Abstract
Polyurethanes (PUs) are an exceedingly heterogeneous group of plastic polymers, widely used in a variety of industries from construction to medical implants. In the past decades, we have witnessed the accumulation of PU waste and its detrimental environmental impacts. PUs have been identified as one of the most toxic polymers leaching hazardous compounds derived both from the polymer itself and the additives used in production. Further environmental impact assessment, identification and characterization of substances derived from PU materials and establishing efficient degradation strategies are crucial. Thus, a selection of eight synthetic model compounds which represent partial PU hydrolysis products were synthesized and characterized both in terms of toxicity and suitability to be used as substrates for the identification of novel biocatalysts for PU biodegradation. Overall, the compounds exhibited low in vitro cytotoxicity against a healthy human fibroblast cell line and virtually no toxic effect on the nematode Caenorhabditis elegans up to 500 µg mL−1, and two of the substrates showed moderate aquatic ecotoxicity with EC50 values 53 µg mL−1 and 45 µg mL−1, respectively, on Aliivibrio fischeri. The compounds were successfully applied to study the mechanism of ester and urethane bond cleaving preference of known plastic-degrading enzymes and were used to single out a novel PU-degrading biocatalyst, Amycolatopsis mediterranei ISP5501, among 220 microbial strains. A. mediterranei ISP5501 can also degrade commercially available polyether and polyester PU materials, reducing the average molecular number of the polymer up to 13.5%. This study uncovered a biocatalyst capable of degrading different types of PUs and identified potential enzymes responsible as a key step in developing biotechnological process for PU waste treatment options.
1. Introduction
Polyurethanes (PUs) are ranked the sixth most common synthetic polymer used [1], with a growing market value estimate of over USD 50 billion for 2021 [2]. The production of PUs involves reacting diisocyanates with polyols to obtain thermoplastics, thermosets, or foams [3]. PUs are widely used as coatings, insulators, foams, elastic fibers, textiles in carpet underlayment, thermal isolation, car seats, mattresses, etc. [4]. High and wide usage leads to waste accumulation, which more than often ends up in landfills and the environment. The bulk of PU waste is incinerated for energy recovery and releasing toxic compounds [5], while 29.7% is recycled and 30.8% is still landfilled [6]. PU and PU microplastic particles are identified as one of the more toxic polymers [7,8]. Widely used PU mattresses have been shown to continually release a number of volatile organic compounds [9]. Thermal recycling of mattresses also releases toxic isocyanates [10] while PU coatings leach ecotoxic compounds based on 4,4′-methylenediphenyl diisocyanate (4,4′-MDI) and toluene diisocyanate (TDI), which are known carcinogens [11]. On the other hand, certain types of PU are well established for use as medical devices, with numerous studies confirming their biocompatibility and non-toxicity [12], thus underlining the great diversity of PU polymers.
Current recycling strategies for PU waste revolve around mechanical and chemical recycling. Mechanical approaches transform PU waste into granules, flakes, or powder, which can be used be used in new products, i.e., PU carpet underlayment [13]. It is a fairly cost-effective and environmentally acceptable process; however, it is a form of ‘downcycling’ as materials of lower quality and value are produced. For the recovery of monomers that can be incorporated into new polymers, chemical recycling is a more sensible route. Hydrolysis, alcoholysis, aminolysis, phosphorolysis [6], and glycolysis [14] have been used in the recycling of PU in pilot-scale plants [13]. However, the high energy consumption and environmental impacts of chemical recycling decrease its role in sustainable development; however, some of these problems could be bypassed by biocatalysis. The benefits of biocatalysis have clearly been shown in the case of polyethylene terephthalate (PET), which has led to the development of biotechnological depolymerization of post-consumer PET bottles with the efficient recovery of terephthalic acid [15]. Therefore, research efforts to develop superior enzymes and modify microorganisms for the degradation of other plastic and mixed plastic waste materials are ongoing. Based on the type of polyols used, PUs are classified into polyester PUs and polyether PUs. Depending on the type of diisocyanate used, PUs can be aromatic or aliphatic [16]. The structure of thermoplastic PU polymers consists of hard (highly crystalline regions made up of isocyanates, chain extenders with urethane bonds) and soft segments (amorphous regions mainly consisting of polyols with ester or ether bonds) [17] (Figure 1a).

Figure 1.
Polyurethane (PU) (a) polymer representation; (b) proposed model substrates synthesized in this study derived from hydrolysis of PU hard segment and PU-related compounds for detection of novel biocatalysts.
Hard/soft segment composition and isocyanate type govern the biodegradability of PU, with polyester Pus being more prone to microbial degradation than polyether Pus. Soft segments are more accessible for enzymatic attack, ester bonds have higher biodegradability than urethane bonds [18], and aromatic isocyanate-based PUs are considered more difficult to biodegrade than the aliphatic ones [17]. Despite decades of research, an efficient biocatalytic system for PU hydrolysis has not been reported yet [1,19]. From a number of microorganisms reported to degrade PU, only a handful of enzymes/microorganisms have been shown to hydrolyze the urethane bond [20]. A database of known and confirmed plastic-degrading enzymes—Plastic-Active Enzymes Database (PAZy) [21,22]—contains 10 PU-active enzymes; however, most of them act only on the ester bonds of the polymer [23]. Given the complex structure and variety of bonds that can be found in PU, the research focus is shifting from individual enzymes to multiple enzyme systems. One such study employed an amidase and esterase in a bid to simultaneously degrade urethane and ester linkages in four different thermoplastic PUs and proved that combining enzymes leads to increased urethane bond hydrolysis [24]. Alternative approaches, such as using Tenebrio molitor larvae and their gut microbiota for the degradation of PU, have been employed as well [25]. However, the lack of efficient and robust urethane bond degrading biocatalysts is still the main bottleneck in the development of biotechnological PU waste recycling systems [26].
The most widespread substrate for assessing PU degradation is a colloidal dispersion, Impranil, an anionic aliphatic polyester PU of proprietary structure [27]. A change in optical density (clearing), whether in solution, on agar plates, or combined with dyes, is considered evidence of PU degradation [28]. Impranil-clearing assays cannot give information on the type of enzyme responsible for the activity and whether ether or urethane bonds were hydrolyzed, and, most importantly, the amount of Impranil clearing does not always correlate to the amount of degradation [29]. However, Impranil has proved useful in research describing the adsorption of peptides to plastic [30]. When investigating enzyme mechanisms, urethane bond-containing small molecules present far more promising approaches with p-toluenesulfonamide- [24] and p-nitrophenol- [31] tagged molecules being used for assessing urethanase activity.
In this work, we synthesized eight PU model substrates based on widely used phenyl isocyanate and toluene diisocyanate containing both urethane and ester bonds (Figure 1b). These substrates were used for the screening and identification of novel PU-degrading biocatalysts while allowing for the study of the mechanism of the action. PU model substrates were designed to represent polymer partial degradation products, valuable for assessing potential environmental risks associated with PU materials and their degradation. Accordingly, their toxicity and environmental impact were investigated and compared to known PU-associated pollutants, including adipic acid and 2,4- toluenediamine (2,4-TDA). Amycolatopsis mediterranei ISP5501 was identified as a urethane bond-degrading strain using the PU-7 model substrate and was further confirmed to degrade both polyester and polyether PU materials. Potential enzymes involved were identified by the genome analysis.
2. Results and Discussion
2.1. PU-Model Compounds Synthesis and Characterization
Based on the structural characteristics of different PU materials, we designed eight PU model compounds (Figure 1b). Six of them were synthesized and structurally characterized for the first time (PU-2, PU-3, PU-4, PU-5, PU-6, and PU-8), while two of them were reported earlier (PU-1 and PU-7), but lacked proper spectral characterization [32,33,34]. NMR spectra of the full set of PU model compounds are provided as Figures S1–S16.
Model substrates were synthesized following three main reactions: (1) reaction of hydroxyalkyl esters with phenyl isocyanate and toluene diisocyanate; (2) reaction of 2-hydroxyethyl phenyl-carbamate with hexanoyl chloride or adipic chloride; and (3) hydrogenolysis. Compounds PU-1, PU-2, and PU-3 were obtained by reaction of hydroxyalkyl esters with phenyl isocyanate, while compounds PU-6, PU-7, and PU-8 were obtained by reaction of hydroxyalkyl esters with toluene diisocyanate. These reactions are typical reactions of obtaining so-called urethane ester monomers whose polymerization gives polymeric materials applicable as impregnating and adhesion agents, crosslinking agents, non-toxic dental materials, coatings, and waveguide protection materials [35]. Compounds PU-4 and PU-5 were obtained by reacting 2-hydroxyethyl phenyl-carbamate and hexanoyl chloride or adipic chloride. The yield of all products was highly dependent on the reaction conditions and required optimization of the purification process to achieve both high yield and purity. Solubility in a selection of common organic solvents was tested for each of the PU model compounds, with all being well soluble in DMSO as solvent of choice for toxicity evaluations (Table S1). As expected, PU-8 was the least soluble, especially in polar solvents, including methanol and ethanol. PU-6, PU-7, and PU-8 represent urethane bond-containing partial degradation products of TDI-based PUs with different polyol segments (Figure 1b) and are realistic targets for further enzyme degradation and substrates for urethanase enzyme detection. PU-4 and PU-5 containing both urethane and ester bonds were used for the study of enzyme cleaving preferences.
The biodegradability of urethane bonds in small molecules is higher than in PU polymers [1], but the diverse structures of PU model substrates allow for examining urethane bond cleaving in different molecular contexts and thus bridge the gap from small molecule model substrates to the bulk polymer hydrolysis. Previously, a model substrate composed of 14 units of adipic acid, seven units of 1,4-butanediol (BDO), seven units of ethylene glycol (EG), and one unit of 2,4-TDA was used for assessment of microbial PU hydrolysis and subsequent monomer utilization [36].
2.2. Cyto- and Ecotoxicity of the PU-Model Compounds
The cytotoxicity of carbamate compounds is well documented in the literature [37]; therefore, the antiproliferative effect of PU model substrates was tested on a healthy lung fibroblast MRC-5 cell line as the inhalation may be the route of entry for compounds released during polyurethane degradation [38]. Low toxicity of the compounds was observed under the conditions tested. Even at a relatively high concentration of 10 µg mL−1, all tested compounds supported 90% to 100% cell survival, while in higher concentrations for compounds PU-3, PU-6, PU-7, and PU-8, cell survival rates were between 60% to 80% (Figure 2a). For compounds PU-3, PU-6, and PU-8, greater cell death may be caused by physical interference with cells, as they showed lower solubility and were in a state of suspension when added to aqueous solution of cell propagation medium.
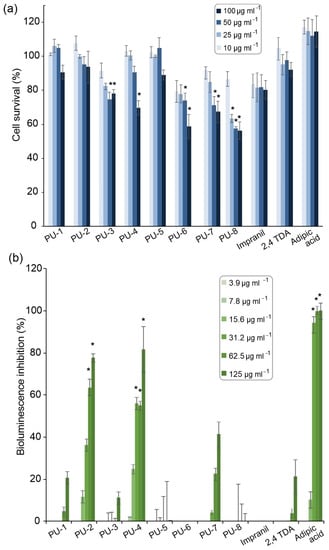
Figure 2.
Quantification of in vitro MRC-5 cells survival in the presence of PU model compounds at concentration range (a). Inhibitory effect of PU model substrates on A. fischeri bioluminescence at concentration range (b). The effect was compared to untreated control using a t-test, * p ≤ 0.01.
As a model system for ecotoxicity evaluation of organic chemicals, A. fischeri is usually employed for the evaluation of aquatic toxicity to marine and freshwater organisms. The inhibition of bioluminescence is a non-specific and sensitive toxicity assay [39]. PU-5, PU-6, and PU-8 displayed no inhibitory effects on A. fischeri bioluminescence, while PU-1 and PU-3 caused only a minimal decrease in bioluminescence at the highest concentration tested and can be considered non-toxic as well. Comparing all investigated PU model compounds, PU-2 and PU-4 proved to have the highest aquatic toxicity, with EC50 values of 53 µg mL−1 and 45 µg mL−1, respectively (Figure 2b), and with EC50 values between 100 and 10 µg mL−1 could be considered as moderately toxic [40]. In contrast, the EC20 value of 2,4-TDA was 116 µg mL−1, with even lower concentrations reported in the literature (50 µg mL−1) [41]. These results indicate that PU degradation intermediates can be more toxic than diamines released during complete PU hydrolysis and need to be taken into account when assessing the environmental impact of PU degradation strategies. Adipic acid, another product of partial PU hydrolysis, had the lowest EC50 value (18 µg mL−1) and was the only one to completely inhibit A. fischeri bioluminescence at higher concentrations. To the best of our knowledge, there are no reports investigating the influence of adipic acid on A. fischeri, with Šepič et al. reporting EC50 values of 140 and 128 µg mL−1 after acute tests with crustacea Daphnia magna and Thamnocephalus platyurus, respectively [42]. PU and polyvinyl chloride (PVC) leachates of unknown composition were previously found toxic to D. magna [43].
C. elegans is a multicellular model organism that combines both the advantages of using whole animals and in vitro systems for toxicity assessment, with comparative studies confirming the replicability of results in mammalian model organisms [44]. Given its terrestrial habitat and ubiquitous distribution, C. elegans may also prove useful in investigating the environmental impact of plastic degradation products [45]. The PU model substrates did not cause C. elegans death in any of the concentrations tested (results not shown). Only TDA at a concentration of 500 µg mL−1 caused 70% mortality; however, it is highly unlikely that TDA can reach such high concentrations in the environment.
Worth mentioning is the fact that Impranil as polymeric dispersion proved to be less toxic than monomeric PU degradation products in all toxicity tests, underlining the need for toxicity analysis when investigating and optimizing polymer degradation processes. In a study exploring the baseline toxicity, oxidative stress induction, cytotoxicity, and endocrine activity of plastic extracts, PUs were identified as having the highest toxicity along with poly(vinyl chloride) (PVC) [7]. Other studies explored the toxicity of specific PU degradation products, such as the corresponding diamines derived from two of the most common diisocyanates, with both MDA [46] and TDA [36,47] classified as carcinogens.
2.3. Degradation of PU Model Substrates by Known Esterases and Proteases
The label-free nature of the PU model substrates allows for the examination of the enzyme mechanisms without potential enzyme bias towards a chromophore of fluorophore [48]. The applicability of such a set of substrates was demonstrated for the screening and characterization of novel PETases [49] and the identification of bisphenol-A polycarbonate-degrading bacteria [50]. The potential of PU model substrates to be used as bond-specific screening molecules was assessed using hydrolases from different families. Recombinant FoCut5a, HiC, and IsPETase are already proven to be capable of cleaving ester bonds of different polyester materials. Specifically, FoCut5a can degrade PET model substrates and polycaprolactone (PCL) powder (Dimarogona et al., 2015), and HiC cutinase can fully degrade PET films of low crystallinity (Ronkvist et al., 2009), while IsPETase is a well-known PET hydrolase (Yoshida et al., 2016). Recombinant DaPUase was from the bacterium Comamonas acidovorans, a microorganism able to utilize polyester PU as the sole carbon source (Akutsu et al., 1998). Lastly, the two commercial proteases (BacProt and StrepProt) were chosen because they can potentially act on both ester and urethane bonds. DaPUase, FoCut5a, and IsPETase are enzymes that show maximum activity at temperatures around 30 °C, in contrast to BacProt, HiC, and StrepProt with temperature optimum around 50 °C. As a result, the hydrolysis of PU-5 compound was analyzed after treatment with both esterases and proteases at 30 °C and at 50 °C (Figure S17). PU-5, a model substrate containing two terminal phenyl groups (Figure 1b), was chosen as the model substrate since the degradation could be efficiently monitored via a standard HPLC equipped with a UV-detector.
The main hydrolysis product after treating PU-5 with FoCut5a, IsPETase, HiC, and DaPUase was PU-1, indicative of ester bond cleaving (Figure S17a). Smaller amounts of PU-4 were also detected, further confirming that the ester bond was cleaved since both PU-1 and PU-4 are products of ester bond cleavage. The tested enzymes could not recognize the urethane bond under conditions tested, probably because of the stereochemical inhibition caused by the aromatic ring. In the sample containing no enzyme, an additional product was detected, which can be attributed to autohydrolysis of the substrate during incubation. Interestingly, protease digestion also resulted in PU-1 as the main degradation product, cleaving only the ester and not the urethane bond of the substrate. Serine proteases have been already reported to hydrolyze PU, showing simultaneously proteolytic and esterase activity [51]. BacProt is classified as subtilase, which belongs to the serine proteinase family, while StrepProt is a mixture of at least three proteases, including an extracellular serine protease. It seems that even these proteases show preference towards the ester bond over the urethane bond (Figure S17b). It has been reported that subtilisin and subtilisin-like proteases preferentially cleave hydrophobic non-aromatic residues [52].
2.4. Identification of New PU-Degrading Biocatalysts
A total of 220 microbial strains were screened for their Impranil-clearing activity on agar plates. Eighteen strains showed good zones of Impranil clearance (Figure 3) and were identified by 16S rDNA sequencing (Table S3), with the majority of strains showing the highest sequence similarity to Streptomyces species, a genus whose biocatalytic potential has gained interest in recent years [53]. The unknown structure of Impranil complicates any effort to investigate enzyme mechanisms using this substrate, but it does have a very important role in high-throughput pre-screening efforts [54]. Three strains with the ability to degrade Impranil, namely Streptomyces sp. TIT2, Pseudomonas sp. 44, and Amycolatopsis mediterranei ISP5501—and one Bacillus sp. BPM12, which had no Impranil-clearing activity but showed high esterase activity in standard assays (unpublished data)—were selected for biocatalytic reactions using PU-7 as a substrate (substrate with two urethane bonds at positions two and four; Figure 1b).
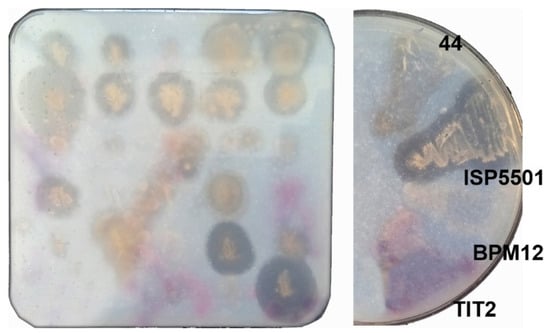
Figure 3.
Pre-screening of bacterial strains on Impranil-containing agar plates. Based on the clearing zone, four bacterial strains (Pseudomonas sp. (44), Amycolatopsis mediterranei (ISP5501), Bacillus sp. (BPM12), and Streptomyces sp. (TIT2)) were selected for whole-cell biocatalytic reactions with PU-7.
Possible degradation products were not easily observed using a standard HPLC approach, therefore UHPLC-MS screen of whole-cell biotransformation reactions afforded the detection of urethanase activity in a high-throughput manner and to differentiate cleaving activities of amidases/proteases and esterases. MS spectra of reactions were compared to PU model substrate control reactions containing no biocatalyst (Figure 4). Cells of A. mediterraneiei ISP5501 were the only ones to produce a variety of expected degradation products (Table S2, Figure S18). Masses of ten predicted urethane bond hydrolysis products were detected when A. mediterraneiei ISP5501 was used as a biocatalyst (Figure 4, Table S2). Both the aromatic and corresponding aliphatic hydrolysis products were detected. TDA and smaller aliphatic degradation products were not detected, either because they were near the detection limit (<100 m/z) or because they could have been used as carbon and energy source and assimilated by ISP5501 during biocatalysis. Based on the detected masses, the urethane bond could have been cleaved by two proposed methods. A distinction between cleaving the urethane bond in the amide vs. the ester part could be made since both amine and carbamic acid moiety-containing degradation products were detected. Based on this, ISP5501 could harbor more than one urethane-cleaving enzyme since urethane bond cleaving by esterase leads to the formation of carbamic acid (Figure 4a), while amidases/proteases hydrolysis yields amides (Figure 4b,c). The products of the activity of both types of enzymatic activities were also detected (Figure 4d,e).
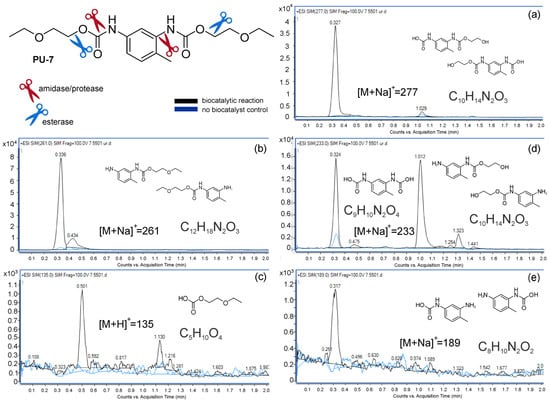
Figure 4.
Biocatalytic reactions using PU-7 as substrate and resting whole cells of A. mediterranei ISP5501 as biocatalyst. Control reactions containing no biocatalysts are presented as a blue line. Products were identified using MS. The esterase activity generated two compounds of the molecular formula C10H14N2O3 (a), the amidase activity generated two compounds of the molecular formula C12H18N2O3 (b), and one compound of the molecular formula C5H10O4 (c), while the activity of both types of enzymes generated two compounds of molecular formula C10H14N2O3 and one of C9H10N2O4 (d) and two compounds of the molecular formula C8H10N2O2 (e).
The proposed mechanisms of urethane bond cleavage by esterases in literature are conflicting. Liu et al. suggest the formation of a carbamic acid and an alcohol [1], while Magnin et al. argue that urethane bond cleaving will result in an amide and an alcohol with the release of a carbon dioxide molecule due to the instability of carbamic acid [23]. Our results suggest urethane bond cleaving with esterases does produce stable carbamates that can be detected by MS. Furthermore, it is interesting to note that masses of ether bond hydrolysis products were detected as well, opening up the opportunity for using ISP5501 in the degradation of highly recalcitrant polyether PU.
A. mediterranei, traditionally linked with medicinal importance and industrial-scale production of rifamycin [55], is a taxon with considerable presence in polymer degradation studies. Research regarding the applicability of Amycolatopsis species in plastic degradation identified several efficient poly(L-lactic acid) (PLA) depolymerases [56] and a study exploring the phylogenetic distribution of plastic-degrading enzymes identified that the largest number of PLA depolymerases come from Amycolatopsis genus [57]. More than 10 PETase-like enzymes have also been identified in Amycolatopsis species [58], making this genus a promising source of novel plastic-degrading biocatalysts and a tool for plastic waste management in a circular economy.
2.5. PU Polymer Degradation by A. mediterranei ISP5501
The potential of the A. mediterranei ISP5501 to degrade PU polymer was further studied on different PU materials. Using whole cells as biocatalysts, the average molecular number Mn of polyether PU decreased by 7.0 ± 0.9%, while in the presence of Impranil the Mn was reduced by 13.5 ± 0.3% compared to those of the initial untreated material (Table 1). The increased degradation observed in the second case can be attributed to the fact that hydrolytic enzymes such as esterases, cutinases, and lipases, are induced in the presence of Impranil (Zhang et al., 2022). Polydispersity index (PDI), defined as Mw/Mn (where Mw = weight average molecular weight and Mn = number average molecular weight) increased due to biocatalytic reaction (Table 1). The enzymes taking part in PU hydrolysis preferentially broke down the smaller carbon chains of PU having exo-activity (i.e., cleaving from the ends), as implied by the minor Mw reduction. Nevertheless, the enzymatic degradation of PU powder was not so extensive for mass loss to occur. This is in accordance with previous findings whereby the enzymatic treatment of polyether PU results in minor mass loss, probably because of material recalcitrance under tested conditions [24,59].

Table 1.
Molecular weight and number (Mw, Mn) and polydispersity index of PU material after A. mediterranei ISP5501 whole-cell biocatalysis.
The enzymes responsible for PU hydrolysis are proteases/amidases and esterases, while several reports mention ureases as potential enzymes for poly(ether urea) PU breakdown [60,61]. Interestingly, none of the aforementioned activities were detected in the A. mediterranei culture supernatant, suggesting that the responsible enzymes can be possibly membrane-bound, similar to bacterial strains, such as C. acidovorans and Pseudomonas capeferrum, and a fungal Penicillium strain, which have been reported to possess membrane-bound esterases showing high hydrolytic activity against PU [62,63,64]. For this reason, A. mediterranei ISP5501 was grown in liquid cultures supplemented with Impranil, and the extracellular and/or intracellular protein fractions were utilized for PU degradation. As shown in Table 2, the intracellular fraction decreased Mn by 10.6 ± 0.3%, while in the case of fraction mixture the corresponding decrease was slightly lower. It is noteworthy that the extracellular fraction caused imperceptible PU degradation, implying that the intracellular fraction, which is also enriched with membrane proteins released after cell lysis, possesses all the necessary enzymes for urethane bond cleavage. The esterase activity reached 19.9 mU mL−1 in the intracellular fraction (pNPB assay), whereas no such activity was detected extracellularly. The proteolytic activity (azocasein assay) was not detected in any of the fractions, although this alone does not provide enough evidence for ruling out protease activity, as protease/amidase products were detected using whole cells and PU-7 as substrate (Figure 4).

Table 2.
Molecular weight and number (Mw, Mn) and polydispersity index of PU material after treatment with extracellular and intracellular protein fractions of A. mediterranei ISP5501.
To further correlate PU degradation with specific enzymatic activities, the genome of A. mediterranei was sequenced and investigated. The A. mediterranei ISP5501 genome consists of a 10198110 bp long scaffold, which is in accordance with reference genomes including Gene bank accession NC_022116.1 and is 99.4% complete based on BUSCO analysis. The predicted proteome of ISP5501 consists of 9386 proteins. We classified these proteins into their respective protein families based on the functional annotations of PGAP and InterProScan and we focused on families associated with PU depolymerization. These families include esterases, ureases, proteases, amidases, and other α/β hydrolases (Table 3), the largest of which are proteases in all subcellular locations and in total. Based on the enzyme assays performed, the enzymes predicted as membrane-bound are of great interest. In the nine membrane esterases found, there are three lipases, while three membrane amidohydrolases and six α/β hydrolases were also indicated. Next, we searched for members of these enzyme families in the A. mediterranei RefSeq genomes and did not observe any noticeable difference between the ISP5501 and the other genomes regarding these enzyme families (Table S4).

Table 3.
PU depolymerization-associated enzyme families in the predicted proteome of A. mediterranei ISP5501.
The BLAST searches using known PU-active enzymes as templates identified six possible PU-active enzymes in the ISP5501 proteome. One of them is the extracellular AML cutinase, further corroborating that this enzyme could be responsible for urethane bond cleaving. Two extracellular esterases were also identified as homologous to the PET and PU-active triacylglycerol lipase from Thermomonospora curvata (Tcur_1278), an enzyme with hydrolytic activity against poly(ε-caprolactone) [65]. Both esterases (pgaptmp_003900, pgaptmp_004139) have a carbohydrate-binding ricin B lectin domain and have a protein sequence identity of 54.5% and 45.6% with Tcur_1278, respectively. Using the LED HMM profiles, these three candidates were classified as members of family 49, superfamily 1. This family contains PETase-like homologs that consist of the core α/β-hydrolase domain without additional structural modules like lids [66]. We also identified three intracellular carboxylesterases homologous to the polyurethane esterase from Delftia acidovorans (PudA) with a protein sequence identity range of 31.0–36.5%. Recently a cutinase from A. mediterranei AML (UniProt No- A0A0H3DES9) was investigated for its plastic-degrading potential. The enzyme couldn’t degrade PLA or PET; however, it could degrade PCL and poly(1,4-butylene succinate) [67]. The search of A. mediterranei ISP5501 genome for known etherases yielded intracellular etherase pgaptmp_009080 N-acetylmuramic acid 6-phosphate etherase, which is not likely to be involved in PU degradation due to the specific mechanism of activity on the lactyl side chain N-acetylmuramic acid 6-phosphate.
The antiSMASH analysis of the ISP5501 genome indeed detected the rifamycin biosynthetic gene cluster encoding five type I polyketide synthases that constitute the core biosynthetic genes [68]. The genome of ISP5501 also encodes a putative polyhydroxyalkanoate (PHA) PhaC polymerase (pgaptmp_006620), the key enzyme involved in PHA biosynthesis [69], which is in line with all previous observations that this taxon has a metabolic network both for extensive utilization of various carbon sources and also for effective funneling of metabolic intermediates into the secondary antibiotic synthesis process. Therefore, one can envisage upcycling of polymeric materials into compounds of high value, such as antibiotics.
3. Conclusions
Plastic pollution is a pressing environmental problem the scale of which is still largely unknown. PUs have been identified as one of the most toxic types of plastic and, due to a lack of efficient recycling strategies, present an ongoing issue. In this study, we synthesized eight PU model compounds representing partial hydrolysis products and screening molecules for the identification of novel PU-degrading biocatalysts. When tested on lung fibroblast cells, A. fischeri and C. elegans, PU degradation products proved more toxic and ecotoxic than PU polymers themselves, a fact that needs to be taken into account when assessing potential PU waste management strategies. The model compound PU-5 proved useful for bond-specific screening, differentiating between ester and urethane bond hydrolysis and aiding in the discovery of urethane bond-specific biocatalysts. Additionally, a novel urethane bond-degrading bacterium, A. mediterranei ISP5501, was identified using the PU-7 model compound. A. mediterranei ISP5501 is capable of degrading both ester- and ether-based Pus, a highly sought-after feature for PU depolymerization. Given the fact PUs are a heterogenous group of polymers, the use of robust biocatalysts capable of degrading different types of PUs is essential for developing efficient PU degradation processes as a first step to achieve their effective and sustainable conversion into valuable compounds.
4. Materials and Methods
4.1. Reagents
2,4-Toluene diisocyanate, phenyl isocyanate, adipic acid, 2,4-TDA, thiazole orange dye, and all other chemicals and reagents were purchased from Sigma-Aldrich (St. Louis, MO, USA) unless stated otherwise. PU polymer Impranil DNL SD (Impranil) was obtained from Covestro (Leverkusen, Germany) and PU polymer Laripur LPR7560 was purchased from Coim group (Milano, Italy).
4.2. PU Model Compounds Synthesis
PU models based on phenyl isocyanate and toluene diisocyanate were synthesized (Figure 1b) using the previously published procedure of reactions of hydroxyalkyl esters with phenyl isocyanate [35] with some modifications. All model compounds were isolated and purified using chromatographic separation methods, and purified compounds were characterized using NMR (Varian/Agilent NMR 400 MHz (1H at 400 MHz, 13C at 100 MHz), Palo Alto, Santa Clara, CA, USA). Samples were dissolved in three different deuterated solvents (CDCl3, CD3OD3, and DMSO-d6). Chemical shifts (δ) are expressed in ppm and coupling constant (J) in Hz. PU-1 and PU-7 were previously known compounds, but not so well structurally characterized [32,33]. For all compounds, detailed preparation procedures and full spectra assignation can be found in the supporting information (Figures S1–S16).
4.3. Ecotoxicity Assessment of PU Model Compounds
4.3.1. Cytotoxicity Evaluation (MTT Assay)
PU-1 to PU-8 along with Impranil, adipic acid, and 2,4-TDA were assessed for their ability to inhibit the proliferation of human lung fibroblast cell line (MRC-5; American Type Culture Collection (ATCC)) by MTT assay (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazoliumbromide). Pre-grown (24 h) cell monolayers (1 × 104 cells per well) in RPMI 1640 medium supplemented with 100 μg mL−1 streptomycin, 100 U mL−1 penicillin, and 10% (v/v) fetal bovine serum (FBS) containing the tested compounds at concentrations ranging from 12.5 to 100 μg mL−1 were incubated in a humidified atmosphere of 95% air and 5% CO2 at 37 °C for 48 h. The cell viability (extent of MTT reduction) was measured spectrophotometrically at 540 nm using a plate reader (Epoch 2000, BioTek, Winooski, VT, USA), and the cell survival was expressed as a percentage of the control (untreated cells). Cytotoxicity was expressed as the concentration of the compound inhibiting cell growth by 50% (IC50).
4.3.2. Aliivibrio Fischeri Toxicity Tests
All stock solutions (50 mg mL−1) were dissolved in DMSO. Stock solutions were diluted in 2% NaCl up to 500 µg mL−1. PU-3, PU-5, and PU-8 formed a suspension when mixed with NaCl; therefore, all of the 500 µg mL−1 sample solutions were briefly centrifuged for 30 s at 10,000 rpm prior to application. The supernatants were transferred to a 1.5 mL tube and used for the preparation of serial dilutions by diluting each starting concentration by 50% (500–7.81 µg mL−1). Also, the medium for freeze-dried bacteria and reference substance was prepared according to ISO 11348-3 standard. The medium was prepared in a volumetric flask (500 mL) by adding 20 g L−1 NaCl, 2.03 g L−1 MgCl2 × 6 H2O, 0.3 g L−1 KCl and deionized water up to 500 mL. K2Cr2O7 (105.8 mg L−1 prepared in 2% NaCl) was used as a reference substance. DMSO (0.6% prepared in 2% NaCl) was used as control. All solutions were stored at 4 °C until use.
The inhibitory effect on the light emission of A. fischeri was determined with BioFix® Lumi-10 (Macherey-Nagel GmbH & Co. KG, Duren, Germany) according to ISO 11348 standard. Freeze-dried bacteria (A. fischeri NRRL B-11177, Macherey-Nagel GmbH & Co. KG, Duren, Germany) were firstly reconstituted and stored at 4 °C for 10 min using reconstitution solution and additionally stored at 4 °C for 10 min after adding medium for freeze-dried bacteria. Bacteria were incubated at 15 °C with 1 mL of 2% NaCl with different concentrations of PU model substrates and Impranil. The bioluminescence was monitored after 15 min and 30 min of incubation with the test solution.
4.3.3. In Vivo Toxicity on Caenorhabditis Elegans
Synchronized worms (L4 stage) were suspended in a medium containing 95% M9 buffer (3.0 g of KH2PO4, 6.0 g of Na2HPO4, 5.0 g of NaCl, and 1 mL of 1 M MgSO4 × 7 H2O in 1 L of water), 5% LB broth (10 g L−1 tryptone, 5 g L−1 yeast extract, and 10 g L−1 NaCl), and 10 μg mL−1 of cholesterol. The experiment was carried out in 96-well flat-bottomed microtiter plates (Sarstedt, Nümbrecht, Germany) in the final volume of 100 μL per well. Suspension of nematodes (25 μL containing 25–35 nematodes) was transferred to the wells of a 96-well microtiter plate, where 50 μL of the medium was previously added. Next, 25 μL of a solvent control (DMSO) or 25 μL of a concentrated solution was added to the test wells. The final concentrations of the compounds were 500, 100, 50, 25, and 10 µg mL−1. Subsequently, the plates were incubated at 25 °C for 2 days. The fraction of dead worms was determined after 48 h by counting the number of dead worms and the total number of worms in each well, using a stereomicroscope (SMZ143-N2GG, Motic, Wetzlar, Germany). As a negative control experiment, nematodes were exposed to the medium containing 1% (v/v) DMSO.
4.4. Recombinant Proteins and Enzymatic Degradation of PU Model Substrates
Recombinantly expressed enzymes were a cutinase from Fusarium oxysporum (FoCut5a) [70], a cutinase from Humicola insolens (HiC) [71], an outer membrane esterase from Comamonas acidovorans ΤΒ-35 (DaPUase) [72], and a PET hydrolase from Ideonella sakaiensis (IsPETase) [73]. The expression vector pET-22b(+) (Novagen, St. Louis, MO, USA) was used for FoCut5a, DaPUase, and IsPETase expression, while pET-26b(+) (Novagen, St. Louis, MO, USA) was used for HiC. When cell growth reached OD600 0.6–0.8, protein expression was induced by the addition of 0.2 mM isopropyl 1-thio-β-D-galactopyranoside (IPTG), and cultures were further incubated for 20 h at 16 °C. After induction, the cultures were centrifuged at 4000× g for 15 min at 4 °C and the cell pellet was resuspended in 20 mL of 50 mM Tris-HCl buffer containing 300 mM NaCl (pH 8.0). Subsequently, cells were disrupted using an ultrasonic processor (VC 600, Sonics and Materials, Newtown, CT, USA) applying 4 cycles of 60 s sonication (50% Duty Cycle), at 40% amplitude. After disruption, the cell-free extract was collected by centrifugation at 10,000× g for 20 min at 4 °C and the supernatant was filtered through a 0.45 μm filter. Proteins were purified by immobilized metal-ion affinity chromatography (IMAC). The purity of the isolated enzyme was confirmed by sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) and protein concentration was determined by measuring the absorbance at 280 nm, based on the calculated molar extinction coefficient. Fractions containing the purified enzyme were dialyzed overnight at 4 °C against a 20 mM Tris–HCl buffer (pH 8.0).
The enzymatic hydrolysis of the PU-5 model substrate was performed using recombinant enzymes (FoCut5a, HiC, DaPUase, and IsPETase), and commercial protease preparations (protease from Bacillus licheniformis (BacProt) (EC 232-752-2) and Streptomyces griseus (StrepProt) (EC232-909-5), both purchased from Sigma-Aldrich (St. Louis, MO, USA)). Enzymatic reactions containing 10 mg mL−1 of PU substrate and 0.5 μM of each enzyme were performed in 1 mL phosphate buffer pH 7.5, in an Eppendorf Thermomixer Comfort (Eppendorf, Hamburg, Germany) at 30 °C or 50 °C depending on the temperature optimum of the enzyme and 1200 rpm for 24 h. Prior to analysis, 500 μL of methanol was added to the reaction mixture. Afterward, every sample was vortexed and centrifuged at 5000× g at 10 °C. The reaction products were analyzed by HPLC on an Agilent 1260 Infinity II instrument (Agilent Technologies, Santa Clara, CA, USA) using a C-18 reverse-phase Nucleosil®100-5 (Macherey-Nagel, Düren, Germany) under isocratic conditions using a mobile phase consisting of 39.5% methanol, 59.5% water, and 1% triethylamine at a flow rate of 0.8 mL min−1 at 25 °C for 30 min. PU model substrates and degradation products were detected using an Agilent 1260 Infinity II UV Variable Wavelength Detector (G7114B, Agilent Technologies, Santa Clara, CA, USA) at 278 nm.
4.5. Screening and Identification of Microorganisms
An in-house bacterial collection consisting of bacteria isolated from various environmental sites and strains obtained from the Agricultural Research Service (ARS) Culture Collection (Peoria, IL, USA) were screened for their PU-degrading potential using Mineral Salt Medium (15 g L−1 agar, 9 g L−1 Na2HPO4 × 12 H2O, 1.5 g L−1 KH2PO4, 1 g L−1 NH4Cl, 0.2 g L−1 MgSO4 × 7H2O, 0.2 g L−1 CaCl2 × 2H2O, 0.1% trace elements solution, 0.025% N-Z amine) agar plates supplemented with 6 g L−1 of Impranil as carbon source. Cultures were incubated for 3–4 weeks at 30 °C. The formation of clearing zones around microorganisms was considered a positive result. Amycolatopsis mediterranei ISP5501 strain was stained with thiazole orange after growth on solid media. Briefly, cells were scraped into sterile PBS and diluted to approximately 1 × 106 cells mL−1, washed twice with PBS, fixed with paraformaldehyde solution (4%, v/v), and stained with 10 μM of thiazole orange in PBS at 25 °C for 20 min in the dark. Cells were visualized using a fluorescence microscope (Olympus BX51, Applied Imaging Corp., San Jose, CA, USA), under 60 and 100× magnification.
Selected trains were identified by 16S rRNA gene sequencing. The 16S region was amplified via PCR (FastGene TAQ PCR Kit, Nippon Genetics, Düren, Germany) using standard 1496R and 27F primers. Amplicons were sequenced by Macrogen Europe BV (Amsterdam, The Netherlands). Sequences were analyzed using MEGA (Molecular Evolutionary Genetics Analysis) [74] software and the microorganisms were identified using BLAST.
4.6. Whole-Cell Biocatalytic Reactions and Mass Spectrometry Analysis of Degradation Products
Strains used to assess PU model substrate degradation were grown in diluted LB (50%, v/v) with the addition of Impranil (final concentration 0.4%, v/v) at 30 °C and 180 rpm for 72 h. Cells were harvested by centrifugation at 5000× g and 4 °C for 15 min and washed twice with MSM medium. A total of 100 µL of cell suspension was added to 3 mL MSM medium supplemented with PU model substrates (final concentration 1 g L−1) and the whole-cell reaction was incubated for 3 days at 30 °C and 180 rpm.
Reaction products of whole-cell biocatalytic reactions were analyzed using UHPLC-MS/MS. Samples were centrifuged at 12,000× g for 15 min and methanol (to a final concentration of 40%) was added. Next, the samples were filtered through a 0.22 µm PTFE filter. A library of predicted ester and urethane bond degradation products ions was constructed for MS screening (Table S2). The analysis was performed on Agilent 1290 Infinity UHPLC with a 6460 Triple Quad MS detector (Agilent Technologies, Santa Clara, CA, USA). For HPLC analysis, a modified method for monitoring 2,4-TDA [75] was used. An amount of 5 μL of the samples were injected and passed through Agilent Zorbax Eclipse Plus C18 column (Agilent Technologies, Santa Clara, CA, USA) (2.1 × 50 mm, 1.8 µm) at a flow rate of 0.4 mL min−1 at 25 °C. Mobile phases were A: water with 1% formic acid; and B: acetonitrile with 1% formic acid with a gradient: 0–1 min 40–95% B; 2.5 min 95% B. Electrospray source was operated in positive ion mode with the following common parameters: nitrogen drying gas temperature 300 °C, nitrogen sheath gas temperature 300 °C, nitrogen drying gas flow 10 L min−1, nitrogen sheath gas flow 7.5 L min−1, nebulizer pressure 45 psi, capillary voltage 3500 V and nozzle voltage 500 V. Spectra were acquired and analyzed using Agilent Technologies MassHunter software (Version 10.0).
4.7. Degradation of PU Powder Material
LB medium supplemented with 0.4% (v/v) of Impranil (5 mL) was inoculated with A. mediterranei ISP5501 strain and incubated at 30 °C on orbital shaker 180 rpm for 48 h. This culture (1 mL) was used for the inoculation of 100 mL half-strength LB supplemented with 0.4% (v/v) of Impranil. Cultures were incubated at 30 °C and 180 rpm for 72 h and subsequently centrifuged at 5000× g for 10 min at 4 °C. The supernatant (extracellular fraction) was collected, filter sterilized, and dialyzed against PBS buffer overnight, while the cell pellet of each culture was resuspended in 5 mL PBS buffer. The cells were disrupted using the ultrasonic processor VC 600 (Sonics and Materials, Newtown, CT, USA) applying 4 cycles of 60 s sonication (50% Duty Cycle), at 40% amplitude. After sonication, the disrupted cells were centrifuged at 10,000× g for 20 min at 4 °C, and the supernatant was filtered through a 0.45 µm filter (intracellular fraction). Total protein mix (mixed fraction) was obtained by mixing the extracellular and intracellular fractions at a ratio of 3:1 (v/v).
Enzyme reactions were performed in an Eppendorf Thermomixer Comfort (Eppendorf, Hamburg, Germany) by incubating 10 mg of polyether PU with 0.26 mg of the extracellular, intracellular, or mixed fraction in 1 mL of 100 mM phosphate buffer (pH 7), at 30 °C for 72 h. After 72 h, PU powder was isolated by centrifugation, and washed with 2% (w/v) SDS solution, followed by a double rinse with ultrapure water. Finally, the powder was again isolated by centrifugation and freeze-dried under a vacuum before its properties were determined. In control samples, PU powder was treated in the same manner, but each of the protein fractions has been previously boiled for 15 min.
Polyether PU (LPR7560, Laripur) in the form of pellets was cryo-milled in a Pulverisette 14 (Fritsch Corp., Idar-Oberstein, Germany), resulting in particle diameter smaller than 500 μm before it was used as a substrate. PU powder was used as substrate with A. mediterranei ISP5501 whole cells, whereby 100 mg (0.1% w/v) of PU powder was added to cultures, while in some cases 0.4% (v/v) Impranil was also supplemented in the medium. ISP5501 liquid cultures were incubated for 72 h under the aforementioned conditions and subsequently centrifuged at 1000× g for 10 min at 4 °C, allowing the PU powder to precipitate. After discarding the supernatant, the powder was washed and isolated as mentioned before. Abiotic controls (without bacterial cells) were set up for validating experiment results.
PU material characterization was performed using differential scanning calorimetry (DSC) and thermogravimetric analysis (TGA), as previously described [76]. The determination of the molecular masses of virgin and treated PU powder was performed with gel permeation chromatography (GPC) using two PLgel MIXED-D 5 μm columns (300 × 7.5 mm) in Agilent 1260 Infinity II instrument (Agilent Technologies, Santa Clara, CA, USA), as described previously [76].
4.8. Assessment of A. mediterranei ISP5501 Esterase and Proteinase Activities
The esterase activity of A. mediterranei ISP5501 was determined in each protein fraction using p-nitrophenyl butyrate (pNPB) as a substrate. The activity assay for each of the fractions was performed using pNPB at 1 mM concentration in 0.1 M phosphate-citrate buffer at pH 6. Reactions were initiated by adding 20 µL of each of the intracellular or extracellular fractions to 230 µL of the substrate and the release of p-nitrophenol was monitored by measuring absorbance at 410 nm in a SpectraMax-250 microplate reader (Molecular Devices, Sunnyvale, CA, USA) equipped with SoftMaxPro software (version 1.1, Molecular Devices, Sunnyvale, CA, USA) set at 35 °C. Proteolytic activity was determined using azocasein as substrate after modifying the protocol of Samal et al. [77]. In specific, proteolytic activity was estimated after mixing 25 µL of the enzyme with 0.4 mg azocasein and 175 µL of 0.1 M Tris-HCl buffer pH 8. The reactions were incubated for 20 min at 40 °C. After incubation, 200 µL of 0.1 M trichloroacetic acid (TCA) was added to the reaction mixture, and afterward, the reactions were centrifuged at 3000× g for 2 min. Then, 200 µL of the supernatant was removed and mixed with 200 µL of 0.5 M NaOH. The absorbance of the final mixture was measured at 440 nm in a SpectraMax-250 microplate reader (Molecular Devices, Sunnyvale, CA, USA) equipped with SoftMaxPro software (version 1.1, Molecular Devices, Sunnyvale, CA, USA). Urease activity was determined after mixing 500 mM urea and 0.002% phenol red in 10 mM K2HPO4 solution pH 6.2. Reactions were initiated by adding 25 µL of sample in 225 µL of the reaction mixture. The increase in the absorbance at 570 nm was recorded using a SpectraMax-250 microplate reader [78,79]. Apart from protein fractions, whole-cell samples (full cultures) and culture supernatants were also assayed for esterase, protease, or urease activity. Protein concentration was estimated according to Lowry et al. using BSA as standard solutions [80].
4.9. A. mediterranei ISP5501 Whole-Genome Sequencing, Genome Assembly, and Annotation
A 350-bp insert size library was prepared and sequenced in paired-end mode (read length, 150 bp) by Novogene Europe on a NovaSeq 6000 (Illumina, San Diego, CA, USA) instrument and a total of 5,908,038 paired reads were generated. Raw reads were preprocessed with TrimGalore v0.6.5 and cutadapt v2.9 [81]. The Illumina adapter sequences were removed (with a stringency of 3), bases with a quality score less than 10 were trimmed, and reads smaller than 100 bases or with no pair were discarded. De novo genome assembly was performed with Spades v3.13.0 [82] and was further scaffolded and gaps filled through an improvement pipeline. Genome completeness was assessed with BUSCO v5.1.2 using the Actinobacteria phylum single-copy orthologs from OrthoDB v10 [83].
Gene prediction and functional annotation were performed with the NCBI Prokaryotic Genome Annotation Pipeline (PGAP, release 2022-04-14) [84]. The genome was searched for secondary metabolite biosynthetic gene clusters with antiSMASH v.6.1.1 [85]. Predicted proteins were mapped to KEGG pathways using BlastKOALA [86] and their subcellular localization was predicted with the GP4 pipeline for gram-positive bacteria [87]. Proteins predicted as extracellular or of unknown localization with GP4 were further searched for transmembrane proteins with DeepTMHMM [88] since GP4 cannot predict the Actinobacteria outer membrane proteins. The annotated genome assembly has been deposited in GenBank under accession number CP100416.
The predicted proteome of ISP5501 was searched for homologs of PU-active enzymes. All available sequences from biochemically characterized PU-active enzymes were downloaded from PAZy [22] and were used as templates for BLAST searches. The alignments were filtered for protein sequence identity > 30% and for >50% alignment coverage of both the template and the target sequence. Lastly, hidden Markov model profiles of alpha/beta-hydrolase families that contain PU-active enzymes were downloaded from the Lipase Engineering Database (LED) [89] and used to classify the enzymes found with BLAST. Next, we searched for broader enzyme families related to PU hydrolysis. These families include amidases, esterases, proteases, ureases, and other α/β hydrolases. Proteins were assigned to enzyme families with InterProScan v5.56-89.0 and PGAP. All 7 available A. mediterranei proteomes from RefSeq were used for comparison with ISP5501 genome.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/catal13020278/s1, Details of synthetic procedures and NMR spectra (Figures S1–S16); PU-5 degradation products after enzymatic treatment (Figure S17); A. mediterranei ISP5501 visualized under a fluorescent microscope (Figure S18); solubility of PU model compounds in a selection of common organic solvents (Table S1); list of predicted PU-7 degradation products (Table S2); identification of 22 Impranil-degrading bacterial strains by 16S sequencing (Table S3); PU depolymerization associated enzyme families in A. mediterranei genomes (Table S4).
Author Contributions
B.P.: methodology, validation, investigation, writing—original draft, and writing—review and editing; S.S.B.: methodology, investigation, and writing—original draft; D.M.: methodology, investigation, and writing—original draft; T.I.-T.: methodology, investigation, and writing—original draft; B.L.: methodology, investigation, writing—original draft, and visualization; V.B.: conceptualization, validation, and writing—review and editing; V.M.: conceptualization, validation, and writing—review and editing; M.G.: methodology, validation, and writing—review and editing; K.M.: investigation, writing—original draft, and visualization; G.T.: investigation, writing—original draft, and writing—review and editing; R.S.: investigation, methodology, and writing—original draft; E.T.: methodology, resources, investigation, writing—review and editing, and supervision; J.N.-R.: conceptualization, methodology, validation, resources, writing—review and editing, supervision, and funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the European Union’s Horizon 2020 Research and Innovation Programme under grant agreement No. 870292 (BioICEP) and by the National Natural Science Foundation of China (Nos. 31961133016, 31961133015, and 31961133014).
Data Availability Statement
Data is contained within the article or supplementary material.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Liu, J.; He, J.; Xue, R.; Xu, B.; Qian, X.; Xin, F.; Blank, L.M.; Zhou, J.; Wei, R.; Dong, W. Biodegradation and up-cycling of polyurethanes: Progress, challenges, and prospects. Biotechnol. Adv. 2021, 48, 107730. [Google Scholar] [CrossRef] [PubMed]
- Mordorinteligence. Polyurethane Market—Growth, Trends, COVID-19 Impact, and Forecasts (2022–2027). 2021. Available online: https://www.reportlinker.com/p06316155/Polyurethane-Market-Growth-Trends-COVID-19-Impact-and-Forecasts.html?utm_source=GNW (accessed on 28 September 2022).
- Zhang, Y.-Q.; Lykaki, M.; Markiewicz, M.; Alrajoula, M.T.; Kraas, C.; Stolte, S. Environmental contamination by microplastics originating from textiles: Emission, transport, fate and toxicity. J. Haz. Mat. 2022, 430, 128453. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Gracida-Alvarez, U.R.; Gallant, E.T.; Gillis, P.A.; Marques, Y.A.; Abramo, G.P.; Hawkins, T.R.; Dunn, J.B. Material flows of polyurethane in the United States. Environ. Sci. Technol. 2021, 55, 14215–14224. [Google Scholar] [CrossRef]
- Esperanza, M.; Font, R.; GarcíA, A. Toxic byproducts from the combustion of varnish wastes based on polyurethane in a laboratory furnace. J. Haz. Mat. 2000, 77, 107–121. [Google Scholar] [CrossRef]
- Gadhave, R.V.; Srivastava, S.; Mahanwar, P.A.; Gadekar, P.T. Recycling and disposal methods for polyurethane wastes: A review. Open J. Polymer Chem. 2019, 9, 39–51. [Google Scholar] [CrossRef]
- Zimmermann, L.; Dierkes, G.; Ternes, T.A.; VöLker, C.; Wagner, M. Benchmarking the in vitro toxicity and chemical composition of plastic consumer products. Environ. Sci. Technol. 2019, 53, 11467–11477. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Nag, R.; Cummins, E. Ranking of potential hazards from microplastics polymers in the marine environment. J. Haz. Mat. 2022, 429, 128399. [Google Scholar] [CrossRef]
- Oz, K.; Merav, B.; Sara, S.; Yael, D. Volatile organic compound emissions from polyurethane mattresses under variable environmental conditions. Environ. Sci. Technol. 2019, 53, 9171–9180. [Google Scholar] [CrossRef]
- Garrido, M.A.; Gerecke, A.C.; Heeb, N.; Font, R.; Conesa, J.A. Isocyanate emissions from pyrolysis of mattresses containing polyurethane foam. Chemosphere 2017, 168, 667–675. [Google Scholar] [CrossRef]
- Luft, A.; BröDer, K.; Kunkel, U.; Schulz, M.; Dietrich, C.; Baier, R.; Heininger, P.; Ternes, T.A. Nontarget analysis via LC-QTOF-MS to assess the release of organic substances from polyurethane coating. Environ. Sci. Technol. 2017, 51, 9979–9988. [Google Scholar] [CrossRef]
- Ibarra, J.C.; Uscátegui, Y.L.; Arévalo, F.R.; Díaz, L.E.; Valero, M.F. Influence of diisocyanate chemical structure on the properties, degradation and cytotoxicity of polyurethanes obtained from castor oil. J. Biomat. Tissue Eng. 2018, 8, 279–289. [Google Scholar] [CrossRef]
- Simón, D.; Borreguero, A.; De Lucas, A.; Rodríguez, J. Recycling of polyurethanes from laboratory to industry, a journey towards the sustainability. Waste Manag. 2018, 76, 147–171. [Google Scholar] [CrossRef] [PubMed]
- Marson, A.; Masiero, M.; Modesti, M.; Scipioni, A.; Manzardo, A. Life Cycle Assessment of polyurethane foams from polyols obtained through chemical recycling. ACS Omega 2021, 6, 1718–1724. [Google Scholar] [CrossRef]
- Tournier, V.; Topham, C.; Gilles, A.; David, B.; Folgoas, C.; Moya-Leclair, E.; Kamionka, E.; Desrousseaux, M.-L.; Texier, H.; Gavalda, S. An engineered PET depolymerase to break down and recycle plastic bottles. Nature 2020, 580, 216–219. [Google Scholar] [CrossRef]
- Jin, X.; Dong, J.; Guo, X.; Ding, M.; Bao, R.; Luo, Y. Current advances in polyurethane biodegradation. Polym. Int. 2022, 71, 1384–1392. [Google Scholar] [CrossRef]
- Mahajan, N.; Gupta, P. New insights into the microbial degradation of polyurethanes. RSC Adv. 2015, 5, 41839–41854. [Google Scholar] [CrossRef]
- Skleničková, K.; Abbrent, S.; Halecký, M.; Kočí, V.; Beneš, H. Biodegradability and ecotoxicity of polyurethane foams: A review. Crit. Rev. Environ. Sci. Technol. 2020, 52, 157–202. [Google Scholar] [CrossRef]
- Magnin, A.; Entzmann, L.; Pollet, E.; Avérous, L. Breakthrough in polyurethane bio-recycling: An efficient laccase-mediated system for the degradation of different types of polyurethanes. Waste Manag. 2021, 132, 23–30. [Google Scholar] [CrossRef]
- Nakajima-Kambe, T.; Onuma, F.; Akutsu, Y.; Nakahara, T. Determination of the polyester polyurethane breakdown products and distribution of the polyurethane degrading enzyme of Comamonas acidovorans strain TB-35. J. Ferment. Bioeng. 1997, 83, 456–460. [Google Scholar] [CrossRef]
- PAZy—The Plastics-Active Enzymes Database. Available online: https://pazy.eu/doku.php (accessed on 22 July 2022).
- Buchholz, P.; Zhang, H.; Perez-Garcia, P.; Nover, L.-L.; Chow, J.; Streit, W.R.; Pleiss, J. Plastics degradation by hydrolytic enzymes: The Plastics-Active Enzymes Database-PAZy. Proteins 2021, 90, 1443–1456. [Google Scholar] [CrossRef]
- Magnin, A.; Pollet, E.; Phalip, V.; Avérous, L. Evaluation of biological degradation of polyurethanes. Biotechnol. Advan. 2020, 39, 107457. [Google Scholar] [CrossRef]
- Magnin, A.; Pollet, E.; Perrin, R.; Ullmann, C.; Persillon, C.; Phalip, V.; Avérous, L. Enzymatic recycling of thermoplastic polyurethanes: Synergistic effect of an esterase and an amidase and recovery of building blocks. Waste Manag. 2019, 85, 141–150. [Google Scholar] [CrossRef]
- Liu, J.; Liu, J.; Xu, B.; Xu, A.; Cao, S.; Wei, R.; Zhou, J.; Jiang, M.; Dong, W. Biodegradation of polyether-polyurethane foam in yellow mealworms (Tenebrio molitor) and effects on the gut microbiome. Chemosphere 2022, 304, 135263. [Google Scholar] [CrossRef]
- Chow, J.; Perez-Garcia, P.; Dierkes, R.; Streit, W.R. Microbial enzymes will offer limited solutions to the global plastic pollution crisis. Microb. Biotechnol. 2023, 16, 195–217. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Hu, J.; Yang, S.; Xu, W.; Wang, Z.; Zhuang, P.; Grossart, H.-P.; Luo, Z. Biodegradation of polyester polyurethane by the marine fungus Cladosporium halotolerans 6UPA1. J. Haz. Mat. 2022, 437, 129406. [Google Scholar] [CrossRef]
- Howard, G.; Vicknair, J.; Mackie, R. Sensitive plate assay for screening and detection of bacterial polyurethanase activity. Lett. Appl. Microbiol. 2001, 32, 211–214. [Google Scholar] [CrossRef]
- Biffinger, J.C.; Barlow, D.E.; Cockrell, A.L.; Cusick, K.D.; Hervey, W.J.; Fitzgerald, L.A.; Nadeau, L.J.; Hung, C.S.; Crookes-Goodson, W.J.; Russell, J.N., Jr. The applicability of Impranil® DLN for gauging the biodegradation of polyurethanes. Polym. Degrad. Stabil. 2015, 120, 178–185. [Google Scholar] [CrossRef]
- Islam, S.; Apitius, L.; Jakob, F.; Schwaneberg, U. Targeting microplastic particles in the void of diluted suspensions. Environ. Int. 2019, 123, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Gamerith, C.; Acero, E.H.; Pellis, A.; Ortner, A.; Vielnascher, R.; Luschnig, D.; Zartl, B.; Haernvall, K.; Zitzenbacher, S.; Strohmeier, G. Improving enzymatic polyurethane hydrolysis by tuning enzyme sorption. Polym. Degrad. Stabil. 2016, 132, 69–77. [Google Scholar] [CrossRef]
- Nishio, A.; Mochizuki, A.; Sugiyama, J.I.; Takeuchi, K.; Asai, M.; Yonetake, K.; Ueda, M. Synthesis and characterization of ordered polyurethanes from nonsymmetric diisocyanate and ethylene glycol. J. Polym. Sci. A Polym. Chem. 2000, 38, 2106–2114. [Google Scholar] [CrossRef]
- Hikita, T.; Mochizuki, A.; Sugiyama, J.-I.; Takeuchi, K.; Asai, M.; Ueda, M. Head-to-tail regularity of polyurethanes from p-isocyanatobenzyl isocyanate and ethylene glycol by a distannoxane catalyst. Polym. J. 2001, 33, 547–553. [Google Scholar] [CrossRef]
- Chambers, J.; Reese, C.B. The thermal decomposition of some tolylene bis-carbamates. Brit. Polym. J. 1977, 9, 41–46. [Google Scholar] [CrossRef]
- Lubczak, R.; Lubczak, J. Reactions of hydroxyalkyl esters with phenyl isocyanate. J. Appl. Polym. Sci. 2005, 96, 1357–1367. [Google Scholar] [CrossRef]
- Utomo, R.N.C.; Li, W.-J.; Tiso, T.; Eberlein, C.; Doeker, M.; Heipieper, H.J.; Jupke, A.; Wierckx, N.; Blank, L.M. Defined microbial mixed culture for utilization of polyurethane monomers. ACS Sust. Chem. Eng. 2020, 8, 17466–17474. [Google Scholar] [CrossRef]
- Mishra, S.; Pang, S.; Zhang, W.; Lin, Z.; Bhatt, P.; Chen, S. Insights into the microbial degradation and biochemical mechanisms of carbamates. Chemosphere 2021, 279, 130500. [Google Scholar] [CrossRef]
- Amato-Lourenço, L.F.; Carvalho-Oliveira, R.; Júnior, G.R.; Dos Santos Galvão, L.; Ando, R.A.; Mauad, T. Presence of airborne microplastics in human lung tissue. J. Haz. Mat. 2021, 416, 126124. [Google Scholar] [CrossRef]
- Neale, P.A.; Antony, A.; Bartkow, M.E.; Farre, M.J.; Heitz, A.; Kristiana, I.; Tang, J.Y.; Escher, B.I. Bioanalytical assessment of the formation of disinfection byproducts in a drinking water treatment plant. Environ. Sci. Technol. 2012, 46, 10317–10325. [Google Scholar] [CrossRef]
- Ventura, S.P.; De Morais, P.; Coelho, J.A.; Sintra, T.; Coutinho, J.A.; Afonso, C.A. Evaluating the toxicity of biomass derived platform chemicals. Green Chem. 2016, 18, 4733–4742. [Google Scholar] [CrossRef]
- Dodard, S.; Renoux, A.; Hawari, J.; Ampleman, G.; Thiboutot, S.; Sunahara, G. Ecotoxicity characterization of dinitrotoluenes and some of their reduced metabolites. Chemosphere 1999, 38, 2071–2079. [Google Scholar] [CrossRef]
- Šepič, E.; Bricelj, M.; Leskovšek, H. Toxicity of fluoranthene and its biodegradation metabolites to aquatic organisms. Chemosphere 2003, 52, 1125–1133. [Google Scholar] [CrossRef]
- Zimmermann, L.; Göttlich, S.; Oehlmann, J.; Wagner, M.; Völker, C. What are the drivers of microplastic toxicity? Comparing the toxicity of plastic chemicals and particles to Daphnia magna. Environ. Pollut. 2020, 267, 115392. [Google Scholar] [CrossRef] [PubMed]
- Wittkowski, P.; Marx-Stoelting, P.; Violet, N.; Fetz, V.; Schwarz, F.; OelgeschläGer, M.; SchöNfelder, G.; Vogl, S. Caenorhabditis elegans as a promising alternative model for environmental chemical mixture effect assessment—A comparative study. Environ. Sci. Technol. 2019, 53, 12725–12733. [Google Scholar] [CrossRef]
- Judy, J.D.; Williams, M.; Gregg, A.; Oliver, D.; Kumar, A.; Kookana, R.; Kirby, J.K. Microplastics in municipal mixed-waste organic outputs induce minimal short to long-term toxicity in key terrestrial biota. Environ. Pollut. 2019, 252, 522–531. [Google Scholar] [CrossRef]
- Mcqueen, C.A.; Williams, G.M. Review of the genotoxicity and carcinogenicity of 4, 4′-methylene-dianiline and 4, 4′-methylene-bis-2-chloroaniline. Mutat. Res./Rev. Gen. Toxicol. 1990, 239, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Cregut, M.; Bedas, M.; Durand, M.-J.; Thouand, G. New insights into polyurethane biodegradation and realistic prospects for the development of a sustainable waste recycling process. Biotechnol. Adv. 2013, 31, 1634–1647. [Google Scholar] [CrossRef] [PubMed]
- Diefenbach, X.W.; Farasat, I.; Guetschow, E.D.; Welch, C.J.; Kennedy, R.T.; Sun, S.; Moore, J.C. Enabling biocatalysis by high-throughput protein engineering using droplet microfluidics coupled to mass spectrometry. ACS Omega 2018, 3, 1498–1508. [Google Scholar] [CrossRef]
- Djapovic, M.; Milivojevic, D.; Ilic-Tomic, T.; Lješević, M.; Nikolaivits, E.; Topakas, E.; Maslak, V.; Nikodinovic-Runic, J. Synthesis and characterization of polyethylene terephthalate (PET) precursors and potential degradation products: Toxicity study and application in discovery of novel PETases. Chemosphere 2021, 275, 130005. [Google Scholar] [CrossRef]
- Yue, W.; Yin, C.-F.; Sun, L.; Zhang, J.; Xu, Y.; Zhou, N.-Y. Biodegradation of bisphenol-A polycarbonate plastic by Pseudoxanthomonas sp. strain NyZ600. J. Haz. Mat. 2021, 416, 125775. [Google Scholar] [CrossRef]
- Labow, R.S.; Meek, E.; Santerre, J.P. The biodegradation of poly (urethane) s by the esterolytic activity of serine proteases and oxidative enzyme systems. J. Biomat. Sci. 1999, 10, 699–713. [Google Scholar] [CrossRef]
- Gololobov, M.Y.; Morozova, I.P.; Vojushina, T.L.; Timokhina, E.A.; Stepanov, V.M. Subtilisin from Bacillus subtilis strain 72. The influence of substrate structure, temperature and pH on catalytic properties. Biochim. Biophys. Acta 1992, 1118, 267–276. [Google Scholar] [CrossRef]
- Spasic, J.; Mandic, M.; Djokic, L.; Nikodinovic-Runic, J. Streptomyces spp. in the biocatalysis toolbox. Appl. Microbiol. Biotechnol. 2018, 102, 3513–3536. [Google Scholar] [CrossRef] [PubMed]
- Molitor, R.; Bollinger, A.; Kubicki, S.; Loeschcke, A.; Jaeger, K.E.; Thies, S. Agar plate-based screening methods for the identification of polyester hydrolysis by Pseudomonas species. Microb. Biotechnol. 2020, 13, 274–284. [Google Scholar] [CrossRef]
- Singhvi, N.; Singh, P.; Prakash, O.; Gupta, V.; Lal, S.; Bechthold, A.; Singh, Y.; Singh, R.K.; Lal, R. Differential mass spectrometry-based proteome analyses unveil major regulatory hubs in rifamycin B production in Amycolatopsis mediterranei. J. Proteom. 2021, 239, 104168. [Google Scholar] [CrossRef]
- Pranamuda, H.; Tokiwa, Y. Degradation of poly (L-lactide) by strains belonging to genus Amycolatopsis. Biotechnol. Lett. 1999, 21, 901–905. [Google Scholar] [CrossRef]
- Gambarini, V.; Pantos, O.; Kingsbury, J.M.; Weaver, L.; Handley, K.M.; Lear, G. Phylogenetic distribution of plastic-degrading microorganisms. Msystems 2021, 6, e01112–e01120. [Google Scholar] [CrossRef] [PubMed]
- Joo, S.; Cho, I.J.; Seo, H.; Son, H.F.; Sagong, H.-Y.; Shin, T.J.; Choi, S.Y.; Lee, S.Y.; Kim, K.-J. Structural insight into molecular mechanism of poly (ethylene terephthalate) degradation. Nat. Comm. 2018, 9, 382. [Google Scholar] [CrossRef]
- Christenson, E.M.; Patel, S.; Anderson, J.M.; Hiltner, A. Enzymatic degradation of poly (ether urethane) and poly (carbonate urethane) by cholesterol esterase. Biomaterials 2006, 27, 3920–3926. [Google Scholar] [CrossRef]
- Loredo-Treviño, A.; Gutiérrez-Sánchez, G.; Rodríguez-Herrera, R.; Aguilar, C.N. Microbial enzymes involved in polyurethane biodegradation: A review. J. Polym. Environ. 2012, 20, 258–265. [Google Scholar] [CrossRef]
- Wei, R.; Zimmermann, W. Microbial enzymes for the recycling of recalcitrant petroleum-based plastics: How far are we? Microb. Biotechnol. 2017, 10, 1308–1322. [Google Scholar] [CrossRef]
- Puiggené, Ò.; Espinosa, M.J.C.; Schlosser, D.; Thies, S.; Jehmlich, N.; Kappelmeyer, U.; Schreiber, S.; Wibberg, D.; Kalinowski, J.; Harms, H. Extracellular degradation of a polyurethane oligomer involving outer membrane vesicles and further insights on the degradation of 2, 4-diaminotoluene in Pseudomonas capeferrum TDA1. Sci. Rep. 2022, 12, 1–12. [Google Scholar] [CrossRef]
- Shah, Z.; Krumholz, L.; Aktas, D.F.; Hasan, F.; Khattak, M.; Shah, A.A. Degradation of polyester polyurethane by a newly isolated soil bacterium, Bacillus subtilis strain MZA-75. Biodegradation 2013, 24, 865–877. [Google Scholar] [CrossRef] [PubMed]
- Magnin, A.; Hoornaert, L.; Pollet, E.; Laurichesse, S.; Phalip, V.; Avérous, L. Isolation and characterization of different promising fungi for biological waste management of polyurethanes. Microb. Biotechnol. 2019, 12, 544–555. [Google Scholar] [CrossRef]
- Wei, R.; Oeser, T.; Then, J.; Kühn, N.; Barth, M.; Schmidt, J.; Zimmermann, W. Functional characterization and structural modeling of synthetic polyester-degrading hydrolases from Thermomonospora curvata. AMB Express 2014, 4, 44. [Google Scholar] [CrossRef]
- Bauer, T.L.; Buchholz, P.C.; Pleiss, J. The modular structure of α/β-hydrolases. FEBS J. 2020, 287, 1035–1053. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Henehan, G.T.; Kinsella, G.K.; Ryan, B.J. An extracellular lipase from Amycolatopsis mediterannei is a cutinase with plastic degrading activity. Comp. Struct. Biotechnol. J. 2021, 19, 869–879. [Google Scholar] [CrossRef] [PubMed]
- Schupp, T.; Toupet, C.; Engel, N.; Goff, S. Cloning and sequence analysis of the putative rifamycin polyketide synthase gene cluster from Amycolatopsis mediterranei. FEMS Microb. Lett. 1998, 159, 201–267. [Google Scholar] [CrossRef]
- Chek, M.F.; Hiroe, A.; Hakoshima, T.; Sudesh, K.; Taguchi, S. PHA synthase (PhaC): Interpreting the functions of bioplastic-producing enzyme from a structural perspective. Appl. Microbiol. Biotechnol. 2019, 103, 1131–1141. [Google Scholar] [CrossRef]
- Dimarogona, M.; Nikolaivits, E.; Kanelli, M.; Christakopoulos, P.; Sandgren, M.; Topakas, E. Structural and functional studies of a Fusarium oxysporum cutinase with polyethylene terephthalate modification potential. Biochim. Biophys. Acta-Gen. Subj. 2015, 1850, 2308–2317. [Google Scholar] [CrossRef]
- Ronkvist, Å.M.; Xie, W.; Lu, W.; Gross, R.A. Cutinase-catalyzed hydrolysis of poly (ethylene terephthalate). Macromolecules 2009, 42, 5128–5138. [Google Scholar] [CrossRef]
- Shigeno-Akutsu, Y.; Nakajima-Kambe, T.; Nomura, N.; Nakahara, T. Purification and properties of culture-broth-secreted esterase from the polyurethane degrader Comamonas acidovorans TB-35. J. Biosci. Bioeng. 1999, 88, 484–487. [Google Scholar] [CrossRef]
- Yoshida, S.; Hiraga, K.; Takehana, T.; Taniguchi, I.; Yamaji, H.; Maeda, Y.; Toyohara, K.; Miyamoto, K.; Kimura, Y.; Oda, K. A bacterium that degrades and assimilates poly (ethylene terephthalate). Science 2016, 351, 1196–1199. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Molec. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef]
- Espinosa, M.; Blanco, A.; Schmidgall, T.; Atanasoff-Kardjalieff, A.; Kappelmeyer, U.; Tischler, D.; Pieper, D.; Heipieper, H.; Eberlein, C. Toward biorecycling: Isolation of a soil bacterium that grows on a polyurethane oligomer and monomer. Front. Microbiol. 2020, 11, 404. [Google Scholar] [CrossRef]
- Nikolaivits, E.; Taxeidis, G.; Gkountela, C.; Vouyiouka, S.; Maslak, V.; Nikodinovic-Runic, J.; Topakas, E. A polyesterase from the Antarctic bacterium Moraxella sp. degrades highly crystalline synthetic polymers. J. Haz. Mat. 2022, 434, 128900. [Google Scholar] [CrossRef]
- Samal, B.B.; Karan, B.; Stabinsky, Y. Stability of two novel serine proteinases in commercial laundry detergent formulations. Biotechnol. Bioeng. 1990, 35, 650–652. [Google Scholar] [CrossRef]
- Goldie, J.; Veldhuyzen Van Zanten, S.; Jalali, S.; Hollingsworth, J.; Riddell, R.; Richardson, H.; Hunt, R. Optimization of a medium for the rapid urease test for detection of Campylobacter pylori in gastric antral biopsies. J. Clin. Microbiol. 1989, 27, 2080–2082. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Kawase, M.; Tani, S. Urease inhibitory activity of simple α, β-unsaturated ketones. Life Sci 2003, 73, 2985–2990. [Google Scholar] [CrossRef] [PubMed]
- Lowry, O.; Rosebrough, N.; Farr, A.; Randall, R. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comp. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Manni, M.; Berkeley, M.R.; Seppey, M.; Simão, F.A.; Zdobnov, E.M. BUSCO update: Novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Molec. Biol. Evol. 2021, 38, 4647–4654. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; O’neill, K.R.; Haft, D.H.; Dicuccio, M.; Chetvernin, V.; Badretdin, A.; Coulouris, G.; Chitsaz, F.; Derbyshire, M.K.; Durkin, A.S. RefSeq: Expanding the prokaryotic genome annotation pipeline reach with protein family model curation. Nucleic Acids Res. 2021, 49, D1020–D1028. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; Van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef] [PubMed]
- Grasso, S.; Van Rij, T.; Van Dijl, J.M. GP4: An integrated Gram-positive protein prediction pipeline for subcellular localization mimicking bacterial sorting. Brief Bioinformat. 2021, 22, bbaa302. [Google Scholar] [CrossRef] [PubMed]
- Hallgren, J.; Tsirigos, K.D.; Pedersen, M.D.; Armenteros, J.J.A.; Marcatili, P.; Nielsen, H.; Krogh, A.; Winther, O. DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. bioRxiv 2022. [Google Scholar] [CrossRef]
- Biocatnet, The Lipase Engineering Database (LED). Available online: http://www.led.uni-stuttgart.de/ (accessed on 25 July 2022).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).