. Basic Med. Sci. 2017, 20, 327. [Google Scholar] [PubMed]
- Eyvazi, S.; Bandehpour, M.; Kazemi, B. Study of the production and the application of monoclonal antibodies: ScFv. Res. Med. 2017, 41, 138–151. [Google Scholar]
- Yang, H.; Zhong, Y.; Wang, J.; Zhang, Q.; Li, X.; Ling, S.; Wang, S.; Wang, R. Screening of a ScFv antibody with high affinity for application in human IFN-γ immunoassay. Front. Microbiol. 2018, 9, 261. [Google Scholar] [CrossRef]
- Gaba, M.; Gaba, P.; Singh, S.; Gupta, G. An overview on molecular docking. Int. J. Drug Dev. Res. 2010, 2, 219–231. [Google Scholar]
- Alabduladhem, T.O.; Bordoni, B. Physiology, Krebs Cycle. In StatPearls [Internet]; StatPearls Publishing: Tampa, FL, USA, 2021. [Google Scholar]
- Wiley, D.T.; Webster, P.; Gale, A.; Davis, M.E. Transcytosis and brain uptake of transferrin-containing nanoparticles by tuning avidity to transferrin receptor. Proc. Natl. Acad. Sci. USA 2013, 110, 8662–8667. [Google Scholar] [CrossRef]
- Yamazaki, S.; Shikida, N.; Takahashi, K.; Matsuda, Y.; Inoue, K.; Shimbo, K.; Mihara, Y. Lipoate-acid ligase a modification of native antibody: Synthesis and conjugation site analysis. Bioorgan. Med. Chem. Lett. 2021, 51, 128360. [Google Scholar] [CrossRef]
- Yarian, F.; Kazemi, B.; Bandehpour, M. Identification and characterization of a novel single-chain variable fragment (scFv) antibody against Neisseria meningitidis factor H-binding protein (fHbp). J. Med. Microbiol. 2018, 67, 820–827. [Google Scholar] [CrossRef]
- Zarghampoor, F.; Behzad-Behbahani, A.; Azarpira, N.; Khatami, S.R.; Fanian, M.; Aghdaie, M.H.; Dehbidi, G.R. A single tube overlap extension PCR method for splicing of multiple DNA fragments. Avicenna J. Med. Biotechnol. 2020, 12, 37. [Google Scholar] [PubMed]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Zhang, C.; Li, Y.; Pearce, R.; Bell, E.W.; Zhang, Y. Folding non-homologous proteins by coupling deep-learning contact maps with I-TASSER assembly simulations. Cell Rep. Methods 2021, 1, 100014. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Freddolino, P.L.; Zhang, Y. COFACTOR: Improved protein function prediction by combining structure, sequence and protein–protein interaction information. Nucleic Acids Res. 2017, 45, W291–W299. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, Y. I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef]
- Xu, D.; Zhang, Y. Improving the physical realism and structural accuracy of protein models by a two-step atomic-level energy minimization. Biophys. J. 2011, 101, 2525–2534. [Google Scholar] [CrossRef]
- Krieger, E.; Joo, K.; Lee, J.; Lee, J.; Raman, S.; Thompson, J.; Tyka, M.; Baker, D.; Karplus, K. Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: Four approaches that performed well in CASP8. Proteins Struct. Funct. Bioinformat. 2009, 77 (Suppl. S9), 114–122. [Google Scholar] [CrossRef]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35 (Suppl. S2), W407–W410. [Google Scholar] [CrossRef]
- Sippl, M.J. Recognition of errors in three-dimensional structures of proteins. Proteins Struct. Funct. Bioinform. 1993, 17, 355–362. [Google Scholar] [CrossRef]
- Colovos, C.; Yeates, T.O. Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Sci. 1993, 2, 1511–1519. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger, L.; DeLano, W. PyMOL [Internet]. 2020. Available online: http://www.pymol.org/pymol (accessed on 1 September 2022).
- Adasme, M.F.; Linnemann, K.L.; Bolz, S.N.; Kaiser, F.; Salentin, S.; Haupt, V.J.; Schroeder, M. PLIP 2021: Expanding the scope of the protein–ligand interaction profiler to DNA and RNA. Nucleic Acids Res. 2021, 49, W530–W534. [Google Scholar] [CrossRef] [PubMed]
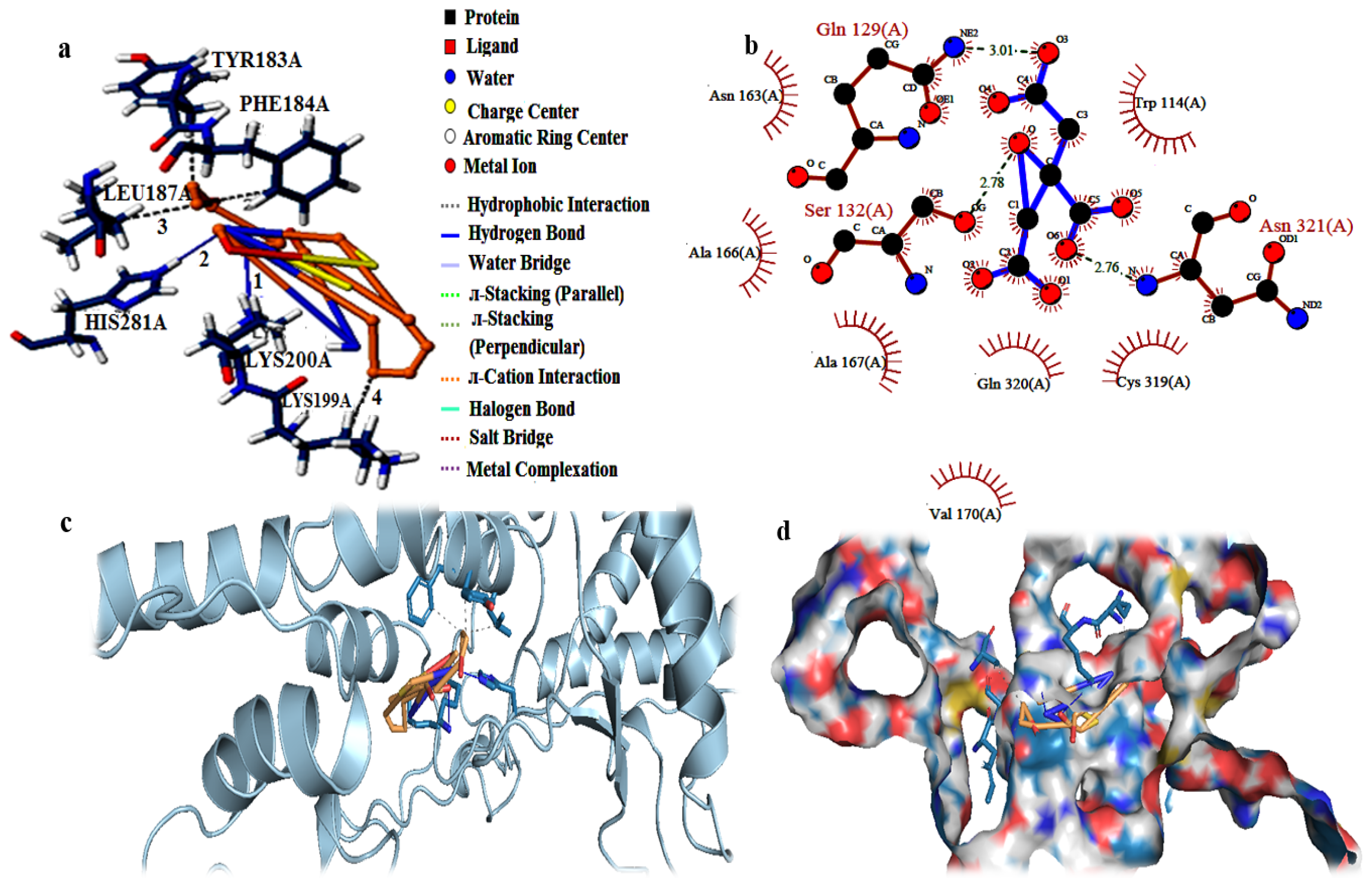
| Complex | Binding Affinity (Kcal/mol) |
|---|---|
| (scFv-Fc)/citrate | −5.5 |
| (scFv-Fc)/PHA | −2.6 |
| (scFv-Fc)/PVA | −2.2 |
| Protein A-G/citrate | −5.2 |
| Protein A-G/SPDP | −6.4 |
| (scFv’s antigen binding site)/citrate | −4.5 |
| Hydrogen Bonds | |||||||||
| Index | Residue | AA | Distance H-A | Distance D-A | Donor Angle | Protein Donor? | Side Chain? | Donor Atom | Acceptor Atom |
| 1 | 200A | LYS | 2.32 | 3.27 | 156.68 | √ | √ | 3297[N3+] | 6745[O3] |
| 2 | 281A | HIS | 2.44 | 3.44 | 169.68 | √ | √ | 4627[Nar] | 6730[N3] |
| Hydrophobic Interactions | |||||||||
| Index | Residue | AA | Distance | Ligand Atom | Protein Atom | ||||
| 1 | 183A | TYR | 3.55 | 6735 | 3006 | ||||
| 2 | 184A | PHE | 3.83 | 6736 | 3030 | ||||
| 3 | 187A | LEU | 3.95 | 6736 | 3081 | ||||
| 4 | 199A | LYS | 3.98 | 6740 | 3272 | ||||
| Fc Cycle 1 | Fc Cycle 2 | scFv | SOEing | |
|---|---|---|---|---|
| Forward | AGCGCCAGCACCAAGGG | TGAAACGGGCTGATGCTGCAAGCGCCAGCACCAAGG | ATATATATCCATGGGACAGGTCCACC | ATATATATCCATGGGACAGGTCCACC |
| Reverse | ATATATATGCGGCCGCCTTGCCGGGGCTCAGGC | ATATATATGCGGCCGCCTTGCCGGGGCTCAGGC | TGCAGCATCAGCCCGTTTC | ATATATATGCGGCCGCCTTGCCGGGGCTCAGGC |
| Chemical Name | PubChem CID | 2D Structure | 3D Structure | Charge |
|---|---|---|---|---|
| Citrate | 31,348 | 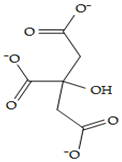 | 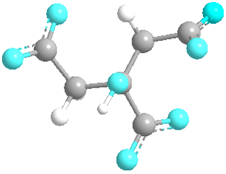 | Anionic |
| Allylamine hydrochloride (PAH) | 82,291 | 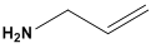 | 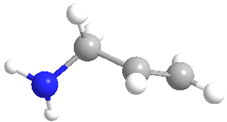 | Cationic |
| Polyvinyl alcohol (PVA) | 11,199 | 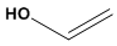 | 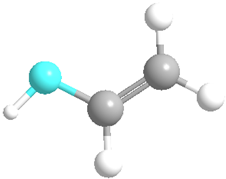 | Neutral |
| N-Succinimidyl 3-(2-pyridyldithio)propionate (SPDP) | 100,682 | 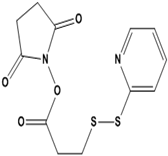 | 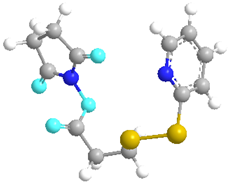 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rad, M.; Ebrahimipour, G.; Bandehpour, M.; Akhavan, O.; Yarian, F. SOEing PCR/Docking Optimization of Protein A-G/scFv-Fc-Bioconjugated Au Nanoparticles for Interaction with Meningitidis Bacterial Antigen. Catalysts 2023, 13, 790. https://doi.org/10.3390/catal13050790
Rad M, Ebrahimipour G, Bandehpour M, Akhavan O, Yarian F. SOEing PCR/Docking Optimization of Protein A-G/scFv-Fc-Bioconjugated Au Nanoparticles for Interaction with Meningitidis Bacterial Antigen. Catalysts. 2023; 13(5):790. https://doi.org/10.3390/catal13050790
Chicago/Turabian StyleRad, Maryam, Gholamhossein Ebrahimipour, Mojgan Bandehpour, Omid Akhavan, and Fatemeh Yarian. 2023. "SOEing PCR/Docking Optimization of Protein A-G/scFv-Fc-Bioconjugated Au Nanoparticles for Interaction with Meningitidis Bacterial Antigen" Catalysts 13, no. 5: 790. https://doi.org/10.3390/catal13050790
APA StyleRad, M., Ebrahimipour, G., Bandehpour, M., Akhavan, O., & Yarian, F. (2023). SOEing PCR/Docking Optimization of Protein A-G/scFv-Fc-Bioconjugated Au Nanoparticles for Interaction with Meningitidis Bacterial Antigen. Catalysts, 13(5), 790. https://doi.org/10.3390/catal13050790








