Abstract
Glycerol is a readily available and inexpensive substance that is mostly generated during biofuel production processes. In order to ensure the viability of the biofuel industry, it is essential to develop complementing technologies for the resource utilization of glycerol. Ethylene glycol is a two-carbon organic chemical with multiple applications and a huge market. In this study, an artificial enzymatic cascade comprised alditol oxidase, catalase, glyoxylate/hydroxypyruvate reductase, pyruvate decarboxylase and lactaldehyde:propanediol oxidoreductase was developed for the production of ethylene glycol from glycerol. The reduced nicotinamide adenine dinucleotide (NADH) generated during the dehydrogenation of the glycerol oxidation product d-glycerate can be as the reductant to support the ethylene glycol production. Using this in vitro synthetic system with self-sufficient NADH recycling, 7.64 ± 0.15 mM ethylene glycol was produced from 10 mM glycerol in 10 h, with a high yield of 0.515 ± 0.1 g/g. The in vitro enzymatic cascade is not only a promising alternative for the generation of ethylene glycol but also a successful example of the value-added utilization of glycerol.
1. Introduction
The manufacturing of biofuels from reproducible feedstocks is increasing throughout the world [1,2,3]. The development of the biofuel industry has generated a large amount of glycerol as an inevitable byproduct [4]. An excess of glycerol production in the biofuel industry leads to a dramatic decrease of the price of glycerol, which has a negative impact on the development of the biofuels industry [5]. Thus, the conversion of glycerol into value-added compounds through chemical or biotechnological routes is now urgently needed for the improvement of the viability of the biofuel industry.
Ethylene glycol is a large-volume commodity that is widely used in many industrial processes, such as glycolic acid production and polyethylene terephthalate synthesis [6]. It is predominantly produced from ethylene oxide through carbonation and subsequent hydrolyzation, or by the hydration of ethylene oxide [7,8]. Direct ethylene glycol production from biomass-derived carbohydrates through chemical catalysis has also been reported [9]. Importantly, ethylene glycol can be produced from glycerol through liquid-phase catalytic hydrogenolysis [10,11]. However, this process usually needs high pressures (4.0–8.0 MPa), high temperatures (453–473 K) and expensive noble metal catalysts (Pt, Ni, Ru, etc.). Several metabolic pathways have been constructed for the biotechnological production of ethylene glycol from renewable substrates such as d-glucose, d-xylose and l-arabinose [12,13,14]. Nevertheless, the production of ethylene glycol from glycerol via a biotechnological route has never been reported, until now.
An in vitro synthetic system is emerging as a promising technique for the synthesis of high-value products from cheap and renewable substrates [15,16]. This biomanufacturing platform leaves out the use of cells, and can thus exclude cell-associated process barriers, such as the transmembrane transport of substances, the toxicity of products, and undesired metabolism pathways. In addition, an in vitro synthetic system exclusively employs purified enzymes, and makes the purification of the target products rather simple to conduct. Some in vitro synthetic systems have been constructed for the generation of valuable chemicals from renewable substrates, such as d-glucose, d-xylose and glycerol [17,18,19]. In this study, we have designed an artificial enzymic cascade for the conversion of glycerol to ethylene glycol that only requires five enzymes. The artificial pathway is also completely redox balanced through self-sufficient reduced nicotinamide adenine dinucleotide (NADH) recycling.
2. Results and Discussion
2.1. Design of the In Vitro Biosystem for Ethylene Glycol Production
Here, an artificial enzymatic reaction cascade was designed for the conversion of glycerol into ethylene glycol (Figure 1). Glycerol is first oxidized to d-glycerate via d-glyceraldehyde by alditol oxidase (Aldo) catalyzed two-step oxidation. The d-Glycerate produced from glycerol is then transformed into 3-hydroxypyruvate by oxidized nicotinamide adenine dinucleotide (NAD+)-dependent glyoxylate/hydroxypyruvate reductase (GRHPR). Then, 3-hydroxypyruvate is decarboxylated to glycolaldehyde by pyruvate decarboxylase (PDC). Finally, the glycolaldehyde is reduced into ethylene glycol by lactaldehyde:propanediol oxidoreductase (FucO), using the NADH produced during the d-glycerate dehydrogenation as the cofactor. The H2O2 generated during the oxidation of the glycerol is decomposed by catalase in order to avoid the spontaneous oxidative of the intermediate products [20]. As a result, one mole of glycerol can generate one mole of ethylene glycol.
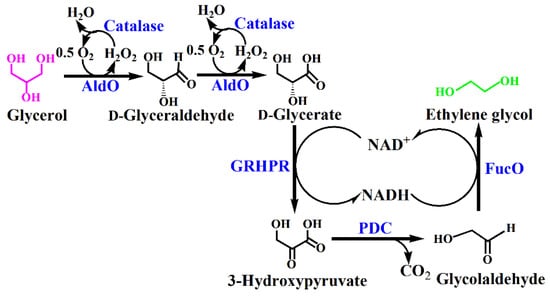
Figure 1.
Scheme of ethylene glycol production from glycerol based on the artificial in vitro enzymatic cascade. The enzymes are alditol oxidase (AldO) from Streptomyces coelicolor A3(2), catalase from Aspergillus niger, glyoxylate/hydroxypyruvate reductase (GRHPR) from Pyrococcus furiosus, pyruvate decarboxylase (PDC) from Zymomonas mobile, and lactaldehyde:propanediol oxidoreductase (FucO) from Escherichia coli.
2.2. Substrate Specificity of AldOmt
AldO is a soluble monomeric flavoprotein containing covalently-bound FAD as its cofactor [21]. This enzyme is first isolated from Streptomyces coelicolor A3(2) and identified as an oxidase primarily active with alditols, and as producing aldoses as products. Interestingly, AldO exhibits regio- and enantio-selective oxidative activity toward 1,2-diols and α-hydroxy carboxylic acids but not the α-hydroxy aldehydes which will be formed as the end products [22]. AldOmt with enhanced oxidative activity towards glycerol was also constructed through directed evolution, and this enzyme has been successfully applied in d-glycerate production from glycerol [23].
The target product of this study, ethylene glycol, is the simplest 1,2-diol, and is also the possible substrate of AldOmt. Thus, the activity of AldOmt toward ethylene glycol was assayed using a Clark-type oxygen electrode, as described in a previous study [24]. As shown in Figure 2a, the AldOmt-dependent oxygen consumption was rather low with the addition of ethylene glycol, while noticeable oxygen consumption was observed when glycerol was added (Figure 2c). The estimated specific activities of AldOmt for glycerol and ethylene glycol were 0.53 ± 0.03 U/mg and 0.018 ± 0.004 U/mg, respectively (Figure 2b). The oxidative activities of AldOmt toward glycerol and ethylene glycol were also determined by high-performance liquid chromatography (HPLC) analysis. As shown in Figure 2c,d, the depletion of glycerol could be observed obviously, while very little consumption of ethylene glycol was detected.
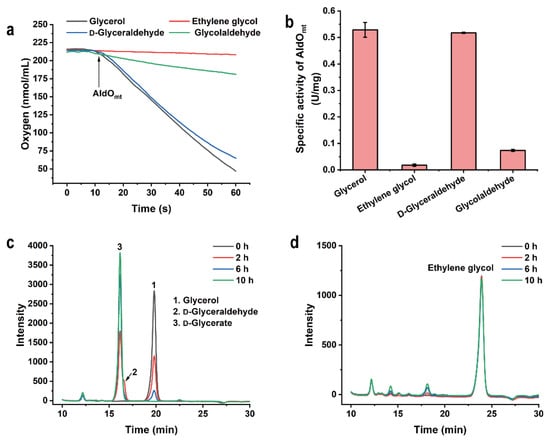
Figure 2.
Substrate specificity of AldOmt. (a) The determination of the oxygen consumption during the AldOmt-catalyzed oxidation of the target substrates. (b) The specific activity of AldOmt toward glycerol, d-glyceraldehyde, glycolaldehyde, and ethylene glycol, respectively. The error bars indicate the standard deviations from three parallel replicates. (c,d) HPLC analysis of AldOmt-catalyzed glycerol (c) or ethylene glycol (d) oxidation. The reactions were carried out in a reaction mixture containing 50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES)-NaOH buffer (pH 7.4), 10 mM glycerol or 10 mM ethylene glycol and 0.2 mg/mL AldOmt, at 30 °C, with a shaking speed of 150 rpm.
AldO oxidizes glycerol into d-glycerate in a two-step reaction via d-glyceraldehyde [23]. Glycolaldehyde, the direct precursor of ethylene glycol in the designed enzymatic reaction cascade, is the analogue of d-glyceraldehyde, and might be oxidized by AldOmt. Thus, the activities of AldOmt toward glycolaldehyde and d-glyceraldehyde were also assayed. As shown in Figure 2a,b, a very much lower oxygen consumption rate was observed when glycolaldehyde, but not d-glyceraldehyde, was added to the reaction mixture, indicating that AldOmt exhibits rather low oxidative activity toward glycolaldehyde. The result of the HPLC analysis further confirmed that AldOmt can hardly catalyze the oxidation of glycolaldehyde (Figure S1).
Based on the data mentioned above, AldOmt can efficiently oxidize glycerol into d-glycerate, but will barely consume the glycolaldehyde and ethylene glycol in the reaction system. Thus, AldOmt is a proper catalyst in the designed enzymatic reaction cascade, and it exhibits little promiscuous activity affecting the production of ethylene glycol.
2.3. 3-Hydroxypyruvate Decarboxylation Catalyzed by PDC
The GRHPR of Pyrococcus furiosus can catalyze the NAD+-dependent dehydrogenation of d-glycerate to produce 3-hydroxypyruvate [18,25]. However, the standard Gibbs energy change (ΔrGθ) of d-glycerate dehydrogenation is estimated to be 20.26 KJ/mol, indicating that this reaction is not thermodynamically favorable. In order to support the GRHPR-catalyzed, thermodynamically unfavorable d-glycerate dehydrogenation, the integration of thermodynamically favorable reactions with 3-hydroxypyruvate or NADH as the substrates is needed.
The PDC of Zymomonas mobilis can catalyze the decarboxylation of pyruvate to produce carbon dioxide and acetaldehyde [26]. In this study, the activity of PDC toward 3-hydroxypyruvate decarboxylation, a thermodynamically favorable reaction (ΔrGθ = −55.23 KJ/mol), was also assayed. As shown in Figure S2, PDC also exhibited decarboxylation activity to catalyze 3-hydroxypyruvate into glycolaldehyde. About 8 mM glycolaldehyde was produced from 10 mM 3-hydroxypyruvate in 4 h by 10 μg/mL PDC.
2.4. Substrate Specificity of FucO
The NADH-dependent reduction of glycolaldehyde can regenerate the NAD+ required for d-glycerate dehydrogenation, and can support the production of ethylene glycol. The lactaldehyde:propanediol oxidoreductase of Escherichia coli (FucO) was reported to reduce glycolaldehyde [27,28]. This enzyme has been overexpressed in E. coli for in vivo ethylene glycol production via a synthetic pathway [29]. In this study, the activity of FucO using glycolaldehyde and NADH as the substrates was also assayed (Figure 3a,b). The specific activity of FucO toward glycolaldehyde reduction was calculated to be 30.9 ± 1.2 U/mg (Figure 3b). The HPLC analysis results showed that FucO rapidly reduces glycolaldehyde into ethylene glycol (Figure 3c).
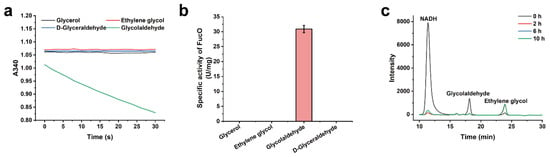
Figure 3.
Substrate specificity of FucO. (a) Time course of reactions of FucO with glycerol, d-glyceraldehyde, glycolaldehyde, or ethylene glycol as the substrate and NADH as the cofactor. (b) Specific activity of FucO toward glycerol, d-glyceraldehyde, glycolaldehyde and ethylene glycol, respectively. The error bars indicate the standard deviations from three parallel replicates. (c) HPLC analysis of the FucO-catalyzed glycolaldehyde reduction. The reaction was carried out in a reaction mixture containing 50 mM HEPES-NaOH buffer (pH 7.4), 10 mM glycolaldehyde, 10 mM NADH and 0.5 mg/mL FucO, at 30 °C, with a shaking speed of 150 rpm.
The activities of FucO toward other reactants in the in vitro system including ethylene glycol, glycerol and d-glyceraldehyde were also assayed. As shown in Figure 3a,b, all of these reactant especially d-glyceraldehyde, the analogue glycolaldehyde, cannot be reduced by FucO. Thus, the NADH generated during d-glycerate oxidation can only be consumed for glycolaldehyde reduction. This property of FucO ensured the self-sufficient NADH recycling of the in vitro system, and avoided the requirement for exogenous reducing agent addition.
2.5. Production of Ethylene Glycol from Glycerol In Vitro
AldOmt, GRHPR, PDC and FucO were overexpressed, purified (Figure S3), and coupled with commercial catalase from A. niger (Sigma-Aldrich, St. Louis, MO, USA) for the production of ethylene glycol from glycerol. As shown in Figure 4, 7.64 ± 0.15 mM ethylene glycol was generated from 10 mM glycerol in 10 h. Some chemists have also attempted to produce ethylene glycol from glycerol by hydrogenolysis. For example, Feng and his colleagues studied the conversion of glycerol to ethylene glycol over Ru-Co/ZrO2 catalysts [30]. However, this catalyst exhibited higher selectivity toward 1,2-propanediol generation than ethylene glycol production. The yield of ethylene glycol was only 0.30 g/g glycerol. Ueda and his colleagues reported the conversion of glycerol to ethylene glycol over Pt-modified Ni catalysts under the conditions of 453 K and 8.0 MPa [10]. The yield of ethylene glycol was 0.479 g/g, which was lower than that of a biocatalytic process utilizing the artificially designed enzymatic cascade (0.515 ± 0.1 g/g) under modest reaction conditions (303 K, 0.1 MPa).
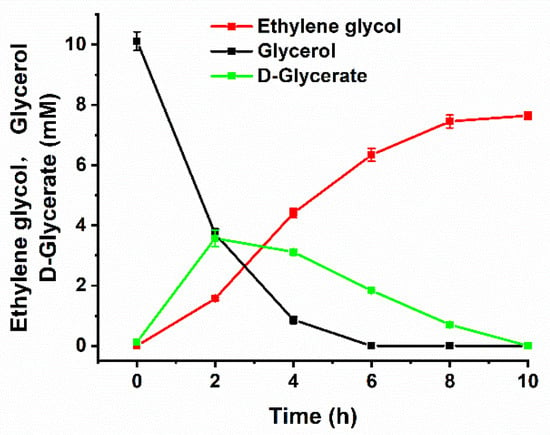
Figure 4.
Conversion of glycerol into ethylene glycol by the artificial enzymatic reaction cascade. The reaction was carried out in a reaction mixture containing 50 mM HEPES-NaOH buffer (pH 7.4), 10 mM glycerol, 1 mM NAD+, 2 mM MgSO4, 0.25 mM thiamine pyrophosphate (TPP), 1 mg/mL AldOmt, 10 mg/mL GRHPR, 5.4 mg/mL PDC, 13 mg/mL FucO and 500 U/mL catalase. The reaction was conducted at 30 °C and a shaking speed of 150 rpm. The error bars indicate the standard deviations from three parallel replicates.
Some biotechnological routes have been developed for the production of ethylene glycol from biomass-derived carbohydrates [12,13,14,19,29,31,32,33,34,35] (Table 1). For example, d-glucose was used as a substrate to produce ethylene glycol via the extension of the serine biosynthesis pathway. The yields of ethylene glycol from d-glucose were only 0.09 g/g by engineered Corynebacterium glutamicum and 0.14 g/g by engineered E. coli [14,31]. Glycolaldehyde, the direct precursor of ethylene glycol, can be produced from d-xylose or xylonic acid through the Dahms pathway [12,29,32]. Due to the carbon loss induced by the coproduction of pyruvate, the theoretical yield of ethylene glycol from d-xylose was 0.413 g/g. Liu and his colleagues disrupted the d-xylose isomerisation pathway and introduced Dahms pathway in E. coli, and obtained a yield of 0.29 g/g ethylene glycol from d-xylose [12]. A higher yield of 0.40 g/g ethylene glycol from d-xylose was obtained by further improving the redox balance of the Dahms pathway, overexpressing FucO and YjhG, and deleting the by-products encoding genes aldA and arcA [29]. Recently, a cell-free bioreaction scheme was also reported to generate ethylene glycol from d-xylose, with a yield of 0.227 g/g [19]. In this study, we designed an artificial enzymatic cascade to produce ethylene glycol from glycerol with only one carbon atom loss. A high yield of 0.515 ± 0.1 g/g ethylene glycol from glycerol was acquired using the in vitro synthetic system.

Table 1.
Yield of ethylene glycol production from various substrates by metabolically-engineered strains or an in vitro biosystem.
The in vitro synthetic system has appealing advantages, such as a high product yield, easy product separation, and simple process control [16]. It has made great progress in recent years. For example, the production of inositol from starch using the in vitro synthetic system has been implemented on an industrial scale [36]. In this study, we designed an enzymatic cascade for the production of ethylene glycol from glycerol. Although a higher yield of ethylene glycol was obtained using the artificially designed enzymatic cascade, the final concentration and productivity of ethylene glycol cannot meet the requirements for industrial application. The low activity of GRHPR may be the key factor restricting the efficiency of the in vitro enzymatic cascade. Systematic screening or the directed evolution of a new biocatalyst with higher activity toward the dehydrogenation of d-glycerate to 3-hydroxypyruvate will be necessary to further improve the production of ethylene glycol. The optimization of the reaction system may also improve the efficiency of the enzymatic cascade, and this deserves an intensive study [37].
3. Materials and Methods
Materials: reduced/oxidized nicotinamide adenine dinucleotide (NADH/NAD+), isopropyl-β-d-1-thiogalactopyranoside (IPTG), thiamine pyrophosphate (TPP), 3-hydroxypyruvate, d-glycerate and glycolaldehyde were purchased from Sigma-Aldrich (St. Louis, MO, USA). Restriction endonuclease and T4 DNA ligase were purchased from Thermo Scientific (Waltham, MA, USA). FastPfu DNA polymerase was purchased from TransGen Biotech Ltd. (Beijing, China). The other chemicals used in this study were analytical pure grade and commercially available.
The cloning and expression of fucO: the strains, plasmids and primers used in this study are listed in Supplementary Materials Table S1. E. coli DH5α was used for the plasmid construction, and E. coli BL21(DE3) was used for the overexpression of the recombinant proteins. The encoding gene of FucO was amplified from the genomic DNA of E. coli K12 with the primers fucO-f/fucO-r (Supplementary Table S1), digested with BamHI/HindIII, and cloned into pET28a in order to construct pET28a-FucO. The plasmid pET28a-FucO was then transferred into E. coli BL21(DE3) to obtain E. coli BL21(DE3)/pET28a-FucO. All of the E. coli strains were cultivated in Luria-Bertani (LB) medium at 37 °C and 180 rpm or 16 °C and 160 rpm. If necessary, ampicillin (at a final concentration of 100 μg/mL) or kanamycin (at a final concentration of 50 μg/mL) was added into the medium.
Purification of the enzymes: the catalase from A. niger used in this study was purchased from Sigma-Aldrich (St. Louis, MO, USA). The AldOmt, GRHPR and PDC were purified as described by Li and his colleagues [20]. E. coli BL21(DE3)/pET28a-FucO was grown at 37 °C and 180 rpm in LB medium with 100 μg/mL kanamycin to an optical density of 0.5 at 620 nm. Then, IPTG was added at a final concentration of 1.0 mM in order to induce the expression of FucO. After cultivation for another 4 h at 37 °C, the cells were harvested and washed twice with 20 mM phosphate solution buffer (PBS, pH 7.4) by centrifugation at 6000 rpm for 10 min; the cells were subsequently suspended in binding buffer (20 mM phosphate buffer, 500 mM NaCl, and 20 mM imidazole, pH 7.4) and lysed using a continuous high-pressure cell disrupter working at 4 °C and 900 bar. The cell lysate was centrifugated at 12,000 rpm for 1 h, and the supernatant then was loaded onto a 5 mL HisTrap HP column (GE Healthcare, Sweden). The target proteins were eluted with a gradient ratio of elution buffer (20 mM phosphate buffer, 500 mM NaCl, and 500 mM imidazole, pH 7.4). The purified proteins including AldOmt, GRHPR, PDC and FucO were subjected to sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE), and the results are shown in Supplementary Figure S3.
Ethylene glycol production: the production of ethylene glycol from glycerol was performed in a 5 mL reaction system containing 1 mg/mL AldOmt, 500 U/mL catalase, 10 mg/mL GRHPR, 5.4 mg/mL PDC, 13.0 mg/mL FucO, 10 mM glycerol, 1 mM NAD+, 0.25 mM TPP, 2 mM MgSO4, and 50 mM HEPES-NaOH (pH 7.4) at 30 °C and 150 rpm. Samples (0.2 mL) were withdrawn every 2 h and subjected to analysis by HPLC.
Analytical methods: the concentrations of the purified proteins were measured by the use of the Bradford protein assay (Bio-Rad, Hercules, CA, USA). The activity of the FucO was assayed by recording the absorption of NADH at 340 nm using a UV–visible spectrophotometer (Ultrospec 2100 pro; Amersham Biosciences, Little Chalfont, UK) [20]. All of the reactions were carried out in a 1.0 mL quartz cuvette at 30 °C, and HEPES-NaOH buffer (50 mM, pH 7.4) with 0.2 mM NADH and 2.5 mM substrate (glycerol, d-glyceraldehyde, glycolaldehyde or ethylene glycol). One unit of FucO activity was defined as the amount of protein catalyzing the consumption of one µmol of NADH per minute.
The oxidase activities of AldOmt toward glycerol, d-glyceraldehyde, glycolaldehyde and ethylene glycol were assayed by measuring the rate of oxygen consumption. The reaction mixture contained 10 mM substrate, 50 mM HEPES-NaOH buffer (pH 7.4), and 0.2 mg/mL AldOmt. The oxygen consumption in the reaction system was monitored using a Clark-type oxygen electrode (Oxytherm System; Hansatech Instruments Ltd., Pentney, UK). One unit of AldOmt activity was defined as the amount of protein catalyzing the consumption of one µmol O2 per minute.
The concentrations of glycerol, glyceraldehyde, d-glycerate and 3-hydroxypyruvate were analyzed using an HPLC system (Agilent 1100 series, Hewlett-Packard, Palo Alto, CA, USA) equipped with an anion exchange column (Aminex HPX-87H, 300 × 7.8 mm; Bio-Rad, USA) and a refractive index detector [20]. The mobile phase was 5.0 mM H2SO4 at a flow rate of 0.4 mL/min and at a temperature of 55 °C. The concentrations of the analytes were calculated by converting the peak areas detected by HPLC using external calibration curves.
4. Conclusions
In conclusion, we designed an artificial in vitro biosystem involving five enzymes for the production of ethylene glycol from glycerol. Ethylene glycol was produced from glycerol with a yield of 0.515 ± 0.1 g/g, which is higher than any reported chemical or biotechnological routes to date. The method presented in this work would not only provide a promising process for ethylene glycol production, but would also expand the utilization of the glycerol produced by the biofuel industry.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4344/11/2/214/s1. Figure S1: HPLC analysis of AldOmt-catalyzed d-glyceraldehyde (a) and glycolaldehyde (b) oxidation. The reactions were carried out in a reaction mixture containing 50 mM HEPES-NaOH buffer (pH 7.4), 0.2 mg/mL AldOmt, 10 mM d-glyceraldehyde or 10 mM glycolaldehyde, at 30 °C and 150 rpm. Figure S2: HPLC analysis of PDC catalyzed 3-hydroxypyruvate decarboxylation. The reaction was carried out in 50 mM HEPES-NaOH buffer (pH 7.4) containing 2 mM MgSO4, 0.25 mM TPP, 10 mM 3-hydroxypyruvate and 10 μg/mL PDC, at 30 °C and 150 rpm. Figure S3: SDS-PAGE results of the purified enzymes. M: Marker; 1: AldOmt; 2: GRHPR; 3: PDC; 4: FucO. Table S1: Strains, plasmids and primers used in this study.
Author Contributions
Conceptualization, methodology, formal analysis, investigation, data curation, K.L. and C.L.; software, validation, Y.Z. and S.G.; resources, K.L., J.Y. and W.S.; writing—original draft preparation, K.L., W.M. and C.G.; writing—review and editing, supervision, project administration, C.L. and C.M.; funding acquisition, C.G., C.L. and C.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Key R&D Program of China, grant number 2019YFA0904900, 2019YFA0904800; Shandong Provincial Funds for Distinguished Young Scientists, grant number JQ 201806; Natural Science Foundation of Shandong Provincial, grant number ZR2018PC008, ZR2020MC005; Tianjin Synthetic Biotechnology Innovation Capacity Improvement Project, grant number TSBICIP-KJGG-005; and the China Postdoctoral Science Foundation, grant number 62450078311268.
Acknowledgments
The authors thank Caiyun Sun from Core Facilities for Life and Environmental Sciences (State Key Laboratory of Microbial Technology, Shandong University) for assistance in the HPLC analysis.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Mahmood, W.M.A.; Theodoropoulos, C.; Gonzalez-Miquel, M. Enhanced microalgal lipid extraction using bio-based solvents for sustainable biofuel production. Green Chem. 2017, 19, 5723–5733. [Google Scholar] [CrossRef]
- Hill, J.; Nelson, E.; Tilman, D.; Polasky, S.; Tiffany, D. Environmental, economic, and energetic costs and benefits of biodiesel and ethanol biofuels. Proc. Natl. Acad. Sci. USA 2006, 103, 11206–11210. [Google Scholar] [CrossRef] [PubMed]
- Heeres, A.S.; Picone, C.S.; van der Wielen, L.A.; Cunha, R.L.; Cuellar, M.C. Microbial advanced biofuels production: Over-coming emulsification challenges for large-scale operation. Trends Biotechnol. 2014, 32, 221–229. [Google Scholar] [CrossRef]
- Tan, H.W.; Abdul Aziz, A.; Aroua, M. Glycerol production and its applications as a raw material: A review. Renew. Sustain. Energy Rev. 2013, 27, 118–127. [Google Scholar] [CrossRef]
- Lu, X.; Vora, H.; Khosla, C. Overproduction of free fatty acids in E. coli: Implications for biodiesel production. Metab. Eng. 2008, 10, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Yue, H.; Zhao, Y.; Ma, X.; Gong, J. Ethylene glycol: Properties, synthesis, and applications. Chem. Soc. Rev. 2012, 41, 4218–4244. [Google Scholar] [CrossRef]
- Kawabe, K. Development of highly selective process for mono-ethylene glycol production from ethylene oxide via ethylene carbonate using phosphonium salt catalyst. Catal. Surv. Asia 2010, 14, 111–115. [Google Scholar] [CrossRef]
- Li, Y.; Yue, B.; Yan, S.; Yang, W.; Xie, Z.; Chen, Q.; He, H. Preparation of ethylene glycol via catalytic hydration with highly efficient supported niobia catalyst. Catal. Lett. 2004, 95, 163–166. [Google Scholar] [CrossRef]
- Salusjärvi, L.; Havukainen, S.; Koivistoinen, O.; Toivari, M. Biotechnological production of glycolic acid and ethylene glycol: Current state and perspectives. Appl. Microbiol. Biotechnol. 2019, 103, 2525–2535. [Google Scholar] [CrossRef]
- Ueda, N.; Nakagawa, Y.; Tomishige, K. Conversion of glycerol to ethylene glycol over Pt-modified Ni catalyst. Chem. Lett. 2010, 39, 506–507. [Google Scholar] [CrossRef]
- Yin, A.-Y.; Guo, X.-Y.; Dai, W.-L.; Fan, K.-N. The synthesis of propylene glycol and ethylene glycol from glycerol using Raney Ni as a versatile catalyst. Green Chem. 2009, 11, 1514–1516. [Google Scholar] [CrossRef]
- Liu, H.; Ramos, K.R.M.; Valdehuesa, K.N.G.; Nisola, G.M.; Lee, W.-K.; Chung, W.-J. Biosynthesis of ethylene glycol in Escherichia coli. Appl. Microbiol. Biotechnol. 2012, 97, 3409–3417. [Google Scholar] [CrossRef]
- Cam, Y.; Alkim, C.; Trichez, D.; Trebosc, V.; Vax, A.; Bartolo, F.; Besse, P.; François, J.-M.; Walther, T. Engineering of a synthetic metabolic pathway for the assimilation of (d)-Xylose into value-added chemicals. ACS Synth. Biol. 2015, 5, 607–618. [Google Scholar] [CrossRef]
- Chen, Z.; Huang, J.; Wu, Y.; Liu, D. Metabolic engineering of Corynebacterium glutamicum for the de novo production of ethylene glycol from glucose. Metab. Eng. 2016, 33, 12–18. [Google Scholar] [CrossRef]
- Rollin, J.A.; Tam, T.K.; Zhang, Y.-H.P. New biotechnology paradigm: Cell-free biosystems for biomanufacturing. Green Chem. 2013, 15, 1708–1719. [Google Scholar] [CrossRef]
- Shi, T.; Han, P.; You, C.; Zhang, Y.P.J. An in vitro synthetic biology platform for emerging industrial biomanufacturing: Bot-tom-up pathway design. Synth. Syst. Biotechnol. 2018, 3, 186–195. [Google Scholar] [CrossRef]
- Gao, C.; Li, Z.; Zhang, L.; Wang, C.; Li, K.; Ma, C.; Xu, P. An artificial enzymatic reaction cascade for a cell-free bio-system based on glycerol. Green Chem. 2015, 17, 804–807. [Google Scholar] [CrossRef]
- Li, Z.; Yan, J.; Sun, J.; Xu, P.; Ma, C.; Gao, C. Production of value-added chemicals from glycerol using in vitro enzymatic cascades. Commun. Chem. 2018, 1, 1–7. [Google Scholar] [CrossRef]
- Jia, X.; Kelly, M.R.; Han, Y. Simultaneous biosynthesis of (R)-acetoin and ethylene glycol from D-xylose through in vitro met-abolic engineering. Metab. Eng. Commun. 2018, 7, e00074. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, M.; Jiang, T.; Sheng, B.; Ma, C.; Xu, P.; Gao, C. Enzymatic cascades for efficient biotransformation of racemic lactate derived from corn steep water. ACS Sustain. Chem. Eng. 2017, 5, 3456–3464. [Google Scholar] [CrossRef]
- Heuts, D.P.H.M.; Van Hellemond, E.W.; Janssen, D.B.; Fraaije, M.W. Discovery, characterization, and kinetic analysis of an alditol oxidase from Streptomyces coelicolor. J. Biol. Chem. 2007, 282, 20283–20291. [Google Scholar] [CrossRef]
- Forneris, F.; Heuts, D.P.H.M.; Delvecchio, M.; Rovida, S.; Fraaije, M.W.; Mattevi, A. Structural analysis of the catalytic mechanism and stereoselectivity in Streptomyces coelicolor alditol oxidase. Biochemistry 2008, 47, 978–985. [Google Scholar] [CrossRef]
- Gerstenbruch, S.; Wulf, H.; Mußmann, N.; O’Connell, T.; Maurer, K.-H.; Bornscheuer, U.T.; Mussmann, N. Asymmetric synthesis of d-glyceric acid by an alditol oxidase and directed evolution for enhanced oxidative activity towards glycerol. Appl. Microbiol. Biotechnol. 2012, 96, 1243–1252. [Google Scholar] [CrossRef]
- Sheng, B.; Xu, J.; Ge, Y.; Zhang, S.; Wang, D.; Gao, C.; Ma, C.; Xu, P. Enzymatic resolution by a d-lactate oxidase catalyzed reaction for (S)-2-hydroxycarboxylic acids. ChemCatChem 2016, 8, 2630–2633. [Google Scholar] [CrossRef]
- Montella, C.; Bellsolell, L.; Pérez-Luque, R.; Badía, J.; Baldoma, L.; Coll, M.; Aguilar, J. Crystal structure of an iron-dependent group III dehydrogenase that interconverts l-lactaldehyde and l-1,2-propanediol in Escherichia coli. J. Bacteriol. 2005, 187, 4957–4966. [Google Scholar] [CrossRef]
- Iding, H.; Siegert, P.; Mesch, K.; Pohl, M. Application of α-keto acid decarboxylases in biotransformations. Biochim. Biophys. Acta Protein Struct. Mol. Enzym. 1998, 1385, 307–322. [Google Scholar] [CrossRef]
- Wang, X.; Miller, E.N.; Yomano, L.P.; Zhang, X.; Shanmugam, K.T.; Ingram, L.O. Increased furfural tolerance due to overex-pression of NADH-dependent oxidoreductase FucO in Escherichia coli strains engineered for the production of ethanol and lactate. Appl. Environ. Microbiol. 2011, 77, 5132–5140. [Google Scholar] [CrossRef]
- Zhu, Y.; Lin, E.C. L-1,2-propanediol exits more rapidly than l-lactaldehyde from Escherichia coli. J. Bacteriol. 1989, 171, 862–867. [Google Scholar] [CrossRef]
- Wang, Y.; Xian, M.; Feng, X.; Liu, M.; Zhao, G. Biosynthesis of ethylene glycol from d-xylose in recombinant Escherichia coli. Bioengineered 2018, 9, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Zhang, Y.; Xiong, W.; Ding, H.; He, B. Hydrogenolysis of glycerol to 1,2-propanediol and ethylene glycol over Ru-Co/ZrO2 catalysts. Catalysts 2016, 6, 51. [Google Scholar] [CrossRef]
- Pereira, B.; Zhang, H.; De Mey, M.; Lim, C.G.; Li, Z.; Stephanopoulos, G. Engineering a novel biosynthetic pathway in Escherichia coli for production of renewable ethylene glycol. Biotechnol. Bioeng. 2016, 113, 376–383. [Google Scholar] [CrossRef]
- Zhang, Z.; Yang, Y.; Wang, Y.; Gu, J.; Lu, X.; Liao, X.; Shi, J.; Kim, C.H.; Lye, G.; Baganz, F.; et al. Ethylene glycol and glycolic acid production from xylonic acid by Enterobacter cloacae. Microb. Cell Fact. 2020, 19, 1–16. [Google Scholar] [CrossRef]
- Pereira, B.; Li, Z.-J.; De Mey, M.; Lim, C.G.; Zhang, H.; Hoeltgen, C.; Stephanopoulos, G. Efficient utilization of pentoses for bioproduction of the renewable two-carbon compounds ethylene glycol and glycolate. Metab. Eng. 2016, 34, 80–87. [Google Scholar] [CrossRef]
- Chae, T.U.; Choi, S.Y.; Ryu, J.Y.; Lee, S.Y. Production of ethylene glycol from xylose by metabolically engineered Escherichia coli. AICHE J. 2018, 64, 4193–4200. [Google Scholar] [CrossRef]
- Cabulong, R.B.; Chung, W.-J.; Ramos, K.R.M.; Nisola, G.M.; Lee, W.-K.; Lee, C.R.; Chung, W.-J. Enhanced yield of ethylene glycol production from d-xylose by pathway optimization in Escherichia coli. Enzym. Microb. Technol. 2017, 97, 11–20. [Google Scholar] [CrossRef]
- You, C.; Shi, T.; Li, Y.; Han, P.; Zhou, X.; Zhang, Y.-H.P. An in vitro synthetic biology platform for the industrial biomanufacturing of myo-inositol from starch. Biotechnol. Bioeng. 2017, 114, 1855–1864. [Google Scholar] [CrossRef]
- Rollin, J.A.; Del Martin del Campo, J.; Myung, S.; Sun, F.; You, C.; Bakovic, A.; Castro, R.; Chandrayan, S.K.; Wu, C.-H.; Adams, M.W.; et al. High-yield hydrogen production from biomass by in vitro metabolic engineering: Mixed sugars coutilization and kinetic modeling. Proc. Natl. Acad. Sci. USA 2015, 112, 4964–4969. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).