Abstract
Endoglucanases (EC 3.2.1.4) are important enzymes involved in the hydrolysis of cellulose, acting randomly in the β-1,4-glycosidic bonds present in the amorphous regions of the polysaccharide chain. These biocatalysts have been classified into 14 glycosyl hydrolase (GH) families. The GH7 family is of particular interest since it may act on a broad range of substrates, including cellulose, β-glucan, and xylan, an attractive feature for biotechnological applications, especially in the renewable energy field. In the current work, a gene from the thermophilic fungus Thermothielavioides terrestris, encoding an endoglucanase GH7 (TtCel7B), was cloned in the secretion vector pEXPYR and transformed into the high-protein-producing strain Aspergillus nidulans A773. Purified TtCel7B has a molecular weight of approximately 66 kDa, evidenced by SDS-PAGE. Circular dichroism confirmed the high β-strand content consistent with the canonical GH7 family β-jellyroll fold, also observed in the 3D homology model of TtCel7B. Biochemical characterization assays showed that TtCel7B was active over a wide range of pH values (3.5–7.0) and temperatures (45–70 °C), with the highest activity at pH 4.0 and 65 °C. TtCel7B also was stable over a wide range of pH values (3.5–9.0), maintaining more than 80% of its activity after 24 h. The KM and Vmax values in low-viscosity carboxymethylcellulose were 9.3 mg mL−1 and 2.5 × 104 U mg−1, respectively. The results obtained in this work provide a basis for the development of applications of recombinant TtCel7B in the renewable energy field.
1. Introduction
Lignocellulosic biomass (LB) represents a potential replacement for the finite and highly polluting fossil fuels that are commonly used as energy sources. It may be obtained from plant matter such as wood chips and agricultural waste, which are abundant, widely available, inexpensive, and renewable [1,2,3]. This type of biomass can be converted into a variety of energy forms, including biofuels, electricity, and heat, making it a flexible and sustainable energy source. The LB components, such as cellulose, hemicellulose, and lignin, can be converted into multiple high-value products through processes in biorefineries. This combination of product generation and bioenergy co-production reinforces the concept of an integrated lignocellulosic biorefinery approach, offering a more comprehensive solution for sustainable energy generation and resource utilization [4,5].
Cellulose is the most abundant natural polysaccharide on Earth and the essential structural component of the plant cell walls of lignocellulosic biomass [3,6]. It is a linear homopolymer of D-glucose units linked through β-1,4-glycosidic bonds [7]. The hydroxyl groups of the D-glucose units form strong networks of intra- and intermolecular hydrogen and Van der Waals bonds, resulting in a highly ordered structure known as the crystalline region. Moreover, the structure also presents disordered regions, arrayed irregularly, forming the amorphous regions [8,9].
Complete hydrolysis of cellulose requires the activity of several enzymes acting cooperatively to convert the homopolymer into fermentable sugars. Endo-1,4-β-glucanases (EC 3.2.1.4) are the main enzymes of the cellulolytic system, cleaving β-1,4 glycosidic bonds from cellulose. Endo-1,4-β-glucanases are responsible for releasing cello-oligosaccharides (COS) with different degrees of polymerization (DP), exhibiting a random degradation pattern, and creating new ends for the action of exo-β-1,4-glucanases (cellobiohydrolases or CBHs). CBHs (EC 3.2.1.91 and 3.2.1.176) hydrolyze the cellulose chains by the ends (reduced and non-reduced), releasing cellobiose as the main product. Finally, β-glucosidases (EC 3.2.1.21) convert COS and cellobiose into glucose [10,11,12,13]. The auxiliary activity (AA) enzymes such as lytic polysaccharide monooxygenases (LPMOs) (AA9, formerly GH61) are also important since these proteins can break the cellulose chain through an oxidative mechanism that is copper-dependent (EC 1.14.99.54 and EC 1.14.99.56), as well as carbohydrate-specific oxidoreductases such as the cellobiose dehydrogenase (CDH) (AA3) and cello-oligosaccharide dehydrogenase (AA7) (EC 1.1.99.18 and EC 1.1.99.-, respectively). AA3 and AA7 catalyze the oxidation of the reducing end C1-OH in cellobiose and COS to the corresponding lactones, donating electrons to LPMOs in the process [14].
The interest in endo-β-1,4-glucanases has grown remarkably due to their wide range of industrial applications, such as the food, feed, juice and beverage, textile, laundry, and pulp and paper industries. In addition, they have been heavily studied aiming at the enzymatic degradation of lignocellulosic materials in biorefineries to produce bioethanol [15,16,17,18,19,20]. Endoglucanases are found in 14 GH families: GH5 to GH10, GH12, GH26, GH44, GH45, GH48, GH51, GH74, and GH124. The endo-β-1,4-glucanases from the GH7 family are known for their promiscuous activity, acting on different substrates, including cellulose, β-glucan, lichenin, laminarin, and even xylan [21]. Therefore, there has been a growing attempt to identify and characterize novel endo-β-1,4-glucanases with improved properties for industrial use. It is necessary to overcome the limitations of current endo-β-1,4-glucanases and advance the field of industrial biotechnology through the development of more efficient and effective enzymes.
Thermothielavioides terrestris (formerly Thielavia terrestris) is a thermophilic fungus belonging to the class Sordariomycetes. It is known for its notable ability to grow in temperatures above 45 °C and produce extracellular enzymes, being classified as a good producer of thermostable enzymes of industrial interest, including cellulases and hemicellulases [22]. Among the CAZymes, different hydrolases GH7 (endo- and exo-glucanases) were identified and annotated depending on the strain of T. terrestris analyzed [23]. Nevertheless, the advancement of methods to enhance the yield and efficiency of fungal enzyme production could have substantial impacts on the economy and environment. Therefore, the heterologous protein expression by microbial systems is attractive due to their fast growth on inexpensive substrates, well-known genetics and physiology, and the availability of host strains [24]. Additionally, the use of filamentous fungi as hosts has benefits, such as their ability to secrete large amounts of protein and efficiently fold and modify it post-transduction [25]. Within this framework, in the current work, we report the cloning, production, purification, kinetics, and biochemical and structural characterization of an uncharacterized GH7 endoglucanase of T. terrestris (TtCel7B) expressed in heterologous host Aspergillus nidulans A773.
2. Results and Discussion
2.1. Sequence Analysis and Molecular 3D Modeling of TtCel7B
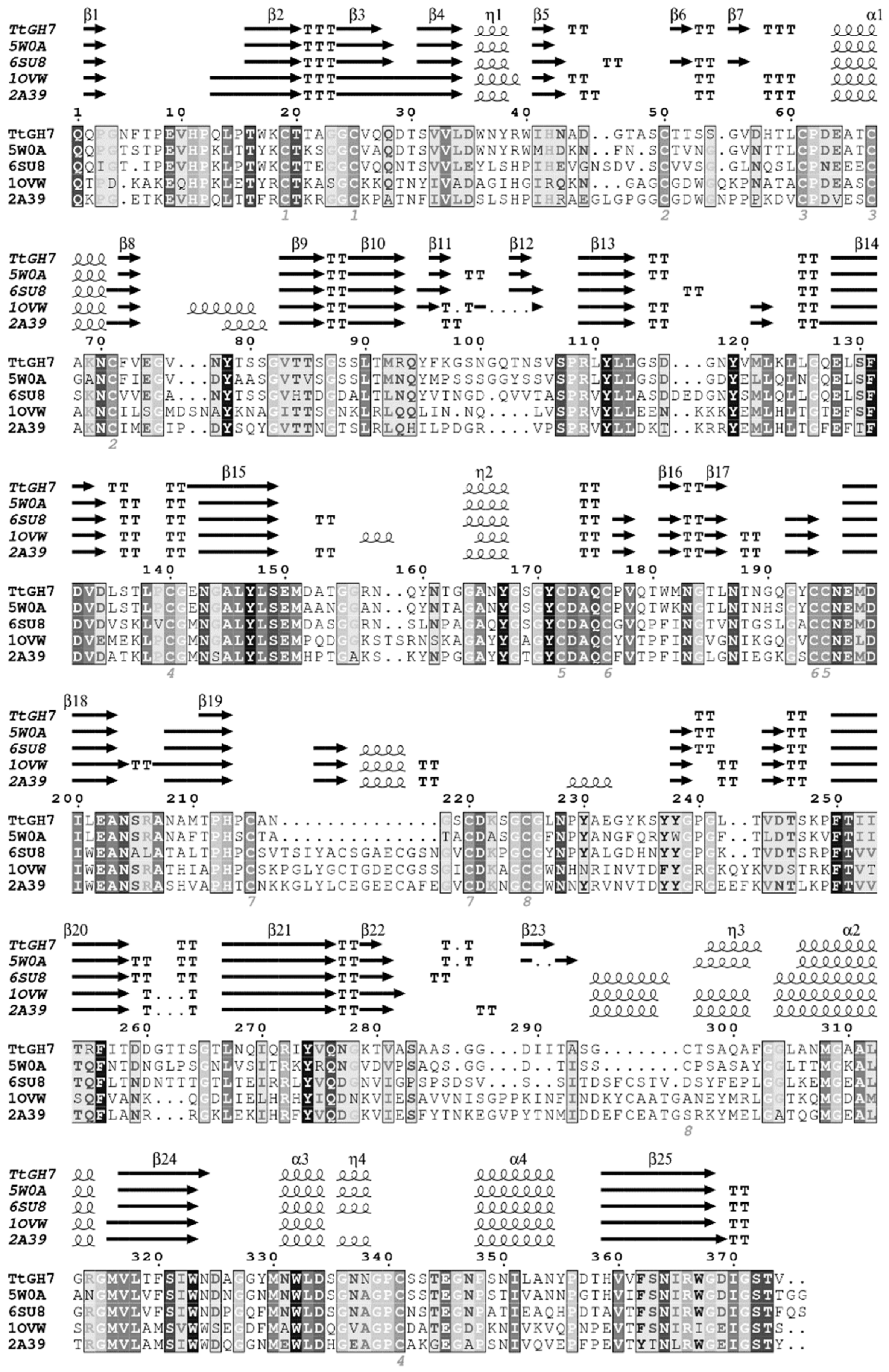
The protein sequence of TtCel7B was identified from CDS localized in T. terrestris chromosome III (AEO67421.1). TtCel7B has 464 amino acids, and domain analysis carried out using Pfam [26] showed the canonical catalytic domain (CCD) of the cellulase GH7 family (residues Gln23 to Thr397) and a C-terminal cellulose-binding module from family 1 (CBM1) positioned between the residues Ala428 and Leu464, united by a 29-peptide linker (residues Val398 to Thr427). A putative secretion signal sequence was annotated using the SignalP server [27] in the amino-terminal region, with a possible cleavage site between the residues 22 and 23. Structure-based amino acid sequence alignment of the CCD of TtCel7B with endoglucanases GH7 deposited in the Protein Data Bank (PDB) showed that putative CCD is composed of twenty-five β-strands, with four α-helices, aligned according to template structures (Figure 1).
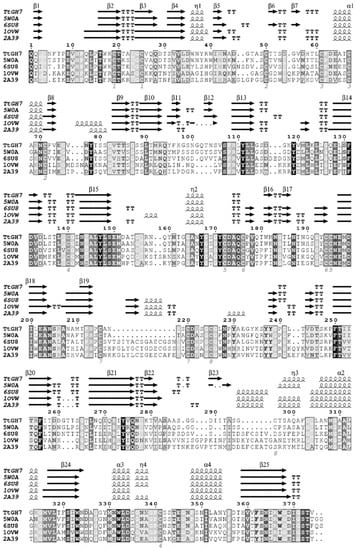
Figure 1.
Multiple sequence alignment and secondary structure estimation of the endoglucanase TtCel7B (TtGH7) of T. terrestris and PDB models from Trichoderma harzianum Cel7B (PDB id 5W0A), Talaromyces emersonii (PDB id 6SU8), Fusarium oxysporum (PDB id 1OVW), and Humicola insolens (PDB id 2A39). Dark- and light-shaded boxes indicate conserved residues. Secondary structure representations: α-helices are represented by α, arrows represent β-strands, and β-turns are marked with TT. The numbered positions indicate the disulfide bonds (Cys residues) predicted. Sequence alignments were performed using Clustal [28], and the figure was prepared with ESPriptS.
Conserved regions and residues were highlighted in boxes in the TtCel7B sequence, including the representative peptide sequence CNEMDIxEAN195–204 (where X represents any amino acid), highly conserved in the endoglucanases GH7 (Figure 1) [29]. Among twenty cysteines residues identified in the TtCel7B sequence, twenty-six were predicted to form eight disulfide bridges (Cys19–25, Cys50–72, Cys61–67, Cys140–341 Cys172–195, Cys176–194 Cys215–220, and Cys225–296), in analogous positions compared with Cel7B of T. harzianum [30], and other endoglucanases used as templates (Figure 1).
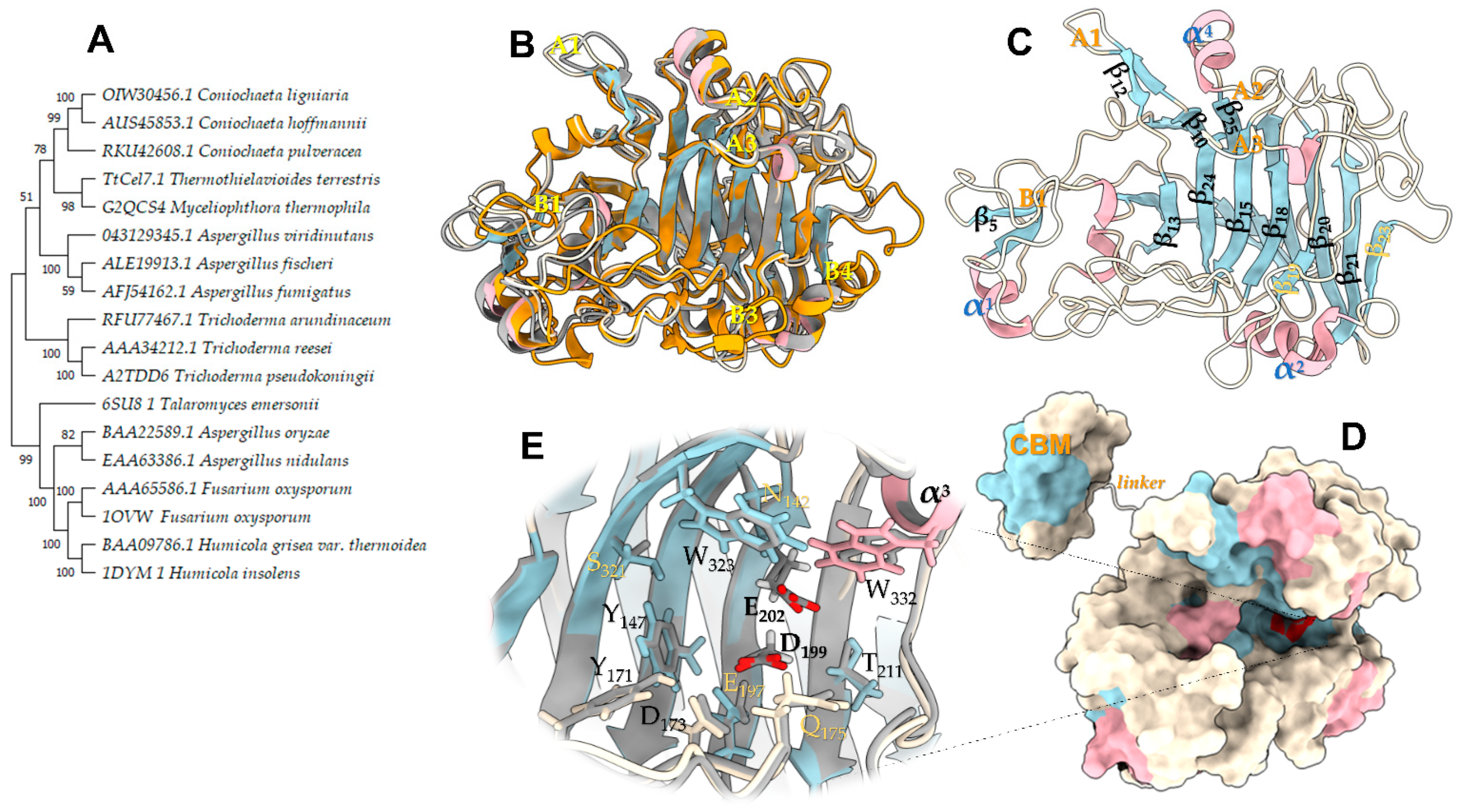
Phylogenetic analysis classified TtCel7B close to the endoglucanases of Thermothelomyces thermophilus (previously Myceliophthora thermophila), Coniochaeta pulveracea, and Aspergillus fumigatus (Figure 2A). The analysis of their sequences showed high identity (74.7, 74.2, and 74.2%, respectively) with these enzymes. Among the GH7 that have a known 3D structure, TtCel7B showed identity of 74, 73, and 59% with GH7 from T. reesei [PDB id 1GLM], T. harzianum [PDB id 5W0A], and Rasamsonia emersonii (T. emersonii) [PDB id 6SU8], respectively. The catalytic domain sequence of TtCel7B was structurally aligned with the Cel7B sequence [PDB id 5W0A], and the model was built without an amino acid sequence corresponding to the signal peptide, peptide linker, and CBM1. The final 3D model proposed with 374 residues was extensively checked and presents a ReFOLD global model quality score of 0.7010 (p-value < 0.001), z-DOPE score of −1.846, and GA341 score of 1.000, indicating a correct fold of structure [31,32,33].
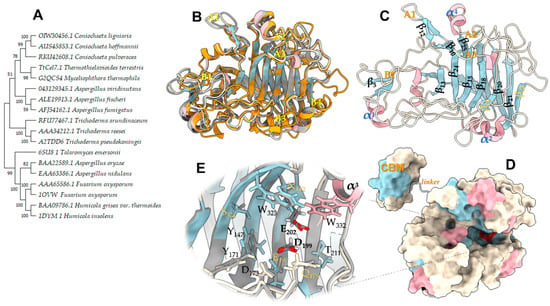
Figure 2.
Phylogenetic analysis and homology model of the endoglucanase TtCel7B. (A) Phylogenetic tree of TtCel7B (TtCel7.1) and close CAZy GH7 proteins. The sequences were retrieved from GenBank, Uniprot, and PDB. (B) A 3D model of TtCel7B of superimposed with template structures from T. harzianum [PDB id 5W0A], R. emersonii [PDB id 6SU8], and F. oxysporum [PDB id 1OVW]. (C) Modeled structure, in the same orientation as in panel (B), with the β-strands, α-helices, and main loops highlighted. (D) Space-filling model of panel (C) linked with the cellulose-binding module (CBM). (E) Zoom-in of the modeled catalytic tunnel. Figure was prepared using UCSF ChimeraX.
The 3D model of TtCel7B was superimposed on the structures of the three GH7 used as templates (Figure 2B). The CCD displays a topology typical to the GH7 family, a β-jelly roll forming a distorted β-sandwich, similar to the structurally homologous templates, including shorter flexible loops covering the catalytic tunnel area (Figure 2B,C) [34,35,36]. The six most relevant loops had been annotated according to nomenclature A1, A2 A3, B1, B3, and B4 in the template structures (Figure 2B). The B1, A1, A2, and A3 loops of TtCel7B are properly aligned with other EG templates. These loops have an important role in the cellulose binding in the catalytic tunnel [37,38]. On the other hand, structural analysis suggests that the B3 and B4 loops are truncated in TtCel7B, as with Cel7B of T. reesei and T. harzianum [PDB id 5W0A] (Figure 1 and Figure 2C) [30]. Putative N-glycosylation sites were annotated according to template ThCel7B [PDB id 5W0A], including the conserved site at Asn183 (Figure 1), an important glycosylation site for the correct orientation of substrates and products in the catalytic tunnel of the enzyme [30]. Other critical residues to the coordination and orientation of long-chain substrates into the catalytic tunnel, such as Tyr171, Tyr147, Trp323, and Trp332 residues [29,35], were adequately aligned with the Cel7B template (Figure 2E). The conserved triad catalytic (Glu202, Asp199, and Glu197) was properly aligned with similar residues, with a distance within ~5.4 Å between side chains of Glu residues (Figure 2E), which suggests that TtCel7B performs the standard retaining catalytic mechanism of hydrolysis, well described for GH7 cellulases [30,38].
2.2. Expression and Purification of Recombinant TtCel7B Endoglucanase
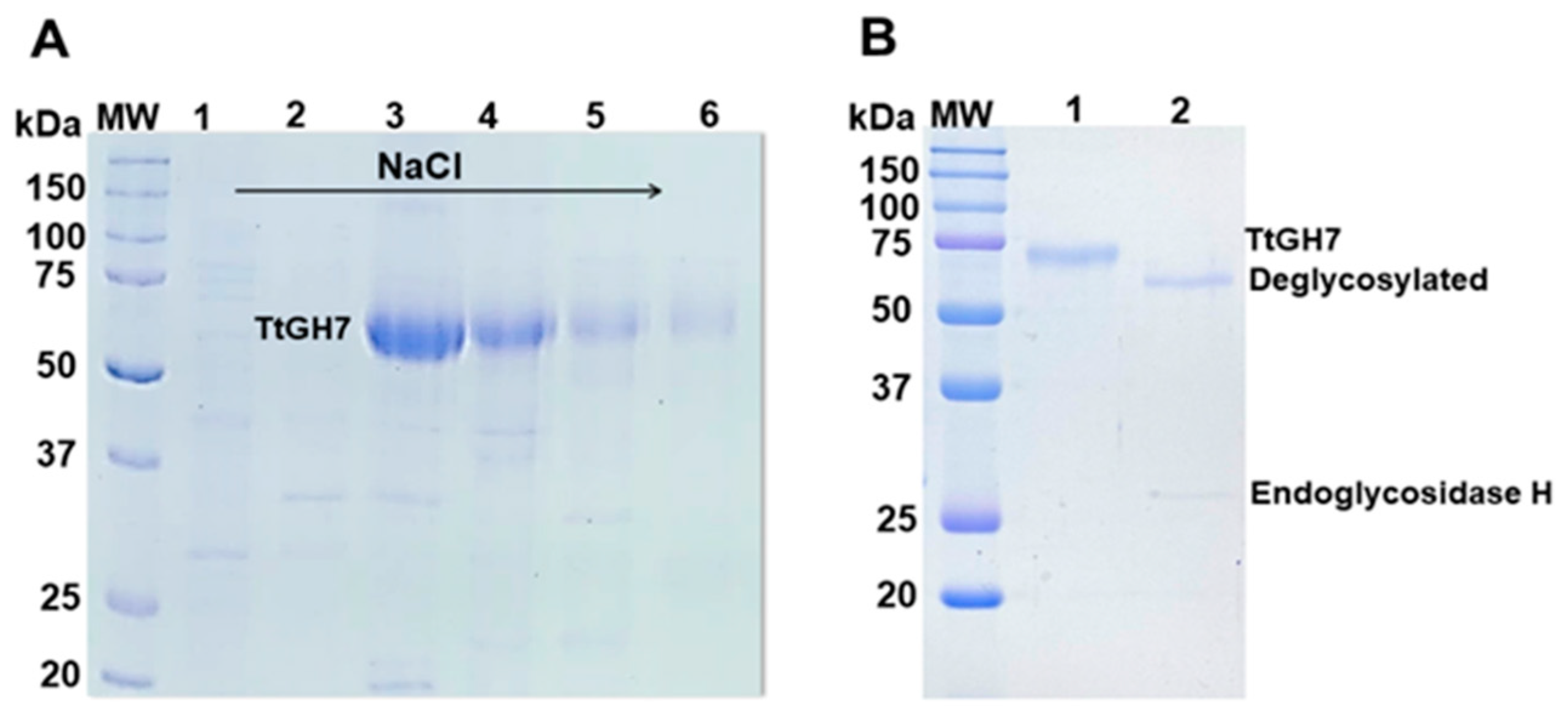
In the current work, we used the vector pEXPYR and the A. nidulans A773 strain as a heterologous host for the expression of TtCel7B. This expression system allows the overexpression along with the high-yield secretion and accumulation of the recombinant client protein with its native signal peptide in the culture supernatant [39]. Thus, the gene encoding endoglucanase TtCel7B was successfully cloned by Gibson Assembly into expression vector pEXPYR and transformed into A. nidulans A773 [39,40]. The resulting recombinant strains were selected by the reversion of the auxotrophic mark that allows growth in the absence of uracil and uridine, and the expression and secretion of TtCel7B in the culture supernatant were evaluated according to previous works [39]. A. nidulans A773 was selected and was cultivated in a minimal medium supplemented with pyridoxine and 5% of maltose for 48 h at 37 °C. The culture supernatant containing TtCel7B was concentrated and purified in two chromatography steps using a Hiprep Q 16/10 Fast Flow column (Figure 3A), followed by size exclusion in a Superdex 75 16/60 column. TtCel7B was successfully purified, as evidenced by a single band in SDS-PAGE, with a molecular weight (MW) estimated at 69.5 kDa (Figure 3B). Its MW value is higher than the theoretical value of 48.7 kDa predicted in silico by the ProtParam software-based amino acid sequence. Thus, mass spectrometry analysis was carried out to confirm the identity of TtCel7B. The fragment mass data obtained were analyzed and compared to theoretical masses predicted by the in silico peptide cleavage of the TtCel7B sequence (Supplementary Table S1), confirming the identity of TtCel7B (AEO67421.1). Therefore, these results indicated that the MW estimated of 69.5 kDa for TtCel7B corresponds to the glycosylated form. Protein glycosylation is usually observed in endoglucanases expressed by filamentous fungus [41], as recently reported for GH7 ThCel7B (T. harzianum) expressed in Aspergillus niger PY11 [42], Af-EGL7 (A. fumigatus) expressed in P. pastoris X-33 [43], ReCel7B (R. emersonii) expressed in Aspergillus oryzae [35], and for MtGH45 (M. thermophila) expressed in A. nidulans A773 [44].
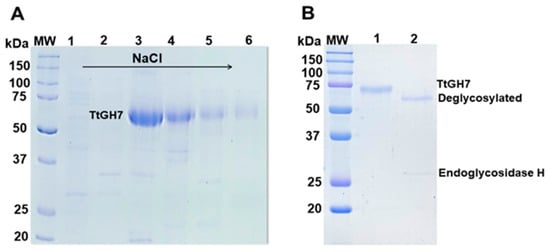
Figure 3.
Purification of the recombinant endoglucanase TtCel7B from T. terrestris expressed in A. nidulans A773. (A) SDS-PAGE fractions (lines 1–6) eluted from the ion-exchange Hiprep Q-FF chromatography column. (B) SDS-PAGE of TtCel7B purified (line 1) from size exclusion Superdex 75 10/300 GL column and analysis of the enzymatic deglycosylation assay (lane 2), showing the purified TtCel7B after digestion treatment with endoglycosidase H. MW: molecular weight standard ladder is shown as kDa.
2.3. Biochemical Characterization
2.3.1. Estimation of Carbohydrate Content of TtCel7B
To estimate the carbohydrate content of purified TtCel7B, deglycosylation assays (N-glycosylation) were performed using the endoglycosidase H from Streptomyces plicatus. SDS-PAGE shows the profile of native and deglycosylated TtCel7B, with a MW estimated of 65.2 kDa, resulting in carbohydrate content of approximately 6% (Figure 3B). These results corroborate the in silico analysis performed by the NetGlyc 1.0 Server and the N-glycosylation sites predicted from the 3D model of TtCel7B (Figure 2). On the other hand, the MW of deglycosylated TtCel7B is still higher than the theoretical value (48.7 kDa), which suggests a high level of O-glycosylation in TtCel7B, probably along the flexible peptide linker and CBM of the enzyme, a region rich in Thr and Ser residues (Figure 1), potential O-glycosylation sites [43,45,46,47]. Therefore, the high yield of secretion and higher molecular weight of recombinant TtCel7B show that the expression system of A. nidulans A773 promoted correct post-translational modifications on the enzyme.
2.3.2. Circular Dichroism (CD) and Determination of Melting Temperature (Tm) of TtCel7B
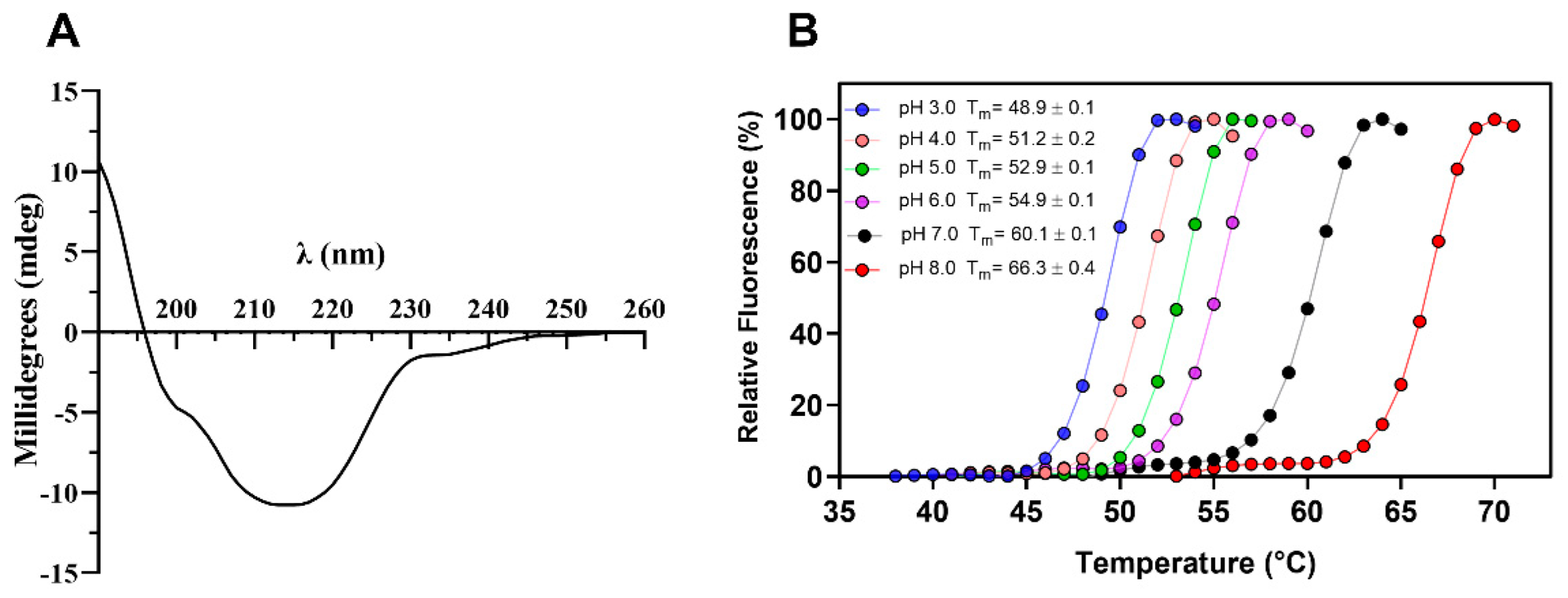
Circular dichroism spectroscopy analysis of TtCel7B showed a typical β-class protein profile spectrum, with the minimum negative band at 214 nm and the absence of a negative band at 222 nm (Figure 4A), suggesting low content of α-helices [42,48,49,50]. Analyses of the CD spectral data with the DichroWeb server [51] showed regular content of β-strands and α-helices of 53% and 8%, respectively. In addition, the Differential Scanning Fluorimetry (DSF) technique was used for the determination of the melting temperature (Tm) of TtCel7B at different pH levels. As shown in Figure 4B, there was a variation in the Tm depending on the pH, with minimal values at pH 3.0 (Tm 48.9 ± 0.1) and maximum at pH 8.0 (Tm 66.3 ± 0.4), indicating a strong effect of pH on the TtCel7B structure (Figure 4B). Similar results for the effect of pH and temperature on the secondary structure were reported for GH7 Cel7A and Cel7B of T. harzianum [42,50].
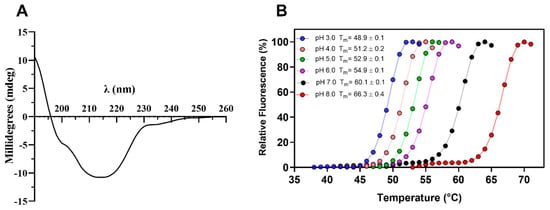
Figure 4.
(A) Secondary structure profile of TtCel7B determined by CD spectroscopy. (B) Thermal stability curves of TtCel7B at different pH and determination of melting temperature (Tm).
2.3.3. Panel Substrates and Effect of Different Compounds on TtCel7B Activity
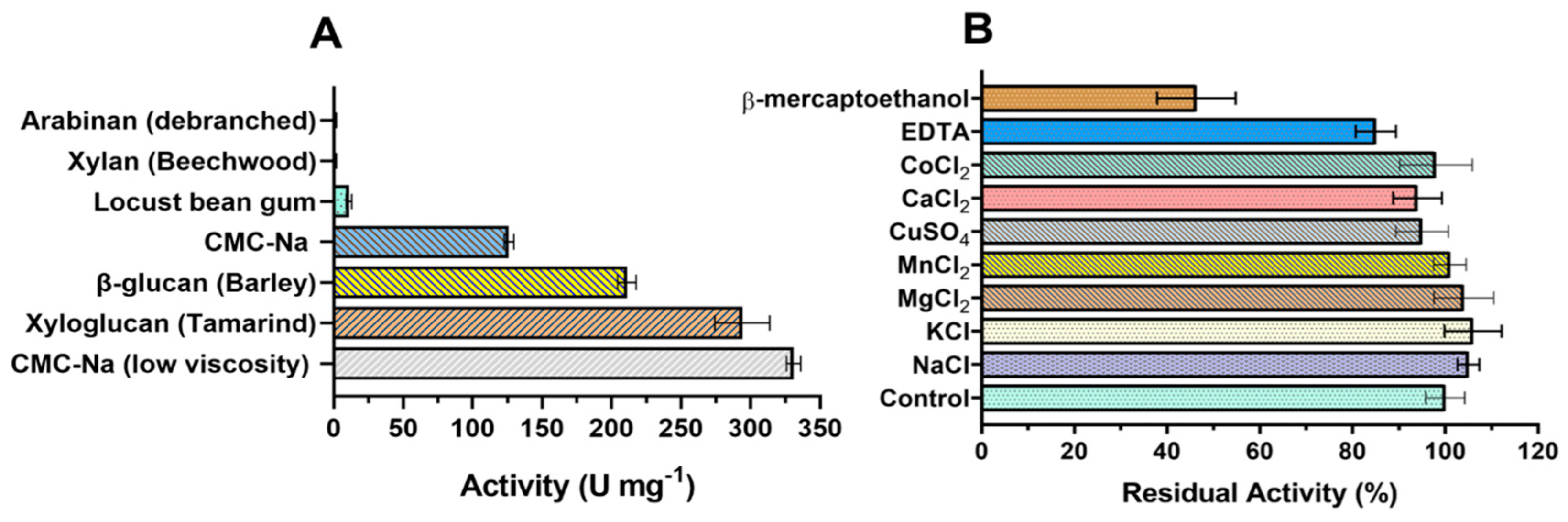
Endoglucanases GH7 are known for their promiscuous activity, acting on different substrates in addition to the cellulose, such as β-glucan, lichenin, xylan, and xyloglucan [21]. To characterize the substrate specificity of TtCel7B, we determined the activity of the enzyme against several polysaccharide substrates. As shown in Figure 5A, TtCel7B hydrolyzed different substrates from CMC to galactomannan (locust bean gum). Although the highest activity was against CMC of low viscosity, 331 ± 5 U mg−1, it also had high activity against xyloglucan from tamarind (294 ± 20 U mg−1) and β-glucan from barley (211 ± 4 U mg−1) (Figure 5A). It is not surprising that TtCel7B had significantly different activities against medium- and low-viscosity CMC, since differences in the degree of polymerization (DP) and degree of substitution (DS) are observed in the medium- and low-viscosity CMC [52,53]. For example, endoglucanase MtEG7 from M. thermophila was shown to be more active in barley β-glucan than CMC, and the possible reason is that CMC is substituted with methoxy side chains, and this could interfere with the enzyme activity against the CMC [54]. TtCel7B showed low activity against locust bean gum, and did not display activity in xylan or arabinan, which indicated higher specificity for polysaccharides β-1,4 and β-1,3-linked substrates containing glucan monomers, such as cellulose, xyloglucan, and β-glucan. Recently, one cellulase from T. terrestris Co3Bag1 was characterized (TtCel7A), showing excellent bifunctional cellulase/xylanase activity against CMC and beechwood xylan, but it also hydrolyzed oat bran, wheat bran, and sugarcane bagasse, releasing glucose and cellobiose as the main products [23]. TtCel7B also displayed activity against a broad range of polysaccharides, an important feature in the designed cocktail enzymatic, present in some GH7 of commercial interest. For example, the substrate specificity of TtCel7B is similar to that of Cel7B of T. reesei [21] and T. harzianum [42], and MtEG7a from M. thermophila [53].
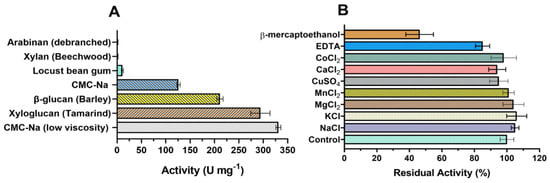
Figure 5.
(A) Substrate specificity of the endoglucanase GH7 from T. terrestris (TtCel7B). (B) Effect of different ions and compounds on TtCel7B activity against medium-viscosity CMC-Na.
The effect of different ions on TtCel7B activity was evaluated at a final concentration of 5 mmol L−1. As observed in Figure 5B, TtCel7B showed more than 90% relative activity for all ions evaluated. Interestingly, the negative effect of EDTA on TtCel7B activity was mild, retaining more than 80% of its relative activity against medium-viscosity CMC-Na (Figure 5B). On the other hand, a negative effect was observed on TtCel7B activity after the addition of β-mercaptoethanol, with a decrease >50% in the relative activity (Figure 5B). These results suggest a critical role of the disulfide bonds in the active structural conformation of TtCel7B. As described above, structural analysis of the 3D model of TtCel7B indicated eight putative disulfide bridges in the TtCel7B structure (Figure 1 and Figure 2).
2.3.4. Effect of Temperature and pH on TtCel7B Activity and Stability
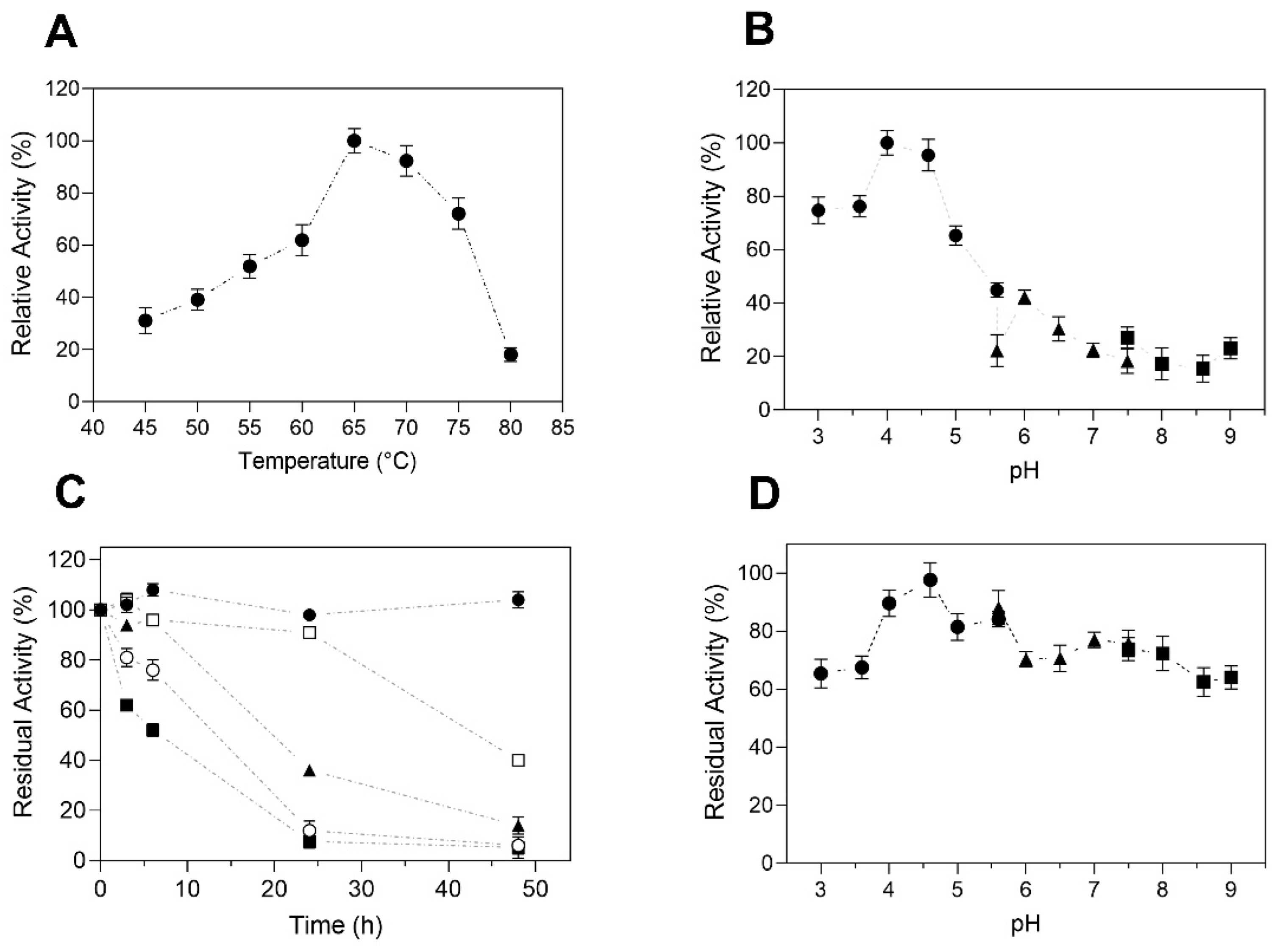
The effect of temperature on TtCel7B activity was determined by the hydrolysis of CMC over the range of 45 to 80 °C. The maximum endoglucanase activity was obtained at 65 °C, decreasing by 30% at 75 °C and more than 70% at 80 °C (Figure 6A). In the thermal stability studies, TtCel7B was most stable at 45 and 50 °C, with residual activities above 90% after 24 h of incubation. At the highest temperature, TtCel7B showed residual activity above 70 and 50% after 6 h of incubation at 60 and 65 °C, respectively (Figure 6C). In relation to the pH effect, TtCel7B was active over a wide range of pH values from 3.0 to 9.0, with maximum activity at pH 4.5 (Figure 6B). However, the activity decreases by more than 50% at a pH above 5.5, indicating a strong effect of pH on TtCel7B activity (Figure 6D). Interestingly, TtCel7B showed residual activities above 60% after being incubated at pH 3.0 to 9.0 for 24 h (Figure 6D). Thus, the activity at high-temperature and low-pH conditions confirmed that TtCel7B corresponds to an acidic thermophilic endoglucanase, with similar characteristics to those reported for MtEG7 of M. thermophila [53] and Cel7B of T. harzianum [42]. Cel7B (expressed on A. niger) showed maximum activity at 55 °C and pH 3.0, and, surprisingly, retained 100% of its residual activity after two months of incubation at 55 °C [42]. MtEG7 (expressed in Pichia pastoris) showed maximum activity at 60 °C and pH 5.0, with residual activities around 50% after 6 h of incubation at 70 and 80 °C [53].
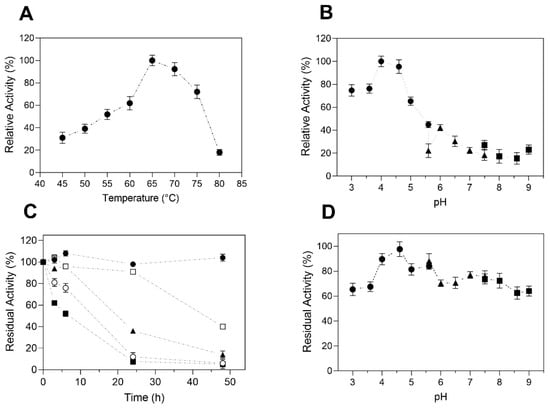
Figure 6.
Effect of temperature and pH on TtCel7B activity and stability. (A) The effect of temperature on activity was determined with CMC as the substrate, at pH 4.0; (B) effect of pH on activity, at 65 °C. Key: (●) citrate buffer, (▲) phosphate buffer, (■) tris-HCI buffer. (C) TtCel7B was incubated for 48 h at temperatures of 45 °C (●), 50 °C (□), 55 °C (▲), 60 °C (○), and 65 °C (■). Residual activity corresponding to the activity before incubation (100%); (D) stability at different pH values for 24 h, with 100% of residual activity corresponding to the activity at pH 4.0. Key: (●) citrate buffer, (▲) phosphate buffer, (■) tris-HCl buffer. Results were expressed as the average of triplicate assays ± the standard error of the mean.
Other recombinant endoglucanases with similar properties expressed in P. pastoris have been reported (Table 1). For example, Af-EGL7 from Aspergillus fumigatus showed maximum activity at 55 °C, at pH 5.0, and retained 100% of its residual activity after 72 h of incubation at 55 °C [43]. For Cel7A from Neosartorya fischerii, the maximum activity was reported at 60 °C, pH 5.0 [55]. Egl7A from T. emersonii presented maximum activity at 70 °C, at a pH of 4.5, and high stability over a range of pH values (1.0–11), with residual activities >70% after 1 h of incubation [56]. CTendo7 from Chaetomium thermophilum showed the best enzyme activity at 55 °C and pH 5.0, showing 61.3% of activity after 60 min at 80 °C, with almost all activity lost after 100 min at 90 °C [47].

Table 1.
General catalytic properties of TtCel7B compared with other endoglucanases.
2.4. Kinetic Parameters
The TtCel7B presented Vmax = 327.2 ± 16.4 U mg–1, KM = 9.49 ± 0.48 mg ml–1, kcat = 358.28 ± 17.9 s–1, and kcat/KM = 37.74 ±1.9 mL mg–1 s–1 using low-viscosity CMC-Na as a substrate (Table 1). Other heterologous GH7 produced in different expression systems have a wide range of kinetic parameter values (Table 1). C. thermophilum (CTendo7) has Vmax and KM of 59.6 ± 8.2 U mg–1 and 79.2 ± 5.8 mg ml–1, respectively; on the other hand, MtEG7a from M. thermophila has Vmax of 622.5 ± 86.4 U mg–1 and of KM 4.5 mg ml–1 on CMC-Na [47,53]. The GH7 from Aspergillus fumigatus (Af-EGL7) expressed in P. pastoris has Vmax and KM of 6193 ± 140 U mg–1 and 24.5 ± 0.6 mg ml–1, respectively, on CMC-Na [43]. On the other hand, the same enzyme expressed on E. coli presented Vmax and KM of 51.9 ± 0.007 U mg–1 and 209.7 ± 0.1 mg ml–1, respectively [57]. This shows that the appropriate expression system could be crucial when aiming at industrial applications, since the same enzyme could have very different catalytic efficiency (kcat/KM) when expressed in different systems (for example, 205.9 ± 0.8 for P. pastoris and 0.2 mL mg–1 s–1 for E. coli to Af-EGL7). T. harzianum endoglucanase I GH7 (Cel7B) has kinetic parameters measured on xyloglucan (Vmax of 0.22 μM s–1 and KM 1.98 ± 0.47 mg ml–1), despite being very well known as an endoglucanase that acts on CMC-Na [42]. Enzymes from the GH7 family can act in different substrates, such as cellulose, β-glucan, lichenin, laminarin, and even xylan, showing promiscuous activity on substrates [42,43,47,53,55]. This characteristic could be interesting for the industrial application of TtCel7B, since this enzyme could be used to produce different products from distinct sources.
3. Materials and Methods
3.1. Strains, Reagents, and Materials
The A. nidulans strain A773 (pyrG89; wA3; pyroA4) was obtained from the Fungal Genetic Stock Center (San Luis, MO, USA) and T. terrestris UAMH 3264 was purchased at the Microfungus Collection of the University of Alberta (Edmonton, AB, Canada).
A Miracloth membrane, 3,5-dinitrosalicilic acid (DNS), carboxymethylcellulose (CMC), and glucose (>95%), were purchased from Sigma-Aldrich (St. Louis, MO, USA). Locust bean, debranched arabinan, β-glucan, and xyloglucan were acquired from Megazyme (Bray, Wicklow, Ireland). Bradford protein assay and Precision Plus protein T.M. standards were purchased from Bio-Rad Laboratories (Hercules, CA, USA). Superdex 75 10/300 GL and Hiprep Q FF Superdex 75 10/300 GL columns were acquired from G.E. Healthcare (Uppsala, Sweden). All other reagents used for the assays were of analytical grade.
3.2. Sequence Analysis and Phylogenetic and Structural Homology Modeling
Multiple protein sequence alignment was carried out with the ClustalW algorithm [28] using sequences obtained from NCBI (National Center for Biotechnology Information) [58], Uniprot (Universal Protein Resource) [59], and PDB (Protein Data Bank) [60]. Selected PDB templates were used to predict the secondary structure and features of the amino acid sequence using the ENDscript server [61]. A phylogenetic tree was built with the neighbor-joining method using the MEGA software (version 11), with bootstrapping of 1000 replicates [62]. The 3D model of TtCel7B was built by homology using the MODELLER software [63]. The templates T. reesei [PDB id 1GLM] and T. harzianum [PDB id 5W0A] were acquired from PDB. Five hundred models were generated and optimized by the VTFM method within MODELLER and one was considered the final model according to the embedded normalized DOPE [64] and GA3415 scores [65]. The quality of the final model was assessed by the programs ReFOLD [32,33] and PROCHECK [66]. The AlphaFold database [67] embedded in UCSF ChimeraX [68] was used to predict the TtCel7B 3D model with a peptide linker and carbohydrate-binding module (CBM) structure.
3.3. Cloning, Expression, and Purification of the Endoglucanase TtCel7B
Extraction of the genomic DNA of T. terrestris was carried out using the kit Wizard Genomic DNA Purification (Promega, Madison, WI, USA), in accordance with the instructions of the manufacturer. Primers were designed based on the coding sequence of TtCel7B and amplified from genomic DNA by polymerase chain reaction (PCR) with the oligonucleotides TtGH7 fwd CATTACACCTCAGCAATGGGCCAGAAGACGCTGCACGGATT and TtGH7 rev GTCCCGTGCCGGTTATTAGAGGCACTGGTAGTACCAGGGGTTCAG (Exxtend Oligo Solutions) using Phusion High-Fidelity DNA Polymerase (New England Biolabs, Ipswich, MA, USA). The amplified genes were used to assemble the pEXPYR vector via the Gibson Assembly Method using the complementary regions (in bold) [69], and the recombinant pEXPYR-TtCel7B construct was used to transform A. nidulans A773, as previously described [39]. The A. nidulans recombinant strains were further selected after growing in the absence of uracil and uridine in the minimal medium, and the expression of TtCel7B was confirmed by SDS-PAGE [39].
Heterologous expression of TtCel7B was carried out according to Damásio et al. (2012) [70] and Segato et al. (2017) [71]. Briefly, a spore suspension (108 spores mL−1) of recombinant strain was inoculated in 500 mL of minimal medium supplemented with 5% maltose (w/v), pH 6.5, and incubated at 37 °C for 48 h. Afterward, the crude extract was filtered in a Büchner funnel with a Miracloth® membrane, followed by concentration and dialysis in 50 mmol L−1 phosphate buffer, pH 6.0, using the QuixStand Benchtop Systems tangential concentrator with a 30 kDa Hollow Fiber Cartridge (GE Healthcare, Chicago, IL, USA). Next, the crude extract was loaded on an anion exchange column (Hiprep Q FF 16/10) equilibrated with 50 mmol L−1 phosphate buffer, pH 6.0, integrated with an ÄKTA Purifier 900 (GE Healthcare) chromatography system. Protein elution was monitored at 280 nm, with a linear gradient from 0 to 1 mol L−1 of NaCl. The protein samples of interest were selected by SDS-PAGE, pooled, and concentrated (30 kDa cutoff—Vivaspin GE Healthcare). The recovered sample was loaded in a size exclusion column (Superdex 75 10/300 GL), equilibrated with phosphate buffer 50 mmol L−1, NaCl 150 mmol L−1, pH 6.0. Protein elution was monitored at 280 nm, and the fractions of interest were selected as described above, and stored at −20 °C.
3.4. Determination of Endoglucanase Activity
Endoglucanase activity was determined by the reducing sugars method using 3,5 dinitrosalicylic acid (DNS) [72]. The assays were carried out with 50 µL of the enzyme, 50 µL of citrate buffer (0.1 mol L−1, pH 5.0), and 100 µL of 1% CMC (w/v). The assay was performed for 10 min at 50 °C and 1300 rpm. After incubation, 200 µL of DNS was added to the assay mixture and boiled at 98 °C for 5 min. Afterward, samples of 100 μL were collected for the quantification of reducing sugars at 540 nm using microplate readers (Spectramax M2, Molecular Devices, San Jose, CA, USA). A standard curve of D-glucose was used to estimate the reducing sugar concentration equivalents. One unit of enzyme activity (U) was defined as the amount of enzyme that catalyzed the release of 1 μmoL of reducing sugar per minute under the assay conditions. Assay controls (blank) were performed without TtCel7B, which was added after the addition of DNS (prior boiling).
3.5. Protein Content, Electrophoresis, and Deglycosylation Analysis
Protein content was determined by the method of Bradford [73], using the Bradford Protein Assay Kit (Bio-Rad) with bovine serum albumin as the standard. Electrophoresis of protein samples (15% SDS-PAGE) was done according to Laemmli [74]. Gels were stained with Coomassie Brilliant Blue R and de-stained with a solution of ethanol/acetic acid/water (5/1/4 v/v/v). Glycosylation of TtCel7B was analyzed with Endoglycosidase H (EndoH) (Roche, Mannheim, Germany), using the manufacturer’s instructions. Briefly, 5 μg of purified TtCel7B was mixed with 250 mU of EndoH in sodium acetate buffer 50 mM pH 5.5 and incubated at 37 °C overnight. The samples were analyzed by SDS-PAGE after treatment and the estimation of the carbohydrate content was performed from the migration difference between the treated and untreated TtCel7B compared to the molecular mass standard.
3.6. Liquid Chromatography–Tandem Mass Spectrometry (LC–MS/MS)
The spot of the purified TtCel7B from the Coomassie-stained SDS-PAGE was excised, reduced, alkylated, and submitted to in situ trypsin gel digestion (Promega). One microliter of tryptic peptides was applied in a C18 column (75 μm × 100 mm) for desalting. The peptides were analyzed by RP-nanoUPLC (nanoAcquity, Waters, Milford, MA, USA) coupled with a Q-TOF Ultimamass Spectrometer (Waters) with a nano-electrospray source at a flow rate of 0.6 μL min−1. The gradient was set to 2–90% (v/v) acetonitrile in 0.1% (v/v) formic acid over 45 min. The equipment was set up on the top three modes. The software MassLynx v.4.1 (Waters) was used to obtain the spectra, and the software Mascot Distiller v.2.3.2.0, 2009 (Matrix Science Ltd., London, UK) was used to convert the raw data files to a peak list format (mgf). The engine Mascot v.2.3 (Matrix Science Ltd.) was used to compare the MS/MS profiles against predicted protein sequences of T. terrestris [75,76].
3.7. Differential Scanning Fluorimetry (DSF) and Circular Dichroism (CD)
Differential Scanning Fluorimetry (DSF) was used for the determination of the melting temperature (Tm) of TtCel7B. In a 96-well PCR plate was added 2 μg of purified TtCel7B, 2 μL of SYPRO Orange solution (50x), and ultrapure water up to 20 μL in each well. The plate was sealed with Sealing Tape for Optical Assays (Bio-Rad #2239444) and centrifuged at 4000× g for 2 min and 4 °C. The plates were heated from 20 to 90 °C in the thermocycler (in increments of 1 °C) with a scanning rate of 1 °C min−1. The thermocycler lid was heated to 100 °C to avoid condensation effects during the experiment. The changes in the fluorescence in the plate wells were monitored simultaneously at 470 and 570 nm, which correspond to the excitation and emission lengths of SYPRO® Orange, respectively. The melting temperature (Tm) was calculated by nonlinear regression using a sigmoidal curve through the Boltzmann equation to adjust fluorescence data.
The analysis of the secondary structure of the proteins was performed by circular dichroism (CD) using a Jasco-810 spectropolarimeter (JASCO Inc., Tokyo, Japan). Protein samples (0.1 mg mL−1) mixed in Tris-HCl buffer (10 mM) were added in a quartz cuvette of 200 µL, with an optical path length of 0.1 mm. The data were collected using a scanning speed of 50 nm/min, spectral bandwidth of 3 nm, and response time of 1 s. Buffer spectra without protein were subtracted in all experiments (blank), with all measurements performed with an average of six accumulations collected, in the range of far UV (190–250 nm).
3.8. Effect of Different Ions and Compounds on TtCel7B Activity
The effect of different ions on TtCel7B activity was evaluated at a final concentration of 5 mmol L−1 in a standard activity assay. The ions added in salt form were CoCl2. 6H2O, CaCl2, CuSO4, MnCl2·4H2O, MgCl2·6H2O, KCl, and NaCl. Reducing agent β-mercaptoethanol and chelating agent EDTA (ethylenediaminetetraacetic acid) also were evaluated at the same final concentration. The relative activities were calculated concerning a control performed under a standard condition of the assay and attributed a relative activity of 100%. Results were expressed as the average of triplicate assays ± the standard error of the mean.
3.9. Effect of Temperature and pH on TtCel7B Activity and Stability
The effect of temperature on TtCel7B activity was determined by the hydrolysis of CMC-Na over the range of 45 to 80 °C, at pH 4.5. The thermal stability was assessed via the incubation of TtCel7B in a water bath for 48 h at temperatures ranging from 45 to 65 °C. The effect of pH on activity was analyzed over the range of 3.0 to 9.0 at 50 °C using 0.05 mol L−1 of the buffers citrate (3.0 to 5.5), phosphate (5.5 to 7.5), and Tris-HCl (7.5 to 9.0). The pH stability of TtCel7B was determined by incubation for 24 h, at 25 °C, at different pH values, with the same buffers used above (3.0 to 9.0). The residual activities were calculated relative to the control, which was treated identically but without incubation and attributed a residual activity of 100%. Results were expressed as the average of triplicate assays ± the standard error of the mean.
3.10. Determination of Kinetic Parameters
The kinetic parameters of maximum velocity (Vmax) and Michaelis–Menten constant (KM) were determined using low-viscosity carboxymethylcellulose (CMC-Na) (Sigma-Aldrich #C5678). The assay was carried out for 10 min in citrate buffer (50 mM) at pH 4.5, 65 °C, and CMC concentrations ranging from 0.02 to 32 mg ml−1. The kinetic parameters were calculated by the software SigrafW [77] from the experimental data using nonlinear regression (Hill equation). The values are presented here as the mathematical mean of three independent measurements from two protein preparations (n = 6).
4. Conclusions
In conclusion, the present work describes the cloning and characterization of an endoglucanase GH7 (TtCel7B) from the thermophilic fungus Thermothielavioides terrestris. The gene was successfully transformed into the high-protein-producing strain Aspergillus nidulans A773, and the purified enzyme showed a molecular weight of 66 kDa. Biochemical assays demonstrated that TtCel7B has a wide range of activity in terms of pH and temperature, with the highest activity at pH 4.0 and 65 °C, and it was also stable over a wide range of pH. Circular dichroism showed high content of β-strands, consistent with the canonical GH7 family β-jellyroll fold found in other GH7. The enzyme showed higher specificity for polysaccharides β-1,4 and β-1,3-linked substrates, such as cellulose, xyloglucan, and β-glucan. The promiscuous activity could be beneficial when aiming at the production of cocktails, since TtCel7B could act in different polysaccharides in lignocellulosic biomass hydrolysis. Finally, the results of this study provide a foundation for the further development of TtCel7B for applications in the renewable energy field.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/catal13030582/s1, Table S1. Amino acid sequence of the endoglucanase (TtCel7B) from T. terrestris. The peptides corresponding to those identified by LC/MS analyses are shown in bold. The mass profiles obtained were used in database MASCOT (Matrix Science, London, UK).
Author Contributions
Conceptualization, methodology, writing—original draft preparation: R.C.A., J.C.S.S., G.S.A. and D.d.A.; software, validation, formal analysis, investigation, visualization: R.C.A., J.C.S.S., G.S.A., D.d.A., L.P.M., R.J.W., M.S.B., G.L.B. and F.S.; data curation: R.C.A., G.S.A., J.C.S.S., D.d.A. and L.P.M.; review, resources and funding acquisition: R.C.A., R.J.W., M.S.B. and F.S.; writing—review and editing, resources, supervision, project administration, funding acquisition: M.d.L.T.M.P. All authors have read and agreed to the published version of the manuscript.
Funding
The authors gratefully acknowledge FAPESP (São Paulo Research Foundation, Grant No. 2018/07522-6, 2019/22284-7, 2021/06679-1, and 2022/042279) and FCT (POCI-01-0145-FEDER-032206)—transnational cooperation project EcoTech, and the National Institute of Science and Technology of Bioethanol, INCT, CNPq 465319/2014-9/FAPESP n° 2014/50884-5) for the financial support. Research scholarships were granted to R.C.A., D.A., L.P.M., and J.C.S.S. by FAPESP (Grant No. 2020/00081-4, 2020/15510-8, 2016/17582-0, and 2019/21989-7, respectively), CNPq 310340/2021-7 to M.L.T.M. Polizeli, and to G.S.A. by CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Finance Code 001).
Data Availability Statement
Not applicable.
Acknowledgments
We acknowledge Mariana Cereira and Mauricio de Oliveira for the technical support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Devi, A.; Bajar, S.; Kour, H.; Kothari, R.; Pant, D.; Singh, A. Lignocellulosic Biomass Valorization for Bioethanol Production: A Circular Bioeconomy Approach. BioEnergy Res. 2022, 15, 1820–1841. [Google Scholar] [CrossRef] [PubMed]
- Haq, I.U.; Qaisar, K.; Nawaz, A.; Akram, F.; Mukhtar, H.; Zohu, X.; Xu, Y.; Mumtaz, M.W.; Rashid, U.; Ghani, W.A.W.A.K.; et al. Advances in Valorization of Lignocellulosic Biomass towards Energy Generation. Catalysts 2021, 11, 309. [Google Scholar] [CrossRef]
- Wang, F.; Ouyang, D.; Zhou, Z.; Page, S.J.; Liu, D.; Zhao, X. Lignocellulosic Biomass as Sustainable Feedstock and Materials for Power Generation and Energy Storage. J. Energy Chem. 2021, 57, 247–280. [Google Scholar] [CrossRef]
- Gomes, D.; Cunha, J.; Zanuso, E.; Teixeira, J.; Domingues, L. Strategies towards Reduction of Cellulases Consumption: Debottlenecking the Economics of Lignocellulosics Valorization Processes. Polysaccharides 2021, 2, 287–310. [Google Scholar] [CrossRef]
- Rajesh Banu, J.; Preethi; Kavitha, S.; Tyagi, V.K.; Gunasekaran, M.; Karthikeyan, O.P.; Kumar, G. Lignocellulosic Biomass Based Biorefinery: A Successful Platform towards Circular Bioeconomy. Fuel 2021, 302, 121086. [Google Scholar] [CrossRef]
- Thapa, S.; Mishra, J.; Arora, N.; Mishra, P.; Li, H.; O′Hair, J.; Bhatti, S.; Zhou, S. Microbial Cellulolytic Enzymes: Diversity and Biotechnology with Reference to Lignocellulosic Biomass Degradation. Rev. Environ. Sci. Biotechnol. 2020, 19, 621–648. [Google Scholar] [CrossRef]
- Sharma, H.K.; Xu, C.; Qin, W. Biological Pretreatment of Lignocellulosic Biomass for Biofuels and Bioproducts: An Overview. Waste Biomass Valorization 2019, 10, 235–251. [Google Scholar] [CrossRef]
- Sun, S.; Sun, S.; Cao, X.; Sun, R. The Role of Pretreatment in Improving the Enzymatic Hydrolysis of Lignocellulosic Materials. Bioresour. Technol. 2016, 199, 49–58. [Google Scholar] [CrossRef]
- Michelin, M.; Teixeira, J.A. Biocatalyst Systems for Xylooligosaccharides Production from Lignocellulosic Biomass and Their Uses. In Biomass, Biofuels, Biochemicals; Singh, S.P., Pandey, A., Singhania, R.R., Larroche, C., Li, Z., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 413–425. ISBN 978-0-12-819820-9. [Google Scholar]
- Sathitsuksanoh, N.; Zhu, Z.; Rollin, J.; Zhang, Y.-H.P. Solvent Fractionation of Lignocellulosic Biomass. In Bioalcohol Production; Waldron, K., Ed.; Woodhead Publishing Series in Energy; Woodhead Publishing: Sawston, UK, 2010; pp. 122–140. ISBN 978-1-84569-510-1. [Google Scholar]
- Chen, F.; Ye, J.; Sista Kameshwar, A.K.; Wu, X.; Ren, J.; Qin, W.; Li, D.-W. A Novel Cold-Adaptive Endo-1,4-β-Glucanase From Burkholderia pyrrocinia JK-SH007: Gene Expression and Characterization of the Enzyme and Mode of Action. Front. Microbiol. 2020, 10, 3137. [Google Scholar] [CrossRef]
- Sharma, A.; Tewari, R.; Rana, S.S.; Soni, R.; Soni, S.K. Cellulases: Classification, Methods of Determination and Industrial Applications. Appl. Biochem. Biotechnol. 2016, 179, 1346–1380. [Google Scholar] [CrossRef]
- Linton, S.M. Review: The Structure and Function of Cellulase (Endo-β-1,4-Glucanase) and Hemicellulase (β-1,3-Glucanase and Endo-β-1,4-Mannase) Enzymes in Invertebrates That Consume Materials Ranging from Microbes, Algae to Leaf Litter. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2020, 240, 110354. [Google Scholar] [CrossRef] [PubMed]
- Freitas, E.N.; Salgado, J.C.; Alnoch, R.C.; Contato, A.G.; Habermann, E.; Michelin, M.; Martínez, C.A.; Polizeli, M.L.T.M. Challenges of Biomass Utilization for Bioenergy in a Climate Change Scenario. Biology 2021, 10, 1277. [Google Scholar] [CrossRef] [PubMed]
- Mandeep; Liu, H.; Shukla, P. Synthetic Biology and Biocomputational Approaches for Improving Microbial Endoglucanases toward Their Innovative Applications. ACS Omega 2021, 6, 6055–6063. [Google Scholar] [CrossRef] [PubMed]
- Yennamalli, R.M.; Rader, A.J.; Kenny, A.J.; Wolt, J.D.; Sen, T.Z. Endoglucanases: Insights into Thermostability for Biofuel Applications. Biotechnol. Biofuels 2013, 6, 136. [Google Scholar] [CrossRef]
- Sahin, S.; Ozmen, I.; Biyik, H. Industrial Applications of Endoglucanase Obtained from Novel and Native Trichoderma atroviride. Chem. Biochem. Eng. Q. 2016, 30, 265–278. [Google Scholar] [CrossRef]
- Yang, T.; Li, X.; Guo, Y.; Peng, S.; Liu, G.; Zhao, J. Effect of Endoglucanases from Different Glycoside Hydrolase Families on Enzymatic Preparation of Cellulose Nanocrystal. Ind. Crops Prod. 2020, 155, 112755. [Google Scholar] [CrossRef]
- Li, X.; Xia, J.; Zhu, X.; Bilal, M.; Tan, Z.; Shi, H. Construction and Characterization of Bifunctional Cellulases: Caldicellulosiruptor-Sourced Endoglucanase, CBM, and Exoglucanase for Efficient Degradation of Lignocellulose. Biochem. Eng. J. 2019, 151, 107363. [Google Scholar] [CrossRef]
- Ontañon, O.M.; Ghio, S.; Marrero Díaz de Villegas, R.; Garrido, M.M.; Talia, P.M.; Fehér, C.; Campos, E. A Thermostable GH8 Endoglucanase of Enterobacter Sp. R1 Is Suitable for β-Glucan Deconstruction. Food Chem. 2019, 298, 124999. [Google Scholar] [CrossRef]
- Vlasenko, E.; Schülein, M.; Cherry, J.; Xu, F. Substrate Specificity of Family 5, 6, 7, 9, 12, and 45 Endoglucanases. Bioresour. Technol. 2010, 101, 2405–2411. [Google Scholar] [CrossRef]
- Berka, R.M.; Grigoriev, I.V.; Otillar, R.; Salamov, A.; Grimwood, J.; Reid, I.; Ishmael, N.; John, T.; Darmond, C.; Moisan, M.-C.; et al. Comparative Genomic Analysis of the Thermophilic Biomass-Degrading Fungi Myceliophthora thermophila and Thielavia terrestris. Nat. Biotechnol. 2011, 29, 922–927. [Google Scholar] [CrossRef]
- López-López, A.; Santiago-Hernández, A.; Cayetano-Cruz, M.; García-Huante, Y.; Campos, J.E.; Bustos-Jaimes, I.; Marsch-Moreno, R.; Cano-Ramírez, C.; Benitez-Cardoza, C.G.; Hidalgo-Lara, M.E. TtCel7A: A Native Thermophilic Bifunctional Cellulose/Xylanase Exogluclanase from the Thermophilic Biomass-Degrading Fungus Thielavia terrestris Co3Bag1, and Its Application in Enzymatic Hydrolysis of Agroindustrial Derivatives. J. Fungi 2023, 9, 152. [Google Scholar] [CrossRef]
- Lopes, A.M.M.; Martins, M.; Goldbeck, R. Heterologous Expression of Lignocellulose-Modifying Enzymes in Microorganisms: Current Status. Mol. Biotechnol. 2021, 63, 184–199. [Google Scholar] [CrossRef] [PubMed]
- Paula, R.G.; Antoniêto, A.C.C.; Ribeiro, L.F.C.; Srivastava, N.; O’Donovan, A.; Mishra, P.K.; Gupta, V.K.; Silva, R.N. Engineered Microbial Host Selection for Value-Added Bioproducts from Lignocellulose. Biotechnol. Adv. 2019, 37, 107347. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The Protein Families Database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef] [PubMed]
- Bendtsen, J.D.; Nielsen, H.; von Heijne, G.; Brunak, S. Improved Prediction of Signal Peptides: SignalP 3.0. J. Mol. Biol. 2004, 340, 783–795. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Kleywegt, G.J.; Zou, J.Y.; Divne, C.; Davies, G.J.; Sinning, I.; Stâhlberg, J.; Reinikainen, T.; Srisodsuk, M.; Teeri, T.T.; Jones, T.A. The Crystal Structure of the Catalytic Core Domain of Endoglucanase I from Trichoderma reesei at 3.6 A Resolution, and a Comparison with Related Enzymes. J. Mol. Biol. 1997, 272, 383–397. [Google Scholar] [CrossRef]
- Sonoda, M.T.; Godoy, A.S.; Pellegrini, V.O.A.; Kadowaki, M.A.S.; Nascimento, A.S.; Polikarpov, I. Structure and Dynamics of Trichoderma harzianum Cel7B Suggest Molecular Architecture Adaptations Required for a Wide Spectrum of Activities on Plant Cell Wall Polysaccharides. Biochim. Biophys. Acta BBA Gen. Subj. 2019, 1863, 1015–1026. [Google Scholar] [CrossRef]
- Lüthy, R.; Bowie, J.U.; Eisenberg, D. Assessment of Protein Models with Three-Dimensional Profiles. Nature 1992, 356, 83–85. [Google Scholar] [CrossRef]
- Shuid, A.N.; Kempster, R.; McGuffin, L.J. ReFOLD: A Server for the Refinement of 3D Protein Models Guided by Accurate Quality Estimates. Nucleic Acids Res. 2017, 45, W422–W428. [Google Scholar] [CrossRef]
- McGuffin, L.J.; Shuid, A.N.; Kempster, R.; Maghrabi, A.H.A.; Nealon, J.O.; Salehe, B.R.; Atkins, J.D.; Roche, D.B. Accurate Template-Based Modeling in CASP12 Using the IntFOLD4-TS, ModFOLD6, and ReFOLD Methods. Proteins Struct. Funct. Bioinform. 2018, 86, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Payne, C.M.; Knott, B.C.; Mayes, H.B.; Hansson, H.; Himmel, M.E.; Sandgren, M.; Ståhlberg, J.; Beckham, G.T. Fungal Cellulases. Chem. Rev. 2015, 115, 1308–1448. [Google Scholar] [CrossRef]
- Schiano-di-Cola, C.; Kołaczkowski, B.; Sørensen, T.H.; Christensen, S.J.; Cavaleiro, A.M.; Windahl, M.S.; Borch, K.; Morth, J.P.; Westh, P. Structural and Biochemical Characterization of a Family 7 Highly Thermostable Endoglucanase from the Fungus Rasamsonia emersonii. FEBS J. 2020, 287, 2577–2596. [Google Scholar] [CrossRef]
- Momeni, M.H.; Payne, C.M.; Hansson, H.; Mikkelsen, N.E.; Svedberg, J.; Engström, Å.; Sandgren, M.; Beckham, G.T.; Ståhlberg, J. Structural, Biochemical, and Computational Characterization of the Glycoside Hydrolase Family 7 Cellobiohydrolase of the Tree-Killing Fungus Heterobasidion irregulare. J. Biol. Chem. 2013, 288, 5861–5872. [Google Scholar] [CrossRef]
- Lin, Y.; Beckham, G.T.; Himmel, M.E.; Crowley, M.F.; Chu, J.-W. Endoglucanase Peripheral Loops Facilitate Complexation of Glucan Chains on Cellulose via Adaptive Coupling to the Emergent Substrate Structures. J. Phys. Chem. B 2013, 117, 10750–10758. [Google Scholar] [CrossRef]
- Borisova, A.S.; Eneyskaya, E.V.; Jana, S.; Badino, S.F.; Kari, J.; Amore, A.; Karlsson, M.; Hansson, H.; Sandgren, M.; Himmel, M.E.; et al. Correlation of Structure, Function and Protein Dynamics in GH7 Cellobiohydrolases from Trichoderma atroviride, T. reesei and T. harzianum. Biotechnol. Biofuels 2018, 11, 5. [Google Scholar] [CrossRef] [PubMed]
- Segato, F.; Damásio, A.R.L.; Gonçalves, T.A.; de Lucas, R.C.; Squina, F.M.; Decker, S.R.; Prade, R.A. High-Yield Secretion of Multiple Client Proteins in Aspergillus. Enzyme Microb. Technol. 2012, 51, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Velasco, J.; Oliva, B.; Mulinari, E.J.; Quintero, L.P.; da Silva Lima, A.; Gonçalves, A.L.; Gonçalves, T.A.; Damasio, A.; Squina, F.M.; Ferreira Milagres, A.M.; et al. Heterologous Expression and Functional Characterization of a GH10 Endoxylanase from Aspergillus fumigatus var. Niveus with Potential Biotechnological Application. Biotechnol. Rep. 2019, 24, e00382. [Google Scholar] [CrossRef]
- Punt, P.J.; van Biezen, N.; Conesa, A.; Albers, A.; Mangnus, J.; van den Hondel, C. Filamentous Fungi as Cell Factories for Heterologous Protein Production. Trends Biotechnol. 2002, 20, 200–206. [Google Scholar] [CrossRef]
- Pellegrini, V.O.A.; Serpa, V.I.; Godoy, A.S.; Camilo, C.M.; Bernardes, A.; Rezende, C.A.; Junior, N.P.; Franco Cairo, J.P.L.; Squina, F.M.; Polikarpov, I. Recombinant Trichoderma harzianum Endoglucanase I (Cel7B) Is a Highly Acidic and Promiscuous Carbohydrate-Active Enzyme. Appl. Microbiol. Biotechnol. 2015, 99, 9591–9604. [Google Scholar] [CrossRef]
- Bernardi, A.V.; Yonamine, D.K.; Uyemura, S.A.; Dinamarco, T.M. A Thermostable Aspergillus fumigatus GH7 Endoglucanase Over-Expressed in Pichia pastoris Stimulates Lignocellulosic Biomass Hydrolysis. Int. J. Mol. Sci. 2019, 20, 2261. [Google Scholar] [CrossRef]
- Berto, G.L.; Velasco, J.; Tasso Cabos Ribeiro, C.; Zanphorlin, L.M.; Noronha Domingues, M.; Tyago Murakami, M.; Polikarpov, I.; de Oliveira, L.C.; Ferraz, A.; Segato, F. Functional Characterization and Comparative Analysis of Two Heterologous Endoglucanases from Diverging Subfamilies of Glycosyl Hydrolase Family 45. Enzyme Microb. Technol. 2019, 120, 23–35. [Google Scholar] [CrossRef]
- Sandgren, M.; Ståhlberg, J.; Mitchinson, C. Structural and Biochemical Studies of GH Family 12 Cellulases: Improved Thermal Stability, and Ligand Complexes. Prog. Biophys. Mol. Biol. 2005, 89, 246–291. [Google Scholar] [CrossRef]
- Amore, A.; Knott, B.C.; Supekar, N.T.; Shajahan, A.; Azadi, P.; Zhao, P.; Wells, L.; Linger, J.G.; Hobdey, S.E.; Vander Wall, T.A.; et al. Distinct Roles of N- and O-Glycans in Cellulase Activity and Stability. Proc. Natl. Acad. Sci. USA 2017, 114, 13667–13672. [Google Scholar] [CrossRef]
- Hua, C.; Li, W.; Han, W.; Wang, Q.; Bi, P.; Han, C.; Zhu, L. Characterization of a Novel Thermostable GH7 Endoglucanase from Chaetomium thermophilum Capable of Xylan Hydrolysis. Int. J. Biol. Macromol. 2018, 117, 342–349. [Google Scholar] [CrossRef]
- Voutilainen, S.P.; Boer, H.; Alapuranen, M.; Jänis, J.; Vehmaanperä, J.; Koivula, A. Improving the Thermostability and Activity of Melanocarpus albomyces Cellobiohydrolase Cel7B. Appl. Microbiol. Biotechnol. 2009, 83, 261–272. [Google Scholar] [CrossRef]
- Corrêa, D.H.A.; Ramos, C.H.I. The Use of Circular Dichroism Spectroscopy to Study Protein Folding, Form and Function. Afr. J. Biochem. Res. 2009, 3, 164–173. [Google Scholar] [CrossRef]
- Colussi, F.; Garcia, W.; Rosseto, F.R.; de Mello, B.L.S.; de Oliveira Neto, M.; Polikarpov, I. Effect of PH and Temperature on the Global Compactness, Structure, and Activity of Cellobiohydrolase Cel7A from Trichoderma harzianum. Eur. Biophys. J. EBJ 2012, 41, 89–98. [Google Scholar] [CrossRef]
- Miles, A.J.; Ramalli, S.G.; Wallace, B.A. DichroWeb, a Website for Calculating Protein Secondary Structure from Circular Dichroism Spectroscopic Data. Protein Sci. 2022, 31, 37–46. [Google Scholar] [CrossRef]
- Rahman, M.S.; Hasan, M.S.; Nitai, A.S.; Nam, S.; Karmakar, A.K.; Ahsan, M.S.; Shiddiky, M.J.A.; Ahmed, M.B. Recent Developments of Carboxymethyl Cellulose. Polymers 2021, 13, 1345. [Google Scholar] [CrossRef]
- Karnaouri, A.C.; Topakas, E.; Christakopoulos, P. Cloning, Expression, and Characterization of a Thermostable GH7 Endoglucanase from Myceliophthora thermophila Capable of High-Consistency Enzymatic Liquefaction. Appl. Microbiol. Biotechnol. 2014, 98, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, J.; Momcilovic, D.; Wittgren, B.; Schülein, M.; Tjerneld, F.; Brinkmalm, G. Enzymatic Degradation of Carboxymethyl Cellulose Hydrolyzed by the Endoglucanases Cel5A, Cel7B, and Cel45A from Humicola insolens and Cel7B, Cel12A and Cel45Acore from Trichoderma reesei. Biopolymers 2002, 63, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Dun, B.; Shi, P.; Ma, R.; Luo, H.; Bai, Y.; Xie, X.; Yao, B. A Novel GH7 Endo-β-1,4-Glucanase from Neosartorya fischeri P1 with Good Thermostability, Broad Substrate Specificity and Potential Application in the Brewing Industry. PLoS ONE 2015, 10, e0137485. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Luo, H.; Shi, P.; Huang, H.; Bai, Y.; Yao, B. A Highly-Active Endo-1,3-1,4-β-Glucanase from Thermophilic Talaromyces emersonii CBS394.64 with Application Potential in the Brewing and Feed Industries. Process Biochem. 2014, 49, 1448–1456. [Google Scholar] [CrossRef]
- Bernardi, A.V.; de Gouvêa, P.F.; Gerolamo, L.E.; Yonamine, D.K.; de Lourdes de Lima Balico, L.; Uyemura, S.A.; Dinamarco, T.M. Functional Characterization of GH7 Endo-1,4-β-Glucanase from Aspergillus fumigatus and Its Potential Industrial Application. Protein Expr. Purif. 2018, 150, 1–11. [Google Scholar] [CrossRef] [PubMed]
- NCBI Resource Coordinators. Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2017, 45, D12–D17. [Google Scholar] [CrossRef]
- Bairoch, A.; Apweiler, R.; Wu, C.H.; Barker, W.C.; Boeckmann, B.; Ferro, S.; Gasteiger, E.; Huang, H.; Lopez, R.; Magrane, M.; et al. The Universal Protein Resource (UniProt). Nucleic Acids Res. 2005, 33, D154–D159. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering Key Features in Protein Structures with the New ENDscript Server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Eswar, N.; Webb, B.; Marti-Renom, M.A.; Madhusudhan, M.S.; Eramian, D.; Shen, M.; Pieper, U.; Sali, A. Comparative Protein Structure Modeling Using Modeller. Curr. Protoc. Bioinforma. 2006, 15, 5.6.1–5.6.30. [Google Scholar] [CrossRef]
- Shen, M.; Sali, A. Statistical Potential for Assessment and Prediction of Protein Structures. Protein Sci. 2006, 15, 2507–2524. [Google Scholar] [CrossRef]
- Melo, F.; Sánchez, R.; Sali, A. Statistical Potentials for Fold Assessment. Protein Sci. 2002, 11, 430–448. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A Program to Check the Stereochemical Quality of Protein Structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure Visualization for Researchers, Educators, and Developers. Protein Sci. Publ. Protein Soc. 2021, 30, 70–82. [Google Scholar] [CrossRef]
- Gibson, D.G.; Young, L.; Chuang, R.-Y.; Venter, J.C.; Hutchison, C.A.; Smith, H.O. Enzymatic Assembly of DNA Molecules up to Several Hundred Kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef] [PubMed]
- Damásio, A.R.L.; Ribeiro, L.F.C.; Ribeiro, L.F.; Furtado, G.P.; Segato, F.; Almeida, F.B.R.; Crivellari, A.C.; Buckeridge, M.S.; Souza, T.A.C.B.; Murakami, M.T.; et al. Functional Characterization and Oligomerization of a Recombinant Xyloglucan-Specific Endo-β-1,4-Glucanase (GH12) from Aspergillus niveus. Biochim. Biophys. Acta 2012, 1824, 461–467. [Google Scholar] [CrossRef] [PubMed]
- Segato, F.; Dias, B.; Berto, G.L.; de Oliveira, D.M.; De Souza, F.H.M.; Citadini, A.P.; Murakami, M.T.; Damásio, A.R.L.; Squina, F.M.; Polikarpov, I. Cloning, Heterologous Expression and Biochemical Characterization of a Non-Specific Endoglucanase Family 12 from Aspergillus terreus NIH2624. Biochim. Biophys. Acta Proteins Proteom. 2017, 1865, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.L. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Perkins, D.N.; Pappin, D.J.; Creasy, D.M.; Cottrell, J.S. Probability-Based Protein Identification by Searching Sequence Databases Using Mass Spectrometry Data. Electrophoresis 1999, 20, 3551–3567. [Google Scholar] [CrossRef]
- Shevchenko, A.; Wilm, M.; Vorm, O.; Mann, M. Mass Spectrometric Sequencing of Proteins from Silver-Stained Polyacrylamide Gels. Anal. Chem. 1996, 68, 850–858. [Google Scholar] [CrossRef] [PubMed]
- Leone, F.A.; Baranauskas, J.A.; Furriel, R.P.M.; Borin, I.A. SigrafW: An Easy-to-Use Program for Fitting Enzyme Kinetic Data. Biochem. Mol. Biol. Educ. Bimon. Publ. Int. Union Biochem. Mol. Biol. 2005, 33, 399–403. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).