Heterologous Expression and Characterization of Plant Lipase LIP2 from Elaeis guineensis Jacq. Oil Palm Mesocarp in Escherichia coli
Abstract
1. Introduction
2. Results
2.1. Gene analysis of LIP2
2.2. Structure Prediction via Homology Modelling
2.3. Screening of Recombinant LIP2 Lipase on Agar Plate
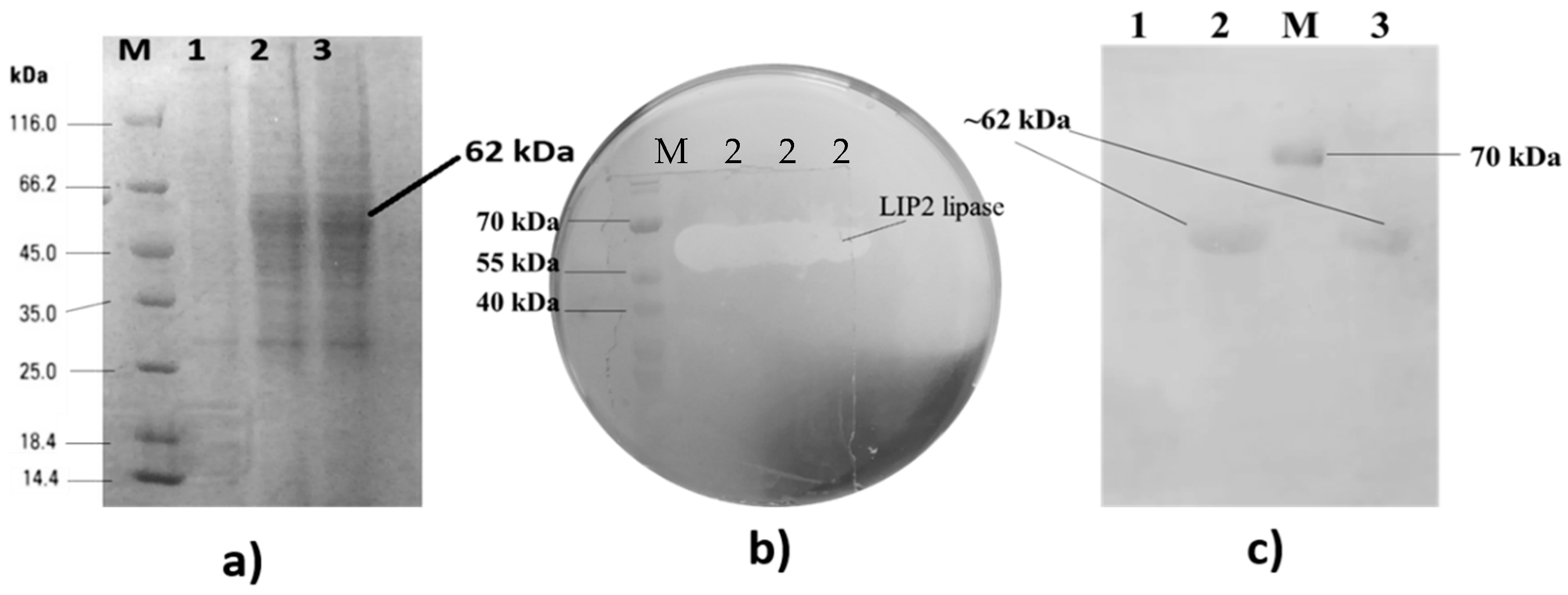
2.4. Detection of LIP2 Lipase Expression by SDS-PAGE
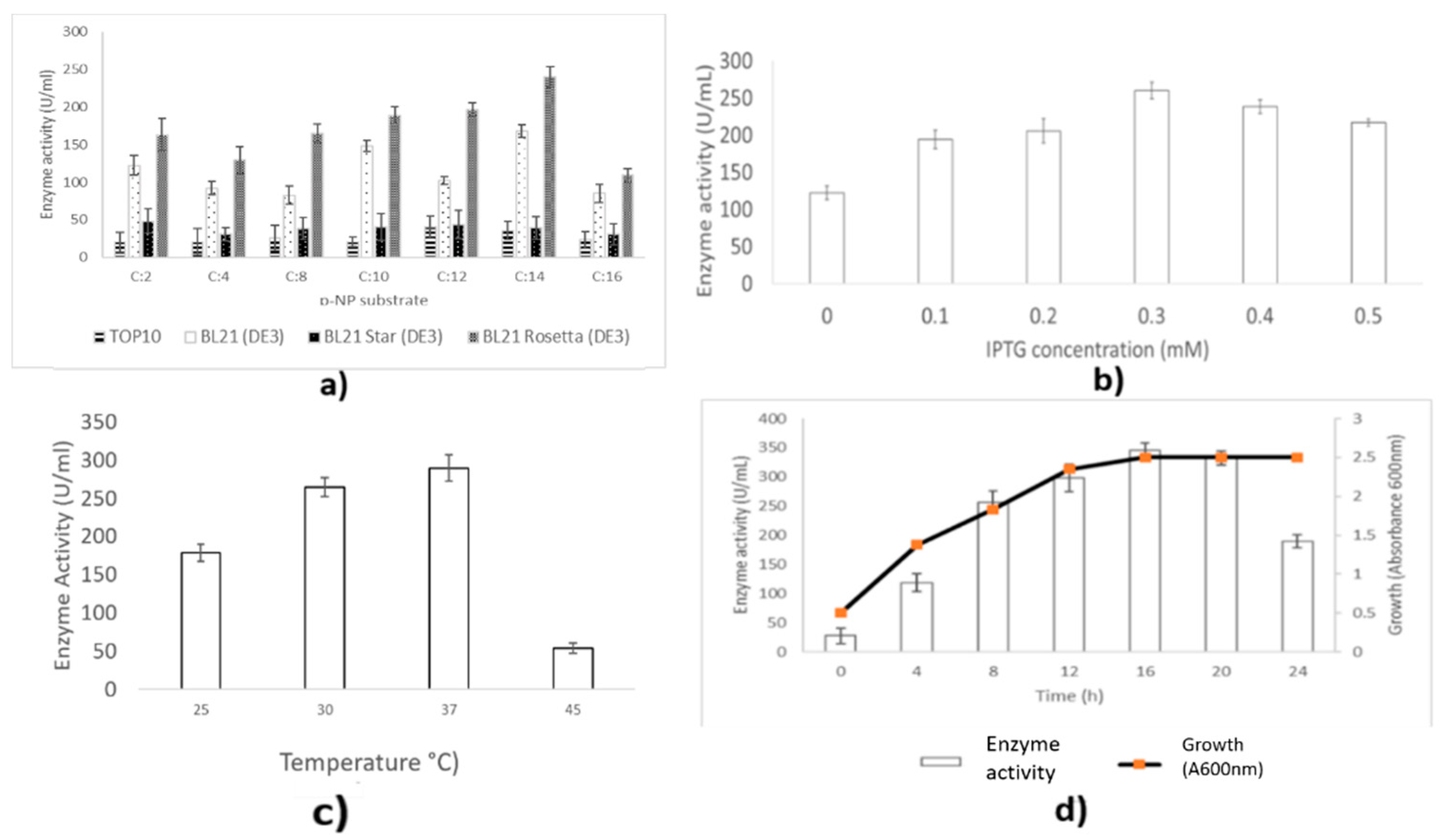
2.5. Optimization of Lipase Secretion
2.6. Purification of LIP2 Lipase
2.7. Characterization of LIP2 via Biochemical Approach
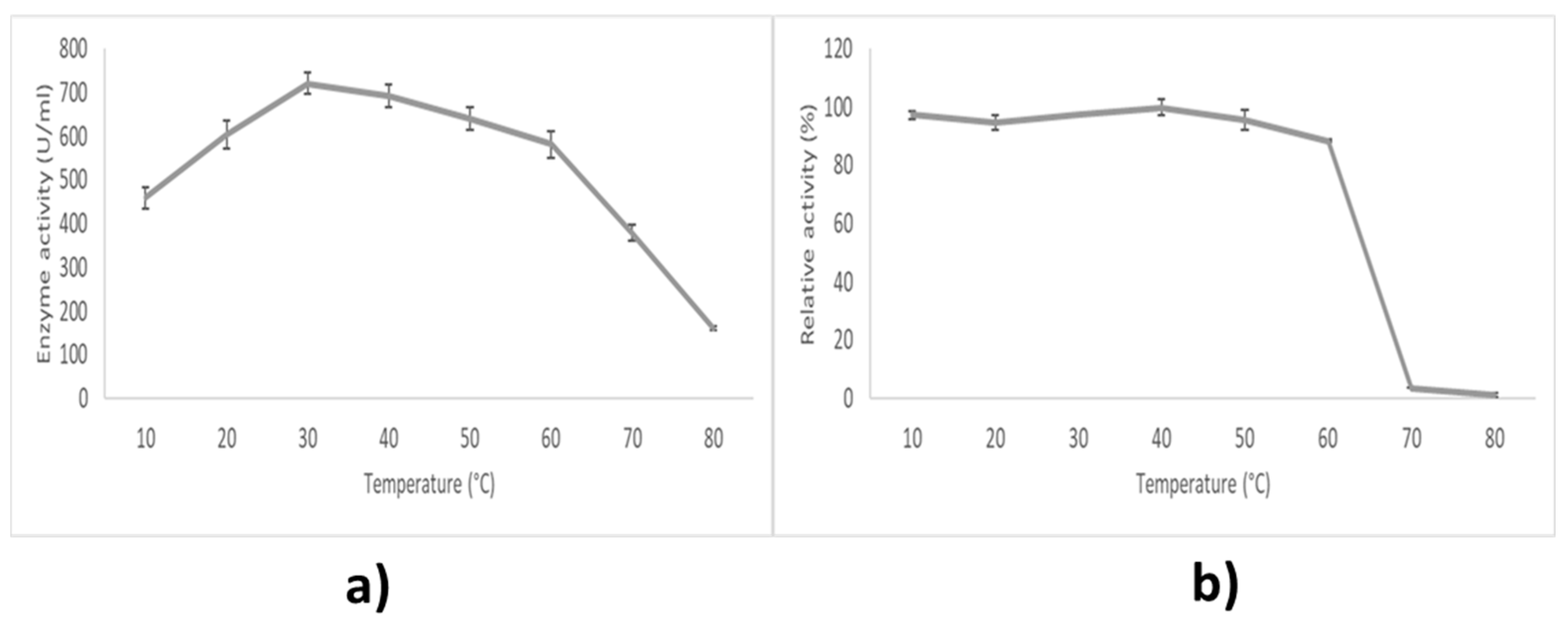
2.7.1. Effect of Temperature on Lipase Activity and Stability
2.7.2. Effect of pH on Lipase Activity and Stability
2.7.3. Effect of Metal Ion on Lipase Activity
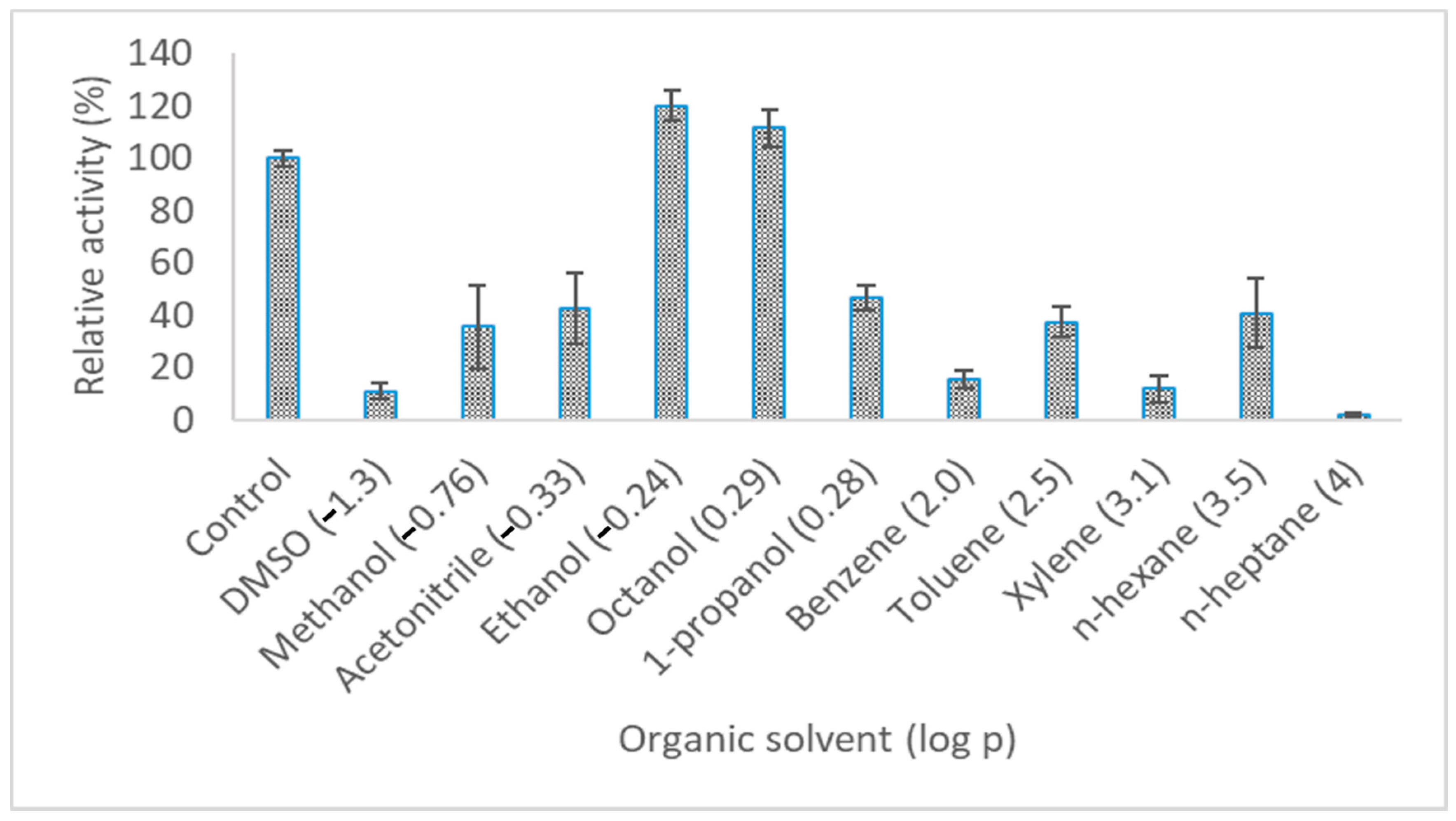
2.7.4. Effect of Organic Solvents on Lipase Activity
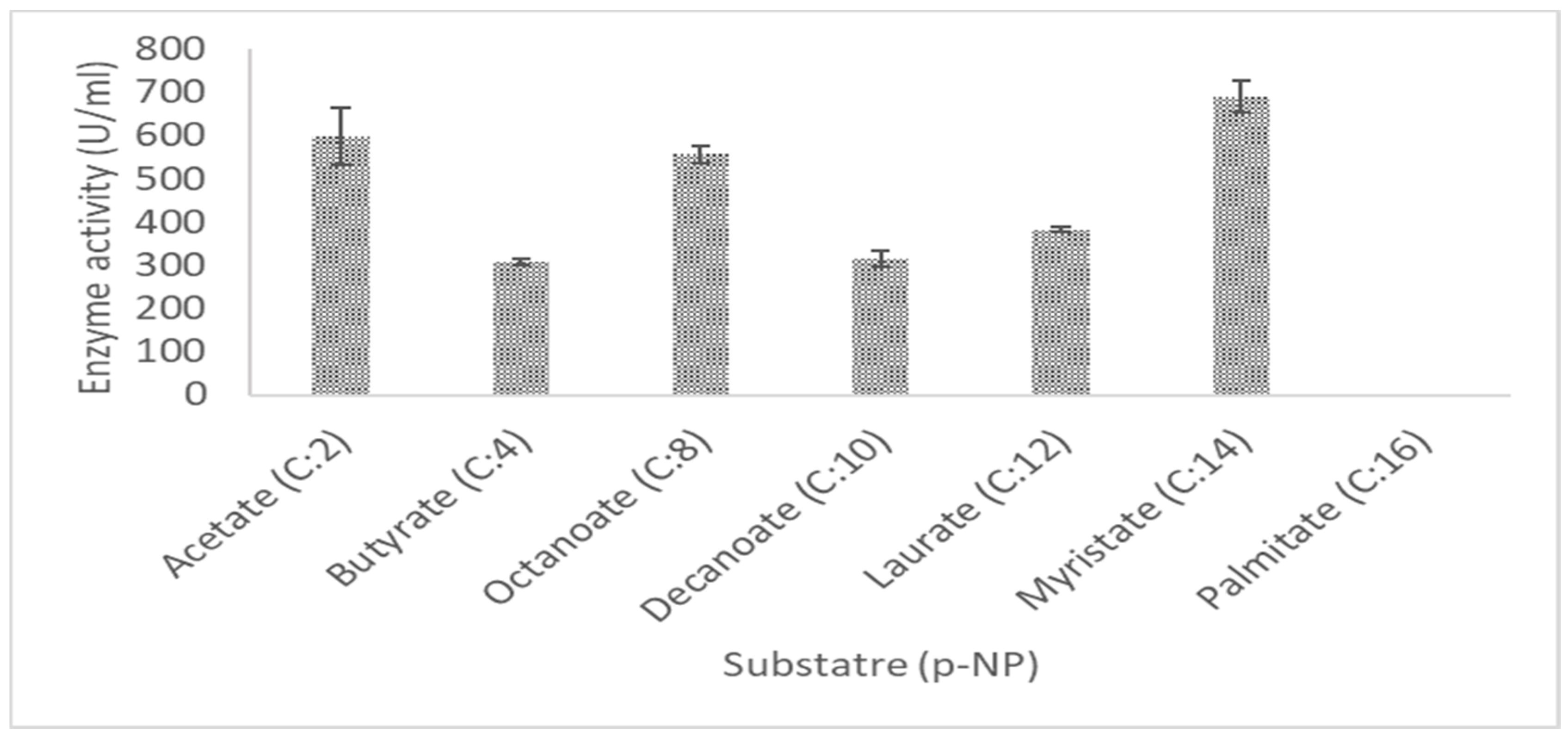
2.7.5. Effect of Substrate Specificity on Lipase Activity
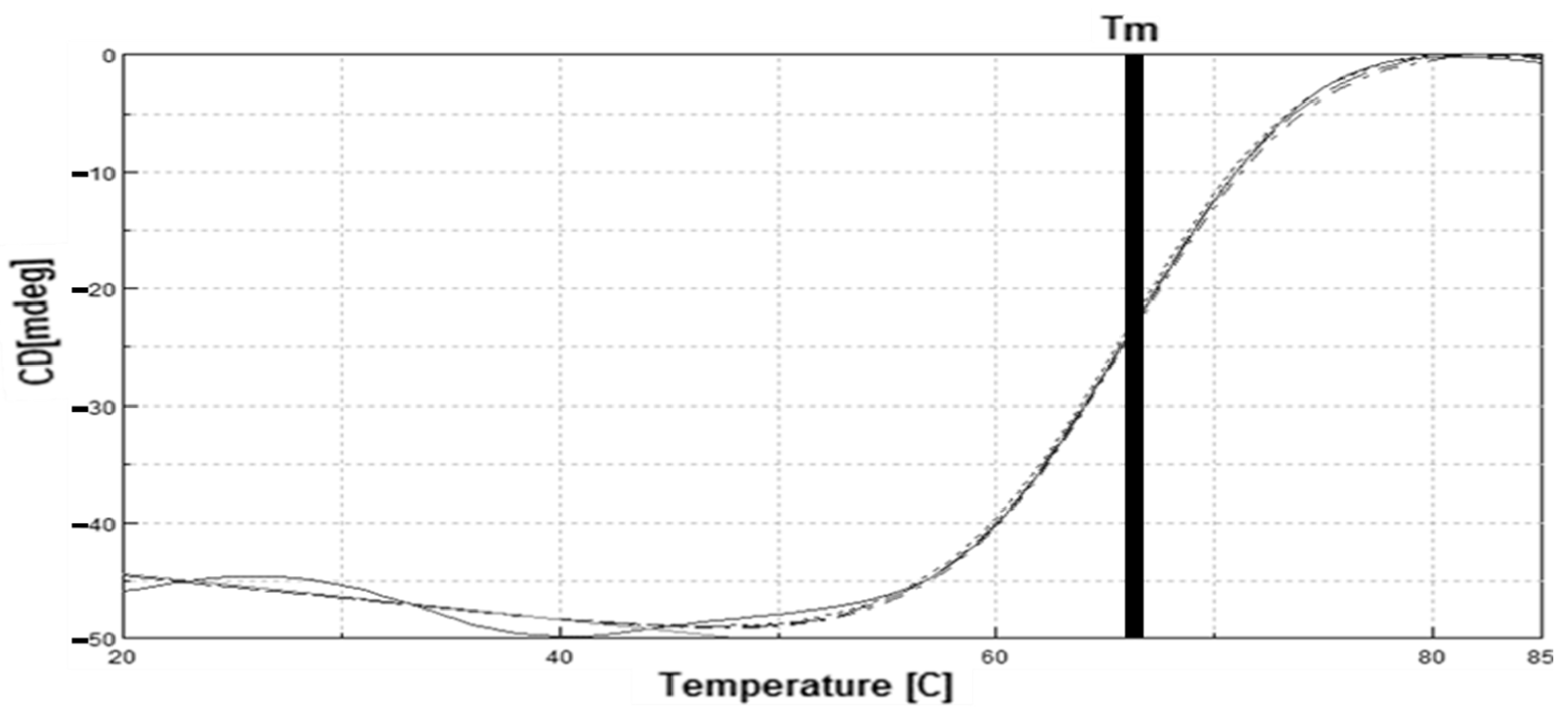
2.8. Circular Dichroism (CD) Spectra Analysis of LIP2 Lipase
2.8.1. Thermal Denaturation Analysis
2.8.2. Determination of Protein Secondary Structure Analysis
3. Discussion
3.1. Predicted LIP2 Structure via Homology Modelling
3.2. Optimization of LIP2 Production
3.3. Purification of LIP2 Lipase
3.4. Characteristics of LIP2 Lipase
4. Materials and Methods
4.1. Structure Prediction via Homology Modelling
4.2. PCR Amplification and Construction of the Expression Systems
4.3. Qualitative Screening of Transformant
4.4. Preparation of Crude Lysate
4.5. Lipase Assay
4.6. Molecular Weight Determination of LIP2 Lipase
4.7. Activity Staining
4.8. His-Tag Staining
4.9. Optimization of LIP2 Lipase Expression
4.9.1. Effect of Host Strains on Lipase Production
4.9.2. Effect of Inducer Concentration on Lipase Production
4.9.3. Effect of Temperature on Lipase Production
4.9.4. Effect of Post-Induction Times on Lipase Production
4.10. Purification of LIP2 Lipase
4.11. Biochemical Characterization of LIP2
4.11.1. Effect of Temperature on Lipase Activity
4.11.2. Effect of Temperature Stability on Lipase Activity
4.11.3. Effect of pH on Lipase Activity
4.11.4. Effect of pH Stability on Lipase Activity
4.11.5. Effect of Metal Ion on Lipase Activity
4.11.6. Effect of Organic Solvent on Lipase Activity
4.11.7. Effect of Substrate Specificity on Lipase Activity
4.12. Thermal Denaturation and Secondary Structure Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Guo, Z.; Xu, X. New opportunity for enzymatic modification of fats and oils with industrial potentials. Org. Biomol. Chem. 2005, 3, 2615–2619. [Google Scholar] [CrossRef] [PubMed]
- Hasan, F.; Shah, A.A.; Hameed, A. Industrial applications of microbial lipases. Enzyme Microb. Technol. 2006, 39, 235–251. [Google Scholar] [CrossRef]
- Seth, S.; Chakravorty, D.; Dubey, V.K.; Patra, S. An insight into plant lipase research—Challenges encountered. Protein Expres. Purif. 2014, 95, 13–21. [Google Scholar] [CrossRef]
- Joshi, R.; Kuila, A. Lipase and their different industrial applications: A review. Braz. J. Biol. Sci. 2018, 5, 237–247. [Google Scholar] [CrossRef]
- Hidayat, C.; Hastuti, P.; Utazmi, S.; Wardhani, A.K.; Pradipta, D.S. Enhancing indigenous lipase activity of germinated Jatropha curcas L. seeds for the enzymatic degradation of phorbol ester. Biocatal. Agric. Biotechnol. 2014, 3, 71–76. [Google Scholar] [CrossRef]
- Melani, N.B.; Tambourgi, E.B.; Silveira, E. Lipases: From production to applications. Sep. Purif. Rev. 2020, 49, 143–158. [Google Scholar] [CrossRef]
- Borrelli, G.M.; Trono, D. Recombinant lipases and phospholipases and their use as biocatalysts for industrial applications. Int. J. Mol. Sci. 2015, 16, 20774–20840. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Ong-Abdullah, M.; Low, E.T.L.; Manaf, M.A.A.; Rosli, R.; Nookiah, R.; Azizi, N. Oil palm genome sequence reveals divergence of interfertile species in old and new worlds. Nature 2013, 500, 335–339. [Google Scholar] [CrossRef]
- Ong, A.L.; Teh, C.K.; Mayes, S.; Massawe, F.; Appleton, D.R.; Kulaveerasingam, H. An improved oil palm genome assembly as a valuable resource for crop improvement and comparative genomics in the arecoideae subfamily. Plants 2020, 9, 1476. [Google Scholar] [CrossRef]
- Ngando, E.G.F.; Dhouib, R.; Carrière, F.; Amvam Zollo, P.H.; Arondel, V. Assaying lipase activity from oil palm fruit (Elaeis guineensis Jacq.) mesocarp. Plant. Physiol. Biochem. 2006, 44, 611–617. [Google Scholar] [CrossRef]
- Sambanthamurthi, R.; Sundram, K.; Tan, Y. Chemistry and biochemistry of palm oil. Prog. Lipid Res. 2000, 39, 507–558. [Google Scholar] [CrossRef]
- Demain, A.L.; Vaishnav, P. Production of recombinant proteins by microbes and higher organisms. Biotechnol. Adv. 2009, 27, 297–306. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Winkler, U.K.; Stuckmann, M. Glycogen, hyaluronate, and some other polysaccharides greatly enhance the formation of exolipase by Serratia marcescens. J. Bacteriol. 1979, 138, 663–670. [Google Scholar] [CrossRef] [PubMed]
- Balaji, L.; Jayaraman, G. Metal ion activated lipase from halotolerant Bacillus sp. VITL8 displays broader operational range. Int. J. Biol. Macromol. 2014, 67, 380–386. [Google Scholar] [CrossRef]
- Gaur, R.; Gupta, A.; Khare, S.K. Purification and characterization of lipase from solvent tolerant Pseudomonas aeruginosa PseA. Process Biochem. 2008, 43, 1040–1046. [Google Scholar] [CrossRef]
- Ma, J.; Zhang, Z.; Wang, B.; Kong, X.; Wang, Y.; Cao, S.; Feng, Y. Overexpression and characterization of a lipase from Bacillus subtilis. Protein Expr. Purif. 2006, 45, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Whitmore, L.; Wallace, B.A. Protein secondary structure analyses from circular dichroism spectroscopy: Methods and reference databases. Biopolymers 2008, 89, 392–400. [Google Scholar] [CrossRef]
- Holmquist, M. Alpha Beta-hydrolase fold enzymes structures, functions and mechanisms. Curr. Protein Pept. Sci. 2005, 1, 209–235. [Google Scholar] [CrossRef]
- Mukherjee, A.K. Hydrophobic-hydrophilic interaction in lipase catalytic triad and possibility of a cofactor mediated catalysis. Int. J. Agric. Food Sci. 2014, 4, 84–89. [Google Scholar]
- Rosano, G.L.; Ceccarelli, E.A. Recombinant protein expression in E. coli: Advances and challenges. Front. Microbiol. 2014, 5, 172. [Google Scholar] [CrossRef]
- Chemler, J.A.; Fowler, Z.L.; McHugh, K.P.; Koffas, M.A. Improving NADPH availability for natural product biosynthesis in E. coli by metabolic engineering. Metab. Eng. 2010, 12, 96–104. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Fu, L.; Li, G.; Jones, J.A.; Linhardt, R.J.; Koffas, M. Production of chondroitin in metabolically engineered E. coli. Metab. Eng. 2015, 27, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Frangioni, J.V.; Neel, B.G. Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal. Biochem. 1993, 210, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Malakar, P.; Venkatesh, K.V. Effect of substrate and IPTG concentrations on the burden to growth of E. coli on glycerol due to the expression of Lac proteins. Appl. Microbiol. Biotechnol. 2012, 93, 2543–2549. [Google Scholar] [CrossRef]
- Aono, R. Cultivation conditions for extracellular production of penicillinase by Escherichia coli carrying pEAP31 on a semi-large scale. Appl. Microbiol. Biotechnol. 1988, 28, 414–418. [Google Scholar] [CrossRef]
- Sørensen, H.P.; Mortensen, K.K. Advanced genetic strategies for recombinant protein expression in E. coli. J. Biotechnol. 2005, 115, 113–128. [Google Scholar] [CrossRef] [PubMed]
- Marbach, A.; Bettenbrock, K. lac operon induction in E. coli: Systematic comparison of IPTG and TMG induction and influence of the transacetylase LacA. J. Biotechnol. 2012, 157, 82–88. [Google Scholar] [CrossRef]
- Kosa, P.; Gavenciakova, B.; Nosek, J. Development of a set of plasmid vectors for genetic manipulations of the pathogenic yeast Candida parapsilosis. Gene 2007, 396, 338–345. [Google Scholar] [CrossRef][Green Version]
- Abigor, D.R.; Opute, F.I.; Opoku, A.R.; Osagie, A.U. Partial purification and some properties of the lipase present in oil palm (Elaeis guineensis) mesocarp. J. Sci. Food Agric. 1985, 36, 599–606. [Google Scholar] [CrossRef]
- Okunwaye, T.; Obibuzor, U.J.; Okogbenin, A.E. Purification and biochemical properties of lipase from raphia palm fruit mesocarp. Afr. J. Biochem. Res. 2015, 9, 73–80. [Google Scholar]
- Henderson, J.; Osborne, D.J. Lipase activity in ripening and mature fruit of the oil palm. Stability in vivo and in vitro. Phytochem. 1991, 30, 1073–1078. [Google Scholar] [CrossRef]
- Samsumaharto, R.A. Partial characterization of lipase from cocoa beans (Theobroma cacao. L.) of clone PBC 159. Indones. J. Chem. 2008, 8, 448–453. [Google Scholar] [CrossRef]
- Yeşiloğlu, Y.; Demirkan, B. Biocatalytic properties of lipase from walnut seed (Juglans regia L.). J. Am. Oil Chem. Soc. 2010, 87, 659–665. [Google Scholar] [CrossRef]
- Su, E.Z.; Zhou, Y.; You, P.Y.; Wei, D.Z. Lipases in the castor bean seed of Chinese varieties: Activity comparison, purification and characterization. J. Shanghai Univ. 2010, 14, 137–144. [Google Scholar] [CrossRef]
- Abdelkafi, S.; Barouh, N.; Fouquet, B.; Fendri, I.; Pina, M.; Scheirlinckx, F.; Carrière, F. Carica papaya lipase: A naturally immobilized enzyme with interesting biochemical properties. Plant. Foods Hum. Nutr. 2011, 66, 34–40. [Google Scholar] [CrossRef]
- Panzanaro, S.; Nutricati, E.; Miceli, A.; De Bellis, L. Biochemical characterization of a lipase from olive fruit (Olea europaea L.). Plant. Physiol. Biochem. 2010, 48, 741–745. [Google Scholar] [CrossRef]
- Gandhi, N.N.; Mukherjee, K.D. Reactivity of medium-chain substrates in the interesterification of tripalmitin catalyzed by papaya lipase. J. Am. Oil Chem. Soc. 2001, 78, 965–968. [Google Scholar] [CrossRef]
- Lee, M.; Kim, J.I.; Na, S.; Eom, K. Metal ions affect the formation and stability of amyloid β aggregates at multiple length scales. Physic. Chem. Chem. Phys. 2018, 20, 8951–8961. [Google Scholar] [CrossRef]
- Khor, H.T.; Tan, N.H.; Chua, C.L. Lipase-catalyzed hydrolysis of palm oil. J. Am. Oil Chem. Soci. 1986, 63, 538–540. [Google Scholar] [CrossRef]
- Pahoja, V.M.; Sethar, M.A. A review of enzymatic properties of lipase in plants, animals and microorganisms. J. Appl. Sci. 2002, 2, 474–484. [Google Scholar]
- Wahab, R.A.; Basri, M.; Rahman, R.N.Z.R.A.; Salleh, A.B.; Rahman, M.B.A.; Leow, T.C. Facile modulation of enantioselectivity of thermophilic Geobacillus zalihae lipase by regulating hydrophobicity of its Q114 oxyanion. Enzym. Microb. Technol. 2016, 93, 174–181. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.C.; Gao, G.J.; Kao, A.L.; Tsai, C.T.; Tsai, Z.C. Two novel lipases purified from rice bran displaying lipolytic and esterification activities. Int. J. Biol. Macromol. 2019, 139, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Cadena, T.; Prada, F.; Perea, A.; Romero, H.M. Lipase activity, mesocarp oil content, and iodine value in oil palm fruits of Elaeis guineensis, Elaeis oleifera, and the interspecific hybrid O × G (E. oleifera × E. guineensis). J. Sci. Food Agric. 2013, 93, 674–680. [Google Scholar] [CrossRef] [PubMed]
- Rahman, R.; Baharum, S.N.; Basri, M.; Salleh, A.B. High-yield purification of an organic solvent-tolerant lipase from Pseudomonas sp. strain S5. Anal. Biochem. 2005, 341, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Malekabadi, S.; Badoei-dalfard, A.; Karami, Z. Biochemical characterization of a novel cold-active, halophilic and organic solvent-tolerant lipase from B. licheniformis KM12 with potential application for biodiesel production. Int. J. Biol. Macromol. 2018, 109, 389–398. [Google Scholar] [CrossRef]
- Kumar, A.; Dhar, K.; Kanwar, S.S.; Arora, P.K. Lipase catalysis in organic solvents: Advantages and applications. Biol. Proc. Online 2016, 18, 1–11. [Google Scholar] [CrossRef]
- Krieger, E.; Vriend, G. YASARA View—molecular graphics for all devices—From smartphones to workstations. Bioinformatics 2014, 30, 2981–2982. [Google Scholar] [CrossRef]
- Ramachandran, G.N.; Ramakrishnan, C.; Sasisekharan, V. Stereochemistry of polypeptide chain configurations. J. Mol. Biol. 1963, 7, 95–99. [Google Scholar] [CrossRef]
- Laemmli, U. Most commonly used discontinuous buffer system for SDS electrophoresis. Nature 1970, 227, 680–686. [Google Scholar] [CrossRef] [PubMed]
- Sommer, P.; Bormann, C.; Götz, F. Genetic and biochemical characterization of a new extracellular lipase from Streptomyces cinnamomeus. Appl. Environ. Microbiol. 1997, 63, 3553–3560. [Google Scholar] [CrossRef] [PubMed]


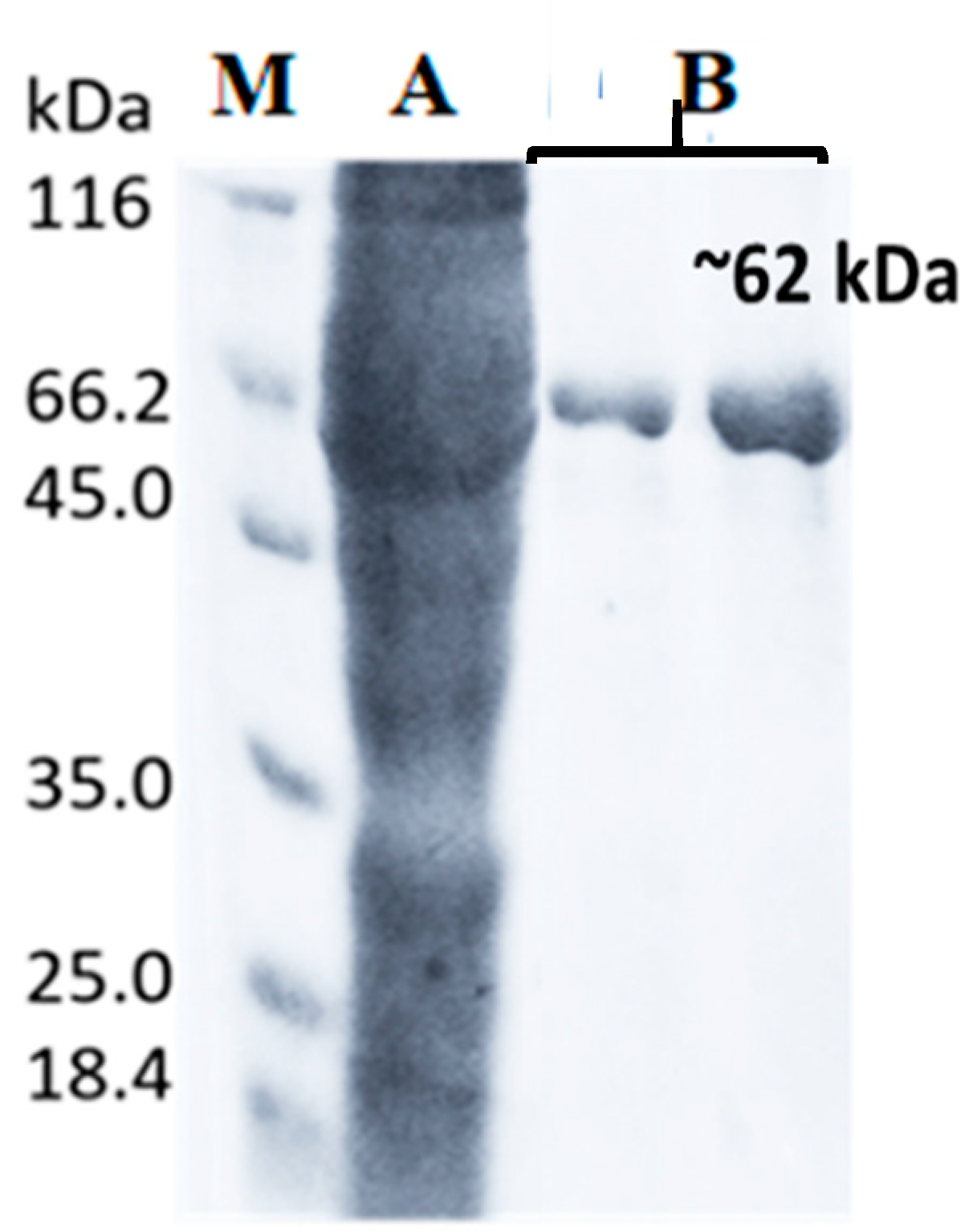


| Step | Vol (mL) | Total Protein (mg) | Total Activity (Units) | Specific Activity (U/mg) | Purification (Folds) | Yield (%) |
|---|---|---|---|---|---|---|
| Homogenate | 20 | 40.6 | 9210.2 | 226.85 | 1 | 100 |
| Ni-Sepharose-HP | 6 | 4.74 | 4029.18 | 850.03 | 3.75 | 43.75 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohd Din, M.H.; Nair, A.; Masomian, M.; Mohamad Ali, M.S.; Raja Abd. Rahman, R.N.Z. Heterologous Expression and Characterization of Plant Lipase LIP2 from Elaeis guineensis Jacq. Oil Palm Mesocarp in Escherichia coli. Catalysts 2021, 11, 244. https://doi.org/10.3390/catal11020244
Mohd Din MH, Nair A, Masomian M, Mohamad Ali MS, Raja Abd. Rahman RNZ. Heterologous Expression and Characterization of Plant Lipase LIP2 from Elaeis guineensis Jacq. Oil Palm Mesocarp in Escherichia coli. Catalysts. 2021; 11(2):244. https://doi.org/10.3390/catal11020244
Chicago/Turabian StyleMohd Din, Mohd Hadzdee, Anusha Nair, Malihe Masomian, Mohd Shukuri Mohamad Ali, and Raja Noor Zaliha Raja Abd. Rahman. 2021. "Heterologous Expression and Characterization of Plant Lipase LIP2 from Elaeis guineensis Jacq. Oil Palm Mesocarp in Escherichia coli" Catalysts 11, no. 2: 244. https://doi.org/10.3390/catal11020244
APA StyleMohd Din, M. H., Nair, A., Masomian, M., Mohamad Ali, M. S., & Raja Abd. Rahman, R. N. Z. (2021). Heterologous Expression and Characterization of Plant Lipase LIP2 from Elaeis guineensis Jacq. Oil Palm Mesocarp in Escherichia coli. Catalysts, 11(2), 244. https://doi.org/10.3390/catal11020244