Abstract
Lignocellulose has economic potential as a bio-resource for the production of value-added products (VAPs) and biofuels. The commercialization of biofuels and VAPs requires efficient enzyme cocktail activities that can lower their costs. However, the basis of the synergism between enzymes that compose cellulolytic enzyme cocktails for depolymerizing lignocellulose is not understood. This review aims to address the degree of synergism (DS) thresholds between the cellulolytic enzymes and how this can be used in the formulation of effective cellulolytic enzyme cocktails. DS is a powerful tool that distinguishes between enzymes’ synergism and anti-synergism during the hydrolysis of biomass. It has been established that cellulases, or cellulases and lytic polysaccharide monooxygenases (LPMOs), always synergize during cellulose hydrolysis. However, recent evidence suggests that this is not always the case, as synergism depends on the specific mechanism of action of each enzyme in the combination. Additionally, expansins, nonenzymatic proteins responsible for loosening cell wall fibers, seem to also synergize with cellulases during biomass depolymerization. This review highlighted the following four key factors linked to DS: (1) a DS threshold at which the enzymes synergize and produce a higher product yield than their theoretical sum, (2) a DS threshold at which the enzymes display synergism, but not a higher product yield, (3) a DS threshold at which enzymes do not synergize, and (4) a DS threshold that displays anti-synergy. This review deconvolutes the DS concept for cellulolytic enzymes, to postulate an experimental design approach for achieving higher synergism and cellulose conversion yields.
1. Introduction
Lignocellulosic feedstocks have huge economical potential as a source of value-added products (VAPs) and biofuel [1,2,3]. Cellulose is the major component of lignocellulose, and it can be used as a source of fermentable sugars that can serve as precursors for VAPs synthesis. Cellulose is sourced from plant material (wood pulp, cotton, or cereals, such as wheat, sugarcane, and rice bagasse), and is a linear polymer that consists of β-D-glucose molecules linked by glycosidic bonds [1,4,5,6]. The linear structure of a cellulose chain is directional, as it consists of a reducing-end glucose that contains an anomeric carbon (C1) and a non-reducing-end glucose, consisting of hydrogen and a hydroxyl group on the C4 carbon (Figure 1). Many cellulose chains bundle via hydrogen bonding to constitute cellulose microfibrils, which consist of crystalline regions intersected by amorphous regions. The β-D-glucose residues in the crystalline regions of the cellulose microfibril are anhydrous; this is because the orientation of the three, free hydroxyl groups of the sugar create strong molecular bonds with glucose molecules in adjacent chains, which make cellulose insoluble in water [2]. Thus, crystalline cellulose is recalcitrant to enzymatic hydrolysis [1,2,7]. As a result, several acid or basic chemicals are generally used to modify the chemical and structural properties of native cellulose to generate amorphous cellulose or cellulose with a reduced degree of crystallinity [2,5,8,9].
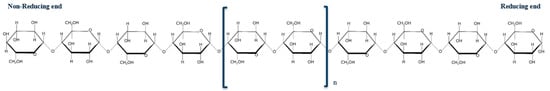
Figure 1.
A representation of a cellulose chain, with reducing and non-reducing ends indicated. The blue brackets with subscript “n” indicate that the degree of polymerization (DP) of the cellulose chain is longer than a DP of 10.
It is well understood that several glycoside hydrolases (GHs) from different families act synergistically to achieve complete hydrolysis of the cellulosic fraction of the biomass feedstock into fermentable sugars [3,7,10,11,12]. There are the following two types of mechanisms of synergy that have been documented to date: (1) simultaneous synergism, whereby all enzymes are added into the reaction at the same time and (2) sequential synergism, whereby enzymes are added into a reaction sequentially, based on their substrate cleavage preferences [11,13]. The simultaneous application of synergistic enzymes is the most preferred technique for degrading biomass feedstocks; for instance, the cellulolytic enzyme homeo-synergism is based on the simultaneous use of the following cellobiohydrolases: CBHI (EC 3.2.1.176) and CBHII (EC 3.2.1.91), endoglucanase (EG, EC 3.2.1.4) and β-glucosidase (β-gl, EC 3.2.1.21). Several research groups have demonstrated synergism between CBHI, CBHII, EG, and β-gl sourced from fungi or bacteria, e.g., Trichoderma reesei and Clostridium thermocellum, during the degradation of cellulose [5,12,13,14,15]. Figure 2 shows the well-established mode of action of cellulases during the synergistic hydrolysis of cellulose. Further analysis of the literature reveals that cellulolytic enzyme synergism can be grouped into the following two classes: (1) synergism between non-catalytic and catalytic cellulolytic enzymes (e.g., expansins and carbohydrate-binding modules (CBMs)), and (2) synergism between catalytic cellulolytic enzymes, e.g., exo–exo, exo–endo, exo–endo–β-gl, or cellulase and lytic polysaccharide monooxygenase (LPMO) synergy [16,17,18,19,20,21]. There are only a few studies that have documented endo–endo synergy, even though EGs constitute the most diverse cellulase group, classified under GH families 5, 6, 7, 8, 9, 10, 12, 44, 45, 48, 51, 74, and not classified sequences (NC) (http://www.cazy.org/Glycoside-Hydrolases.html; accessed on 30 June 2021). It is also well documented that CBHs from different GH families that hydrolyze cellulose on the same cleavage sites do not synergize together, but compete for occupancy on the same binding sites [15,22].
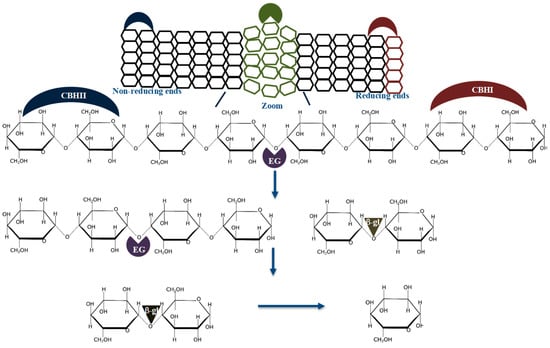
Figure 2.
A schematic representation of cellulose and corresponding cellulolytic enzymes that degrade it. The enzyme cleavage sites on the cellulose are represented by red arrows. Microcrystalline and amorphous (green color) regions of cellulose consist of β-D-glucose linked by β-1,4-glycosidic bonds. Where β-gl, CBHI, CBHII and EG represent β-glucosidase, cellobiohydrolase I, cellobiohydrolase II and endo–glucanase.
Two other enzyme systems play a significant role in cellulose saccharification, these being LPMOs and non-hydrolytic enzymes (expansins and swollenins). Forsberg et al. [23] demonstrated that the LPMOs sourced from bacteria and fungi were originally classified into CBM family 33 and GH61, respectively. According to the CAZy database, the enzymes classified under CBM33 were later assigned to auxiliary activities family 10 (AA10), while those in GH61 were assigned to AA9 (http://www.cazy.org/Auxiliary-Activities.html; accessed on 30 June 2021). The LPMOs in family AA9 and AA10 are copper-dependent enzymes that cleave the glycosidic bonds of crystalline cellulose regions by oxidizing the C1 (EC 1.14.99.54) or C4 (EC 1.14.99.56) and C1/C4 of the glucose residues [23,24] (see Figure 3). The LPMOs in family AA16 are newly classified cellulose-specific enzymes with C1 regiospecificity [25]. Kadowaki et al. [24] demonstrated that some LPMOs sourced from Myceliophthora thermophila, belonging to AA9D, have regiospecificity for C1, and those sourced from Aspergillus nidulans, belonging to AA9 (also known as 22), and from M. thermophila, belonging to AA9J, have regiospecificity for C4. Additionally, the LPMOs sourced from M. thermophila (belonging to AA9I), Thielavia terrestris (belonging to AA9E), and Thermoascus aurantiacus (belonging to AA9A) displayed regiospecificity for both C1 and C4 (referred to as C1/C4). Both bacterial and fungal LPMOs have similar structures and mechanisms of action [23]. The catalytic domain of LPMOs can also be attached to CBMs by a flexible linker [24,26]. Furthermore, Isaksen et al. [27] demonstrated that the fungal LPMO-AA9C, sourced from Neurospora crassa, was capable of cleaving polymeric cellulose and cello-oligosaccharides with a degree of polymerization of about four to six. The LPMOs cleaved the C1 of cellulose or cello-oligosaccharides to produce aldouronic acids; however, when they acted on the C4, they produced gem-diol [26,27]. The LPMOs from families AA9, AA10, and AA16 have been reported to synergize with cellulases to enhance the saccharification of cellulose [25,28,29].
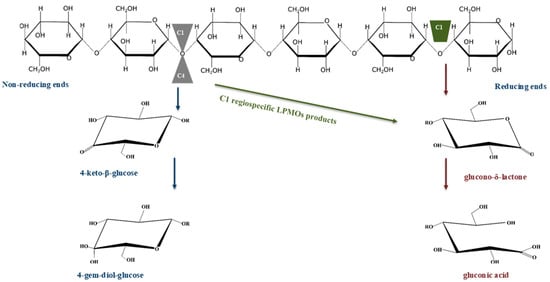
Figure 3.
A schematic representation of cellulose chain and corresponding lytic polysaccharide monooxygenases (LPMOs) that degrade it. LPMOs can cleave glycosidic bonds joining the glucose units of cellulose at C1 (green block), and at either C1 or C4 (grey triangles). The C1 regiospecific LPMOs produce glucono-δ-lactone and gluconic acid (brown arrows), which is stable in solution. In contrast, LPMOs with C1/C4 regiospecificity can produce a mixture of gluconic acid (green and brown arrow) and 4-gem-diol-glucose (blue arrows).
Expansins and expansin-like proteins are non-hydrolytic or non-oxidative enzymes that can bind to the crystalline cellulose and weaken the degree of crystallinity of the polymer by breaking the inter- or intra-molecular hydrogen bonding network [30,31]. Kim et al. [32] demonstrated that Bacillus subtilis-derived expansin sequences were homologous to maize plant expansins, and their 3D structures were also similar. The role of expansins in plants is well established, and plant-derived expansins are classified into two groups, α- and β-expansins [4]. The α- and β-expansins only share 20% amino acid sequence similarity. Interestingly, expansins generally have about 30% sequence similarity to endoglucanases classified under GH45 [4]. Several studies have shown that the expansins synergize with cellulases that hydrolyze the crystalline microfibrils of cellulose [4,30,31,33]. Fungi also possess another non-hydrolytic protein called swollenin, which is similar to the expansins [33]. Swollenins are reported to modify the chemistry and structure of microcrystalline cellulose by reducing its degree of crystallinity, allowing the cellulases to hydrolyze cellulose effectively. Additionally, studies have demonstrated that cellulases and swollenins synergize to improve the saccharification of cellulose [17,33]. In this literature review, we used the DS, a powerful parameter that describes molecular interactions between the enzymes during synergy, to explain hydrolytic cellulolytic enzyme to non-hydrolytic enzyme synergism thresholds, to develop a model to assist in the improvement of cellulose hydrolysis by cocktails.
The DS between enzymes is defined as the observed activity of two or more enzyme combinations divided by the theoretical sum of the activities of the individual enzymes acting on the same substrate. The enzyme combinations are formulated using three protein loading systems, which are protein mass ratio, molar ratio, and activity ratio [11,12,15,20,34]. According to van Dyk and Pletschke [11], the DS can provide important information about the chemical structure of the substrate and enzyme interactions during the hydrolysis of the substrate. Two or more enzymes cooperating during the hydrolysis of the substrate lead to higher soluble sugar production (Figure 2). Additionally, the mass enzyme loading in the reaction may be reduced due to synergism between enzymes, and this could lead to lowered enzyme costs during biomass valorization [5,32,35]. Van Dyk and Pletschke [11] further demonstrated that the DS is a particularly important component for synergy studies, yet not all experiments performed to investigate the GHs or GH-to-LPMO synergism factor in the quantification of the DS.
The lack of investigation of the DS during enzymatic cellulose saccharification is attributed to a focus on hydrolysis yields by researchers. It appears that some enzyme combinations do not display a positive DS, yet these combinations demonstrate higher yields, in terms of soluble sugar production, compared to when the individual enzymes are used to hydrolyze the substrate. However, we propose that investigating the synergism between enzymes should not only be limited to yield improvement, but should also include the DS. This would enable researchers and companies to design better enzyme cocktails for specific feedstocks. This short review will address how we can achieve higher yields and DS by re-evaluating the established synergisms and setting the DS threshold benchmarks, which can lead to higher yields.
2. Defining DS Threshold and Its Impact on Substrate Hydrolysis
It is well accepted that a DS of 1 arbitrary unit (AU) demonstrates no synergism between the enzymes used to formulate a said enzyme cocktail [11,12,15,20,36,37]. These studies suggest that the enzymes do not cooperate during substrate hydrolysis, leading to no improvement in the rate of substrate deconstruction compared to when the mono-components act individually; for example, endoglucanase and exoglucanase enzymes do not synergize, and generally result in a DS of 1 AU, during the hydrolysis of the model amorphous cellulose carboxyl methylcellulose (CMC), because of their uncomplimentary modes of action on the substrate. A value below 1 AU shows that there is no synergism between the enzymes during the hydrolysis of the substrate. This observation suggests that one or more of the enzymes cannot hydrolyze the substrate efficiently, due to competitive binding of two or more enzymes on the same substrate cleavage site (same regiospecific enzymes). Instances that resulted in a DS below 1 AU have been documented between CBHII and EG, processive EGs from GH9 and CBHI or CBHI (from GH48), and CBHI (from GH7) or CBHI (GH7), and an LPMO (AA9E). In all these cases, substrate hydrolysis was impeded, and product formation significantly reduced. Generally, DS values above one are achieved when the enzymes in the cocktails hydrolyze different cleavage sites of the substrate, e.g., CBHI, CBHII, EG, and β-gl degrade cellulose at different cleaving sites, resulting in significantly higher product formation (Figure 2).
It is reported that enzyme synergism and its DS are influenced not only by the substrate, but by other factors covered in many other studies [5,11,37]. The quantification of DS levels is generally based on the enzyme activity or specific activity, the conversion rate and/or the yield of product in relation to substrate quantity [11,12,15,38]. There are no thresholds that distinguish between the patterns of enzymes competing for the same cleavage sites on the substrate and those that do not cooperate at all during substrate hydrolysis. Studies have only focused on reporting that a DS value < 1 results in no synergism. We, therefore, hypothesize that 1 ≥ DS values > 0.5 demonstrates that the enzymes are not cooperating on the substrate, while 0.5 ≥ DS values > 0 could signify competitive binding to the same substrate’s cleavage site (same regiospecificity), and DS values > 1 could signify that there is synergism between non-hydrolytic and hydrolytic cellulolytic enzymes. The following sections of this review attempt to explain and formulate the DS threshold, whereby cellulolytic enzyme synergism can result in a higher yield (DS > 1), or inhibited yield due to non-cooperating (1 ≥ DS value > 0.5) and competing enzymes (0.5 ≥ DS value > 0).
3. Unravelling Enzyme Synergism
We hypothesize that, to formulate effective enzyme cocktails, researchers should not only focus on the yield of soluble sugars, as conventional synergy studies have done for the past several decades, but should focus on both the yield of soluble sugars and the DS between enzymes. This approach will generate holistic information about the reaction, with regards to the interactions between the enzymes in the cocktails, as well as the products formed. Hence, we attempted to establish the DS threshold that results in high yields of soluble sugars. A detailed discussion of various cellulolytic enzyme synergy studies that were conducted with different enzyme combinations are presented below and summarized in Table 1.
3.1. Non-Hydrolytic and Catalytic Cellulolytic Enzyme Synergy
The crystalline nature of cellulose requires a concerted effort of non-hydrolytic enzymes that weaken the hydrogen bonds between the cellulose fibers, or assist with threading and transferring the cellulose fibers to the catalytic cleft of hydrolytic enzymes [4,5]. These non-hydrolytic cellulolytic enzymes are grouped under CBMs, expansins, expansin-like proteins, and swollenins [4,31,39,40,41]. Lee et al. [30] demonstrated that expansins reduce the crystallinity of the cellulose, enhancing the activity of the commercial cellulase preparation Celluclast® 1.5 L 4.8-fold compared to when the cellulase preparation was used alone. A DS value of 4.6 AU between Celluclast® 1.5 L and an expansin was predicted by Lee et al. [30]. Pech-Cervantes et al. [41] demonstrated that the synergy between an expansin-like protein and cellulase did not have a significant impact on the hydrolysis of amorphous cellulose substrate (CMC). However, the expansin-like protein modified the structure of the crystalline cellulose substrate, filter paper, leading to synergy between it and the cellulase, whereby a four-fold improvement in sugar release was recorded. Again, the calculated DS value, using the published data, computed a DS value of 4.2 AU. A similar impact of synergism was reported by Kim et al. [42], where expansin-to-cellulase synergism increased the release of reducing sugars by 240% (corresponding to a calculated DS value of 2.4 AU), compared to the use of cellulase alone. These studies show that there can only be the following two thresholds with regards to expansin-to-cellulase synergism: (1) DS greater than or equal to 1 AU (DS ≥ 1) on crystalline cellulose, and (2) DS equal to one arbitrary unit (DS = 1) on amorphous cellulose. It is important to note that the currently reported synergistic studies were conducted using commercial cellulase preparations, suggesting that there is a need to use the expansins or expansin-like proteins with pure cellulases (CBHs or EGs), so that the mechanism of enzyme interaction can be more closely elucidated.
Another group of enzymes with a similar function to expansins are swollenins (SWO), which are mostly produced by fungi [35,43]. In contrast to expansins, SWOs have cross-activity on amorphous cellulose (CMC), lichenan, mixed-linked glucan (MLG), and laminarin, while on microcrystalline celluloses (e.g., Avicel, cotton, and filter paper), they display non-hydrolytic activity by remodifying its structure [35,39,43]. A SWO displayed synergy with commercial cellulase on Avicel, leading to the release of 30% more reducing sugars [35]. Zhang et al. [39] purified and used CBHI (Cel7A), or endoglucanase (EG) supplemented with β-glucosidase (β-gl) and SWO, to hydrolyze phosphoric acid swollen cellulose (PASC) and cellulose nanocrystals (CNC). Interestingly, the EG, β-gl, and SWO combination displayed higher activity than the Cel7A, β-gl, and SWO combination on PASC, while on CNC, the EG-containing mixture did not show any significant activity, while the Cel7A mixture showed higher activity. Our highest estimated DS value for the EG-containing mixture was 1.2 AU, and the Cel7A-containing mixture displayed 2.0 AU on PASC, suggesting that the Cel7A and SWO synergistic interaction was more productive. Also, the synergism between Cel7A and SWO was effective because both enzymes have proven to be effective on the crystalline substrate (CNC), while EG cleaves amorphous regions of PASC. Our estimated DS values from the published data demonstrate that the EG mixture had an estimated DS value of 1.0 AU (with a 20% conversion rate of CNC), while the Cel7A mixture had the highest DS value of 1.4 AU (with a 60% conversion rate of CNC). These observations agree with a study by Santos and co-workers, which demonstrated that SWOs reduce the crystallinity of cellulose and modify the microfibril structure to create more binding and cleavage sites for cellulolytic enzymes, in particular, CBHs [43].
It is important to note that both expansins and SWOs facilitate the activity of cellulase cocktails or pure mono-component cellulases (CBHI or EG) on regenerated amorphous cellulose (RAC) or PASC, or filter paper and/or Avicel [30,35,39,42,43]. However, further laoding high concentrations of expansins and SWO, beyond a certain threshold, on the substrate does not translate to a linear increment in overall cellulose hydrolysis. The concentration that resulted in higher cellulase and non-catalytic synergism before expansin/SWO binding saturation was 10 mg protein/g biomass [17,35]. At higher loadings of expansin (30 mg protein/g biomass), a reduction in biomass conversion, from a 15% (for expansin at 10 mg/g biomass) to 4.5% conversion rate, was observed [35]. In contrast, at a similar enzyme loading (30 mg protein/g biomass), SWO did not show any significantly different activity compared to the activity displayed by 10 mg protein/mg biomass [17]. These findings support our hypothesis that expansin and SWO synergism with cellulases cannot result in DS values that are below 1 AU, and in instances where there is no synergism, the DS values will be equal to 1 AU (Table 1).
3.2. Synergism between Processive Cellulolytic Enzymes; Exo–Exo or Exo–Endo Processive Synergy
There is strong evidence that supports the hypothesis that some backbone cleaving enzymes always interact synergistically, resulting in a DS that is higher than 1 AU and a soluble sugar yield that is higher than that produced by the sum of the individual enzymes during the hydrolysis of biomass [11]; for instance, cellulase enzymes that cleave microcrystalline cellulose, such as Cel6A and Cel7A (exo–exo synergy), or Cel6A, Cel7A, and EG (exo–endo synergy; see Section 3.5), generally synergize with a DS that is higher than 1 AU [11,15,20]. Exo–exo synergy between cellulases has been demonstrated by Boisset et al. [44] using bacterial microcrystalline cellulose (BMC), while other researchers from our laboratory (Enzyme Science Programme (ESP) at Rhodes University) have used Avicel, PASC, or NaOH-treated Avicel, also called regenerated amorphous cellulose (RAC), as suitable substrates [12,15,34]. Badino et al. [20] showed that the DS between GH7 (Cel7A) and GH6 (Cel6A) enzymes sourced from Hypocrea jecorina was above 2.2 AU, while the glucose yield produced by synergistic actions was four-fold higher than that produced by individual enzymes. A study by Boisset et al. [45] also found similar results using Humicola insolens GH7 (Cel7A) and GH6 (Cel6A), which showed a DS of about 4 AU between enzyme combinations, and converted more than 50% of BMC compared to 15% by Cel6A and 28% by Cel7A alone. Based on the literature and our experiences, the fungal-sourced Cel7A (GH7) and Cel6A (GH6) synergism generally displays a DS above 2 AU, and will always result in a higher yield of soluble sugars than individual enzymes.

Table 1.
Various types of cellulolytic enzymes synergism with recorded/predicted degrees of synergy (DS), as well as the theorized DS thresholds that result in improved conversion yields of cellulose substrates.
Table 1.
Various types of cellulolytic enzymes synergism with recorded/predicted degrees of synergy (DS), as well as the theorized DS thresholds that result in improved conversion yields of cellulose substrates.
| Type Synergy | Cellulolytic Enzymes | Cellulose Substrate | Conversion Yield/Activity Increase | Recorded/Predicted # DS Values | DS-Threshold * | Reference |
|---|---|---|---|---|---|---|
| Non-catalytic and catalytic active cellulolytic enzyme synergy | Expansins: Cel7A or Cel7B | model cellulose II films | <5-fold | N/A | DS value > 1 | [31] |
| Bpexpansins and celluclast™ Bp- or Cm-expansins: celluclast™ | Avicel PASC Filter paper | <2-fold <1-fold <5-fold | 2.5 1.3 7.3 | [18] [19] | ||
| Bsexpansin: cellulase | Filter paper | <2-fold | 2.5 | [42] | ||
| Xcexpansin: Accellerase 1500 | Filter paper | 36% | 1.4 | [46] | ||
| TrSWO: CHI or EGII | Valonia cell walls | N/A | N/A | [33] $ | ||
| Po-SWO: cellulases | Avicel | <2-fold | 2.2 | [17] | ||
| Processive Cellulolytic enzyme synergism (Exo-Exo &Exo-processive Endo a) | HjCel6A: HjCel7A HiCel6A: HiCel7A Cel6A: Cel7A | Avicel Bacterial cellulose Mercerized Avicel | <3-fold 90% <1.2-fold | 2.3 3 1.4 | DS value > 1.2 | [20] [44] [15] |
| Cel9A: Cel48A EG: CBH CcCel9A: CcCel48 HiCel6A: HiCel7A | Filter paper Filter paper Filter paper Bacterial cellulose | 17% <3-fold <1.2-fold 30% | 1.7 2.6 1.7 4.5 | [47] [48] [49] [44] | ||
| Endo-Endo synergism | CcCel9A: CcCel9B CelZ: CelY | CMC CMC | 2-fold 2-fold | 1.1 1.8 | ND | [49] [50] |
| Endo-Beta-glucosidase | CcCel9A: BlgA CgEG1: CgBlg1 CgEG1: CgBlg1 EG: Bgl EG: Bgl c | Filter paper CMC Sigma-cell Filter paper DMOS-SCB | <1.8-fold <90% <80% 1.75-fold 3-fold | 2.1 9 2.8 2.0 3 | DS value > 1.3 | [49] [51] [51] [51] [51] |
| Endo-Exo synergy b | TrCel7A: TrCel5A Cel6: Cel5A ThCel7B: ThCel7A HiCel7A: HiCel45A HiCel6A:HiCel45A HiCel6A:HiCel7A: HiCel45A TrEGII: TrCBHI: TrCBHII CBHI: EGII: CBHI CcCel9A:CcCel9BCcCel48A TrCel7A: TrCel7B | Bacterial cellulose Cellulose-III Filter paer Bacterial cellulose Bacterial cellulose Bacterial cellulose Avicel Cellulose with DP3000 Avicel Filter paper steam-pretreated spruce | N/A 5% 92% 25% 15% 90% 25% N/A 27% <3-fold <2-fold | 1.7 2.16 1.6 2.6 2.1 2.0 2 2.6 1.6 2.6 1.8 | DS value > 2 | [16] [52] [14] [44] [44] [44] [53] [5] [54] [49] [55] |
| Cellulases and LPMO synergism | AaAA16: CBHI Celluclast: mgLPMO10 TtMO9E: Cel6A Celluclast: CelS2 MtLPMO9L: CBHII TrCel7A: TrCel6A: TrCel7B: TtAA9 MtEG5A: MtEG7A: MtLPMO9 | PASC Avicel PASC Filter paper PASC PASC PASC | <1.8-fold 36% 2-fold 4-fold 3-fold 2.8-fold <2.5-fold | 1.8 1.4 1.9 4 3 3 2.6 | DS values > 2 | [25] [26] [37] [23] [56] [29] [57] |
Bp: Bacillus pumilus, Bs: Bacillus subtilis, Xc: Xanthomonas campestris, Cm: Micromonospora aurantiaca ATCC 27029, Tr-SWO: Trichoderma reesei-swollenin, Th: Trichoderma harzianum, Tl: Trichoderma longibrachiatum, Po: Penicillium oxalicum, Hj: Hypocrea jecorina, Hi: Humicola insolens, Cc: Clostridium cellulosi, Ba: Bacillus amyloliquefaciens mg: derived from metagenome, Aa: Aspergillus aculeatus, Mt: Myceliophthora thermophila, Tt: Thermothelomyces thermophilus. # Predicted DS values were estimated directly from published data. * Theoretical DS value thresholds which generally result in higher degrees of synergy. $ Solubility studies which were equivalent to synergistic activity. a Conversional exo–exo synergy can be substituted with processive EG in a new approach of exo-processive endo. b Endo–exo synergy is generally supplemented with β-gl to prevent product inhibition. c The enzymes were immobilized on synthetized Fe3O4 nanoparticles.
In some bacterial systems, such as Clostridium phytofermentans ISDg, a model microorganism for consolidated bioprocessing (CBP), there is a different cellulase system that degrades the crystalline regions of the cellulose [47]. The CBH enzymes employed by C. phytofermentans ISDg are Cel48 and Cel9. Zhang and co-workers suggested that Cel48 attached to the substrate from the reducing end, while Cel9 (processive EG) attached from the non-reducing end. In addition, CBHs that belong to GH 48, such as Cel48, usually contain a family 3 CBM [47]. Anaerocellum thermophilum is another bacterium that has demonstrated that the GH48 and GH9 enzyme system is generally employed to hydrolyze microcrystalline cellulose via exo–exo synergy [58]. Additionally, Irwin and co-workers showed that there was higher synergism between Cel48A and Cel9A sourced from Thermobifida fusca compared to other enzymes from different GH families, such as GH5 and GH6 [59]. Cel9 from Bacillus licheniformis was shown to have a CBM3c linked to the catalytic domain, and displayed the highest activity on PASC, BMC, and filter paper. Cel9 (GH 9) from Clostridium cellulosi possesses five CBMs, namely, CBM3c, three CBMX2s, and CBM3b [60]. Zhang and co-workers proposed that the multi-modular Cel9A cleaves crystalline cellulose fibers via a “wire-walking mode”, whereby the CBMX2s and CBM3b bind to the cellulose substrate, while CBM3c, linked adjacent to the catalytic domain (CD), extends the cleft up to 17 subsites. Thus, it appears that the union of Cel9′s CBM3c and its CD assists in the processive nature of this enzyme on crystalline cellulose. Therefore, Cel9A qualifies as a pseudo CBH, based on its hydrolysis mechanism on microcrystalline cellulose. The exo–exo synergy between Cel48 and Cel9A on filter paper displayed a DS value of 1.5 AU [49]. Another study similarly reported a DS of about 1.5 AU for Cel48 (GH48) and Cel9 (GH9), while the soluble sugars produced from Avicel were four-fold higher than those produced by the individual enzymes [47]. These observations support our hypothesis that there are the following two or more distinct exo-cellulase systems that result in higher DS and yields of soluble sugars: (1) one from the fungal cellulolytic systems (mostly Cel7A and Cel6A), and (2) another from the bacterial cellulolytic systems (Cel 48 and Cel 9A).
Fungal exo–exo synergy is evolutionally designed to depend on the CBHs from GH family 6 and 7, because their fusion always results in a higher DS and yield of soluble sugars [11,16]. We propose that a DS greater than 2 AU should be considered as the threshold that always results in a higher yield of soluble sugars during GH6 and GH7 CBHs synergism (Table 1). Thus, the DS threshold of 2 AU must be used as a benchmark for Cel7A and Cel6A during the formulation of enzyme cocktails. However, a lower DS threshold for the bacterial exo–exo synergy systems, of about 1.5 AU, must be achieved because Cel48A and Cel9A displayed a very high yield of soluble sugars from the crystalline cellulosic substrate at low DS values, compared to the fungal system [47,49]. Lastly, we recommend that the fungal or bacterial exo–exo synergy systems should not be mixed in one formulation of an enzyme cocktail, since this could lead to competitive inhibition (see Section 4 for details on this inhibition).
3.3. Endo–Endo Synergy
Endoglucanases of fungal origin are diverse and complex, as some strains from certain species secrete more than three endoglucanases, which are classified under GH families 5, 6, 7, 9, 12, 44, 45, and 74 [61]. Foreman et al. [61] demonstrated that the filamentous fungus Trichoderma reesei secretes large amounts of Cel7B (EGI), Cel5A (EGII), Cel12A (EGIII), Cel61A (EGIV), and Cel45A (EGV). Even though Foreman and colleagues assigned EGIV or Cel61A to GH family 61, all the enzymes that belong to this family are now classified under auxiliary activity (AA) family 9 (http://www.cazy.org/AA9.html; accessed on 30 June 2021) (see Section 3.6 for details). In addition, Wood et al. [62] purified five distinct endoglucases (EGI to EG-V, with molecular weights of between 25 kDa and 62.5 kDa) from Penicillium pinophilum and hypothesized that each endoglucanase plays a specific role during biomass degradation. Mode of hydrolysis deciphering assays of these EGs revealed that EGI only hydrolyzed cello-oligosaccharides with a degree of polymerization (DP) higher than six, while EGII, EGIII, EGIV, and EGV hydrolyzed oligosaccharides with a DP higher than two [63]. Interestingly, EGII and EGIV only produced cellobiose, even when hydrolyzing cellotriose or cellopentaose; this suggests that the two EGs also performed transglycosylation [62,63,64]. In addition, EG from Aspegillus terreus, which belongs to GH12, could only hydrolyze cello-oligosaccharides with a DP > 4 [65]. The observations regarding the different DPs required for various classes of EGs, by Bhat et al. [63] and Segato et al. [65], suggest that it is vital to test the synergism between the different classes of EGs from different GH families and determine their degrees of synergy to develop a DS threshold for the fungal EG system. The fact that EGVI, or Cel61A, is currently classified into AA family 9 is another reason why the synergism between endoglucases from different families must be considered.
Five endoglucanases of the ruminant bacterial symbionts were detected, and their molecular weights were 42, 50, 52, 53, and 101 kDa [66]. Even though the 52 and 53 kDa EGs were more active compared to three other proteins, the authors did not classify these EGs into different GH families. Some of the bacterial EGs belong to GH family 8, and are specific for the β-1,4-glycosidic bonds of cellulose or β-glucans [67]. The GH8 endoglucanase produces mostly cellotetraose, cellotriose, and cellobiose from polymeric substrates, such as CMC and β-glucan. However, on oligosaccharides such as cellopentaose, the enzyme produced mostly cellotetraose and cellobiose, while cellohexaose hydrolysis resulted in cellotetraose, cellotriose, and cellobiose. Scapin and co-workers [67] showed that the protein structure of EG from GH8 resembles an (α/α)6 barrel and possesses a catalytic cleft with six subsites, which results in a unique mode of action that is specific for substrates with a DP > 5. Hakamada et al. [68] also reported on a 43 kDa EG from GH8 that was sourced from Bacillus circulans KSM-N257, which had a similar mode of action and structural features to the EG reported by Scapin et al. [67], except that it had higher activity towards lichenin, followed by CMC. The Thermobifida fusca strain UPMC 901 was shown to have multiple EGs using zymography, with about four bands equaling 82, 60, 35, and 30 kDa [69]. Some Bacillus sp. also produced diverse EGs with high molecular weights of 80 to 100 kDa. It is apparent that the bacterial endoglucanase system is not well classified into old categories as fungal systems are (which are grouped into EGI to EGV); however, some studies have classified them into new GH families 5, 6, 7, 8, 9, 10, 12, 44, 51, and 74 [70] (http://www.cazy.org/; accessed on 30 June 2021).
The literature shows that EGs from both fungal and bacterial systems are biochemically characterized, but there is a lack of evidence supporting their endo–endo synergy. We propose that endo–endo synergism could be vital for the effective conversion of cellulose or complex oligosaccharides from β-glucan to soluble sugars. Also, endo–endo synergy could reveal more details on the mechanism of action of EGs classified as being members of GH family 51 and 74, which are not well characterized. Lastly, there is a lack of information regarding the role of cellulolytic enzymes from GH 61 (now classified in AA family 9) with EGs in cellulase synergy during lignocellulose saccharification.
3.4. Endoglucanase (EG)-β-Glucosidase (β-gl) Synergy
Even though there is a lack of information regarding the synergism between endoglucanases, there are several studies that demonstrate that EGs and β-gl do synergize during cellulose degradation. This information is vital because it can be used to define the threshold of DS between EG and β-gl interactions, which can result in the generation of higher yields of soluble sugars from biomass hydrolysis. Endoglucanase sourced from Scytalidium thermophilum synergized with β-gl from Humicola insolens and released more than a two-fold higher amount of reducing sugars from DMSO-pretreated sugarcane bagasse [71]. The DS of about 1.6 AU demonstrated that the EG and β-gl synergized; however, the authors only used one enzyme combination (3 EG: 1 β-gl) for their synergy studies. An EG-to-β-gl cocktail was also used to produce cellulose nanocrystals (CNC); in this case, their combinations were not well defined [72]. Additionally, Teixeira et al. [72] used the synergy of Pyrococcus horikoshii endoglucanase and Pyrococcus furiosus β-glucosidase to achieve conversion yields of 60% from wet-disk-milled Celish® KY and 40% from unbleached Kraft wood pulp. The EG and β-gl synergism was superior to the commercial β-gl (referred to as OptimashTMBG) because it converted more than two-fold more of the amorphous regions of the biomass to soluble sugars, compared to commercial β-gl, resulting in shorter CNC. Cairo et al. [51] also demonstrated that the synergism between EG and β-gl resulted in a DS of about 9.79, and about a 10-fold higher conversion rate of CMC to soluble sugars when compared to the use of individual enzymes. Theoretically, true EG and β-gl synergy always results in DS values higher than 1 AU, and yields of soluble sugars that are higher compared to when the enzymes are acting individually. This theoretical observation is supported by the fact that the EGs produce various oligosaccharides that have a DP > 2, and β-gls cleave the EG products to glucose [12,51,73,74]. Thus, we suggest that for as long as a true EG (non-processive EG) hydrolyzes amorphous cellulose, the β-gl will produce glucose from these products, resulting in high DS and yields of soluble sugars. Several studies have demonstrated the co-evolution of the EG and β-gl interaction by co-expression of their genes in engineered fungi or bacteria to produce a protein that displays a dual function of EG and β-gl [73,74]. Thus, we propose that the DS threshold of 1.3 AU will always result in high yields of soluble sugars between true EG and β-gl.
3.5. Exo–Endo and/or β-Glucosidase Synergy (Cellulolytic Enzyme Cocktail)
Based on both theoretical and empirical evidence, CBHs, EGs, and β-gls can synergize during the hydrolysis of biomass (Figure 2), due to their varied substrate specificities. The synergism occurs in cases where the reaction contains the following: (1) CBHI and CBHII, which attack the biomass from the reducing and non-reducing ends of cellulose chains to produce cellobiose, respectively; (2) true EGs, which randomly cleave the amorphous regions of the biomass to produce oligosaccharides with a DP between 2 and 10, or cleave the amorphous regions of the microcrystalline cellulose to produce new chain ends for CBHs; (3) β-gls, which degrade the CBHs and EG-generated products to monosaccharides (glucose) [75]. Boisset et al. [44] demonstrated that CBHI (Cel7A), CBHII (Cel6A), and EG (Cel45A) sourced from Humicola insolens displayed synergism during the hydrolysis of bacterial cellulose ribbons (microcrystalline cellulose). The compositions of Cel6A and Cel7A were varied between 0 and 100%, with dosage variation steps of 20%, while Cel45A was fixed at 1.2%. The cocktail composed of the three enzymes converted more than 60% of the bacterial cellulose ribbons and displayed a DS of about 4.5 AU, while the combination of Cel6A and Cel7A converted only 30% and had a DS of 2 AU [45]. Some of the commercial cocktails, such as Celluclast® 1.5L, consist of Trichoderma reesei cellulases in the following combinations: 55% CBH-7A, 10% CBH-6A, 10% EG-7B, 10% EG-5A, and 1% β-gl [76]. The highest biomass conversion that the authors could achieve was about a 50% yield, only when they added an extra 10% β-gl, but their experimental design did not factor in the DS, which shows the molecular-level interactions between the enzymes. The addition of 50 U/g substrate of a GH12 EG sourced from Gloeophyllum trabeum to 10 U/g substrate of Celluclast® 1.5L resulted in a synergistic effect of about 14.5%, 16.1%, 29.0%, and 13.4% on filter paper, hydrogen peroxide–acetic acid-pretreated pine, corn stover, and rice straw, respectively [77].
The synergy between CBHI (Cel7A) and EG (Cel7B) from T. reesei effectively hydrolyzed steam-pretreated spruce and displayed a DS of about 2 AU throughout the time of the reaction [55]. Woodward et al. [53] demonstrated synergism between cellulases with a final concentration of 20 µg/mL, consisting of 5 µg of EG II/mL, 10 µg of CBH I/mL, and 5 µg of CBH II/mL, and a fixed β-gl concentration of 1.08 units. The DS reached a peak at 2 AU and, at the same point, the soluble sugars reached the highest yield [53]. Bhat and Bhat [64] also suggested that both EGIII and EGV, sourced from Penicillium pinophilum, were required for the complete hydrolysis of crystalline cellulose because they increased the DS values and yields of soluble sugars during synergy with CBHI and CBHII. The DS appears to be time dependent, as was revealed in the study between CBHI (Cel7A) and EG (Cel7B), i.e., at 0–12 h, the enzyme combinations displayed the highest DS of about 2.35 AU, followed by a decrease to 1.6 AU at 12–24 h, and the DS values of 1.6 AU did not change significantly between 24 and 48 h [14]. In the same study, Pellegrini and collaborators demonstrated that the size of the biomass affects the DS between cellulases, because they only hydrolyzed the finer fibers of the filter paper and left the thick fibers untouched. Zhang and Zhang [5] also echoed the same views that the DS between enzymes, which results in higher yields of soluble sugars, is affected by the reaction conditions and the size of the fibers they hydrolyzed. Based on the above information, we propose that the fungal cellulolytic enzyme cocktails have various DS threshold values based on the different model substrates, i.e., 4–10 AU for bacterial cellulose and cotton (DP ≥ 2000) [10,44,78], 1.5–2.5 AU for Avicel/filter paper (DP ~300), and 1.3–15 AU for PASC/RAC (DP ~60) [10,11,14,15].
The above information relates to the fungal cellulolytic enzyme cocktails; however, it is clear that exoglucanase systems of bacterial organisms are different to those of fungal systems (as we proposed in exo–exo synergy; Section 3.2); for instance, Cel6A, Cel48A, Cel6B, and Cel9A sourced from Cellulomonas fimi were all processive on the different types of cellulose biomasses utilized, namely, PASC, Avicel, and crystalline celluloses Iα or III from green algae [22]. It is important to note that Cel6B and Cel9A are classified as endoglucanases, but Uchiyama et al. [22] demonstrated that these enzymes are arguably highly processive, and they used HS-AFM to show that the enzymes were able to move on crystalline cellulose III. Furthermore, combinations of TrCel7A and CfCel6B resulted in high yields of soluble sugars and synergism, while TrCel6A and CfCel6B resulted in anti-synergy. This observation suggests that CfCel6B is a CBH and not an EG, as it competes with CBHII (TrCel6A) for substrate binding sites. Interestingly, CfCel6B and CfCel5B demonstrated a DS of 2.16 AU and an improved yield of soluble sugar at a ratio of 3:1 when the enzyme loading was at a cellulose concentration of 2.5 mg/g [52]. However, when the enzyme concentration of CfCel6B and CfCel5B was increased to 10 mg/g cellulose, the DS values were decreased by 1 AU for all of the following combinations: 3:1, 1:1, and 1:3. The processive and non-processive EGs from GH family 9, and a CBHI from GH family 48, sourced from C. cellulosi CS-4-4, displayed synergy on filter paper when they were supplemented with β-gl from Caldicellulosiruptor sp. [49]. Various molar ratios were tested in this study, and the best combination for the formulated cellulase cocktail was 25:25:10:18 for CcCel9A: CcCel9B: CcCel48A: BlgA, which resulted in a DS of 2.6 and 1.86 mg/mL of glucose release. The literature contains more information regarding fungal cellulolytic enzyme cocktails, but, so far, only a few bacterial cellulolytic enzyme cocktails have been recorded. This suggests that there is still a lot that needs to be done with regard to bacterial cellulolytic enzymes, to establish if their synergy can be superior to that of fungi, or even commercial cocktails. We propose that a DS threshold above 1.5 AU can result in higher yields of soluble sugars for bacterial cellulolytic cocktails, but more studies are required to establish this claim.
3.6. Cellulase and LPMO Synergism
We have pointed out that some of the putative EGs that were initially deposited in GH family 61 were later moved to auxiliary activities (AA) family 9 in the CAZy database (http://www.cazy.org/; accessed on 30 July 2021). Thus, we decided to also investigate the role of AAs, particularly those from AA family 9, 10, and 16, in improving the DS during cellulose hydrolysis and the yield of soluble sugars. LPMOs (also known as AA9, AA10, or AA16) are auxiliary enzymes to cellulases, which cleave the cellulose chains via an oxidative mechanism [21,23,25]. Thus, the mechanism of substrate cleavage by LPMOs is different from cellulases classified under GH families, because they cleave the biomass via hydrolytic mechanisms [5,10]. The DS between these two classes of enzymes can reveal whether they cooperate or act individually during the hydrolysis of the cellulosic biomass.
Synergism between an LPMO and CBHI was demonstrated by core expression of the genes that encode for these proteins in the fungus (Penicillium funiculosum) that produced the LPMO naturally and five engineered strains [21]. It was evident that the secretome from the engineered strains (referred to as PfOAO1 and PfOA3) contained higher Avicelase activity, filter paper activity, and production of hydrogen peroxide, which suggest the presence of cellulases and LPMOs. In addition, the use of PfOAO1 and PfOA3 for the saccharification of acid-pretreated wheat straw increased the conversion to 80% and 75%, respectively. This suggested that the overexpression of CBHI and LPMO in PfOAO1 and PfOA3 increased the synergy between these enzymes. Interestingly, the individual application of three LPMOs from Thermoascus aurantiacus (TaAA9A), Lentinus similis (LsAA9A), and Thielavia terrestris (TtAA9E) on Avicel or PASC resulted in no release of detectable reducing sugars, according to Tokin et al. [37]. In contrast, the LPMOs synergized with TrCel7A and TrCel6A on Avicel or PASC, except for TrCel7A and TtAA9E, which displayed anti-synergy (more details regarding anti-synergy will be provided in the next section). The binary synergy between TrCel6A and TtAA9E resulted in the highest DS of about 2.5 and 2 AU during PASC and Avicel degradation, respectively. The high synergism between these enzymes can be explained by the fact that TtAA9E cleaves cellulose substrates by oxidizing the C1 of sugar moieties. TrCel6A also synergized with TaAA9A or LsAA9A, as their DS values were 2.0 and 1.8 AU on PASC, and 1.5 and 1.3 AU on Avicel, respectively [37]. The CBHI and TaAA9A or LsAA9A displayed a DS of 2.25 and 1.4 AU on PASC, or 1.8 and 1.5 AU, respectively [37]. EG (Cel7B) and MtLPMO9A synergy improved the hydrolysis of Avicel, bacterial cellulose, and sugarcane bagasse (SCB), which resulted in higher glucose production in the presence of Anβ-gl. The DS values between cellulases (TrCel7A, TrCel6A, and TrCel7B) and MtLPMO9A during the degradation of Avicel, BC, and SCB were 2.8, 2.5, and 2.6 AU, respectively. We propose that DS values greater than 1.5 AU should be considered as a threshold that results in higher yields of soluble sugars during the synergism between cellulases and LPMOs from AA9 (LPMO9A) during the degradation of PASC. However, DS values greater than 2 AU between cellulases and LPMO9A should be considered as a threshold that results in the improved degradation of crystalline substrates, such as Avicel. Unfortunately, to date (30 July 2021), the LPMOs from family 16 (AA16) are not yet well characterized, and only a few studies have been conducted; hence, we cannot conclusively provide predictions of the DS threshold [25].
Bacterial LPMOs that possess cellulolytic activity are generally classified under AA family 10 (http://www.cazy.org/AA10.html; accessed on 30 July 2021). Similarly, to the fungal LPMO system, which can oxidize C1 or C4, bacterial LPMOs can cleave glycosidic bonds by oxidizing C1 and C4 of the cellulose chain [23,79] (http://www.cazy.org/AA10.html; accessed on 30 July 2021). Forsberg et al. [79] demonstrated that Streptomyces coelicolor was equipped with LPMOs, which strictly oxidize C1 (ScLPMO10C, also known as CelS2), and a C1/C4 oxidizer (ScLPMO10B). Interestingly, ScLPMO10C and ScLPMO10B synergy improved the degradation of PASC. The enzyme combinations produced 140 µM glucose/gluconic acid compared to 20 µM glucose/gluconic acid produced by the individual enzymes, or 400 µM cellobiose compared to 100 µM produced by the individual enzymes. Even though Forsberg et al. [79] did not measure the DS values for synergism during these studies, our calculations from their glucose/gluconic acid data suggest that a DS value of about 2.3 AU was achieved. In addition, Tuveng et al. [26] also demonstrated that LPMO10 synergized with Cel48A and Cel6B, displaying a DS value of about 1.2 AU during Avicel hydrolysis. It is worth noting that, to date, most studies have focused on fungal cellulase—LPMO synergism—but few have focused on bacterial cellulase—LPMO synergism. Therefore, we cannot speculate on the DS threshold regarding bacterial cellulase—LPMO synergism. These observations suggest that more work still needs to be conducted to establish molecular interactions between bacterial cellulases and LPMOs, or fungal cellulases and the bacterial LPMO system.
4. Unraveling Cellulolytic Enzyme Anti-Synergism
There are the following three cases whereby cellulolytic enzymes do not synergize and the DS values are equal to, or less than, 1 AU (DS = 1 AU): (1) the first case is where the DS values are equal to 1 AU because the cellulases, or cellulases and LPMOs do not cooperate during the reaction, meaning that each enzyme is working on its own; (2) the DS values can be less than 1 AU because of cellulases, or cellulases and non-hydrolytic enzymes that compete for the same substrate binding sites, in order for enzymes to cleave the substrate; (3) the DS values can also be lower than, or equal to, 1 AU because the rate-limiting cellulases/LPMOs and the non-rate-limiting cellulases/LPMOs in the reaction display different rates of activity on their respective components of the substrate (e.g., binary synergism between processive EG and the CBHII). A detailed discussion on the factors that lead to reduced DS thresholds or synergism between cellulosic enzymes continues below, and a summary of this can be found in Table 2.

Table 2.
Factors resulting in anti-synergy between cellulolytic enzymes.
Sometimes the DS is lower than 1 AU, but the yield of soluble sugars produced by the combined cellulases, or cellulases and LPMOs, is higher compared to when the enzymes act individually. This type of anti-synergy occurs between cellulase enzymes such as CBHII (non-rate-limiting enzyme) and EG (rate-limiting enzymes) during the hydrolysis of the crystalline substrate. Anderson et al. [79] used H. insolens EG (Cel45A) and CBHII (Cel6) to show that the rate-limiting Cel45A converted about 1.7% of Avicel, while the non-rate-limiting Cel6A converted 0.1% of Avicel after 52 h. The combination of Cel6A and Cel45A displayed the highest DS of about 0.9 AU after 3 h and 0.5 AU after 52 h [79]. This study demonstrated that the endoglucanase was producing oligosaccharides at a higher rate than CBHII was able to convert the dextrans to cellobiose. The addition of P. brasillianum IBT 20888 β-gl (Cel3A) to a Cel6A/Cel45A mixture did not improve the conversion rate of Avicel (0.4%) or DS (0.5 AU), suggesting that the enzymes were not cooperating during Avicel hydrolysis. Liu et al. [52] demonstrated that the synergy between CBHII (Cel6B) and EGs is largely “endo-driven” because they are involved in creating shortened cellulose chains that can be hydrolyzed by the processive Cel6B. This study corroborates the work conducted by Anderson et al. [79], suggesting that EG is rate limiting, while CBHII is non-rate limiting during their synergic action on cellulose. Lastly, the EG created more sites that could be cleaved by CBHII on the cellulose III allomorph (similar to PASC) compared to Avicel, explaining why the EG and CBHII synergized on PASC, and did not synergize on Avicel [7,79].
Jalak et al. [16] demonstrated that during the hydrolysis of bacterial cellulose by the EG (TrCel5A) and CBHI (TrCel7A), the EG was the rate-limiting enzyme because it removed the amorphous regions on the bacterial cellulose substrate. The amorphous regions of the biomass led to CBHI stalling and becoming redundant; however, the removal of the amorphous regions by EG resulted in higher efficiency of CBHI [16]. A simulation study by Shang and Chu [80] also revealed that the cellulose hydrolyzed by EGs can contain uneven microfiber chains, which can stall CBHI activity, which results in anti-synergy. We argue that EG has the following two distinct functions as a cellulose hydrolysis rate-limiting enzyme during its synergism with CBHs: (1) EG produces the cleavage sites for CBHII on the microcrystalline substrates, and (2) EG removes the amorphous regions for CBHI. These observations suggest that EG is very important during cellulose hydrolysis because, as a rate-limiting enzyme, it determines or improves the efficiency of CBHs (Figure 2; Malgas et al. [3]). It is worth noting that EGI and EGII compete for the same cleavage sites on the cellulose biomass, which leads to inhibition (anti-synergy) towards Avicel hydrolysis [53,81]. Bhat and Bhat [64] reported that the enzyme EGII possesses a CBM, while EGI consists of only a CD. Therefore, the processive nature of EGII leads to anti-synergy due to a “traffic jam” at the site of cleavage between the two EGs. Additionally, Thoresen et al. [12] showed that enzyme cocktails formulated with CsCel48A, microbial Cel6A, and various endoglucanases from different GH families displayed anti-synergy, which resulted in reduced soluble sugars, glucose, and DS values below 1 AU.
In addition, Tokin et al. [37] and Mafa et al. [15] demonstrated that CBHI from GH7 could be inhibited/hindered by cellulolytic enzymes with the same regiospecificity, leading to DS values below 1 AU. The observed DS values below 1 AU were not only due to the same substrate binding sites, but because the enzyme mixture of the CBHI (GH7) and LPMO (AA9E), or CBHI (GH7) and exoglucanase (Exg-D: GH5_38) did not effectively hydrolyze the crystalline substrate (Avicel), and regenerated amorphous cellulose (RAC). To investigate this enzyme obstruction phenomenon, Mafa et al. [15] used a different substrate, β-(1,3-1,4)-mixed linked glucan (MLG), which could be hydrolyzed by CBHI (GH7), CBHII (GH6), and Exg-D (GH5_8). The enzyme mixture of the CBHI and Exg-D continued to show anti-synergy, with DS values below 1 AU. Even though the CBHII and Exg-D mixture did not show synergism on Avicel and RAC, the enzyme mixture showed synergism on MLG, suggesting that substrate chemistry and cleavage site availability were indeed the cause of the reduced synergism between CBHII and Exg-D. Therefore, it is apparent that the enzyme obstruction/inhibition on the same substrate cleavage site could lead to enzymes displaying anti-synergy, with DS values below 1 AU. Zhou et al. [56] also demonstrated that there is some competition between CBHI and LPMO sourced from Myceliophthora thermophila (MtLPMO9L), which belongs to C1-oxidizing LPMOs (Figure 3 and Table 2). Zhou and co-workers demonstrated this competition by performing sequential synergy, where the MtLPMO9L was applied on the substrate first, and, after 5 h, it was removed by centrifugation at 13,000× g; the remaining substrate was boiled to deactivate LPMO, and then 5 mg/mL CBHI was added to the reactions [56]. This approach did not only prevent competition between two enzymes, but it also resulted in a 1.6-fold higher yield during the hydrolysis of Avicel. The findings by Keller et al. [29] also echo the same anti-synergistic patterns between CBHI and LPMO (MtLPMO9A) that were reported by other studies [37,56]. Thus, we propose that 0.6 ≤ DS values < 1.0 demonstrates non-cooperation between enzymes (meaning that each cellulolytic enzyme works independently), while 0.5 ≥ DS values > 0 demonstrates competition of cellulolytic enzymes for the same binding or cleavage sites on the cellulose substrates.
5. Perspectives and Future Recommendations
Cellulolytic enzyme synergism is essential for the deconstruction or modification of cellulose substrates in the biorefinery sector. Hence, research on synergistic interactions has attracted extensive interest from all over the world over the past few decades. Cellulases were shown to be the main enzymes that degrade or modify cellulose substrates; however, an accumulation of information on the modes of action and mechanisms of expansins, SWOs and LPMOs has improved our understanding of cellulolytic enzyme synergism. Hence, we attempted to classify the various synergisms between non-cellulolytic active enzymes and cellulolytic active enzymes, and to theorize their respective DS thresholds. The LPMOs and cellulases, or expansins/SWOs and cellulases synergisms result in enhanced conversion rates of cellulose to soluble sugars, and aldoronic and gem-diol acids. Interestingly, to date, there have been no studies that have investigated the synergy between expansins/SWOs and cellulolytic LPMOs. This suggests that a knowledge gap exists in how expansins, or SWOs and LPMOs (AA9, AA10, and AA16) interact during biomass deconstruction. The enzyme cocktail(s) containing expansins/SWOs, cellulases, and LPMOs could perhaps be superior to the currently available commercial cellulolytic enzyme cocktails. It is vital to understand the different forms of synergy between the enzymes before formulating enzyme cocktails containing expansins/SWOs, cellulases, and LPMOs. This literature review explored the possible types of synergy and grouped them into two major classes. The first class of synergism resulted from non-hydrolytic and catalytically active cellulolytic enzyme interactions, and the second class resulted from the interaction of hydrolytic enzymes. It was clear that the enzymes displayed different interactions during exo–exo, endo–glucosidase, exo–endo, and cellulase–LPMO synergy, and that the varied DS values validated the proposal of different enzyme interactions. As a result of these various DS values, we have theorized DS value thresholds, which indicated the synergism between the cellulolytic enzymes that resulted in higher yields, as a benchmark for each type of synergy in Section 3 and Section 4. Lastly, based on the published data to date, we have also theorized the DS value thresholds for anti-synergism due to non-cooperating enzymes (1 ≥ DS values > 0.6), or competition for the binding/cleavage site (0.5 ≥ DS values > 0) of the cellulose substrates. This literature review can assist studies focusing on producing nano-microcrystalline cellulose through expansins/SWOs and endoglucanases, endo–endo, and endo–β-gl synergism. In addition, the theorized DS value thresholds can be applied to understand the level of synergism between cellulolytic enzymes during the formulation of enzyme cocktails.
We propose a new way of formulating cellulolytic enzyme cocktails effectively, as described below. Firstly, develop the fungal CBH core enzyme set (CES) between CBHI from GH family 7 and CBHII from GH families 5, 6, or 9; or develop the bacterial CBH–CES between CBHI from GH family 48 and CBHII from GH families 5, 6, or 9. It is important to note that CBHIs from family 7 and 48 must be kept separate at all times because their combination in the CES formulations leads to non-productive competition. Perhaps cues may also be found in the CAZy database, which showed that GH family 7 consists mostly of fungal enzymes, about 5220 entries, while the bacterial enzymes only make up 10 entries, and only four entries are unclassified (http://www.cazy.org/GH7_eukaryota.html; accessed on 31 July 2021). GH family 48 consists mostly of bacterial enzymes, constituting 1181 entries, followed by 34 fungal enzyme entries and 4 viral enzyme entries (http://www.cazy.org/GH48_eukaryota.html; accessed on 31 July 2021).
The best performing combinations of fungal or bacterial CBH-CES (supplemented with 10% constant β-gl, relative to CBHs concentrations) must be identified and used in combination with LPMOs to formulate a quaternary CES. This means that C1- and C4-oxidizing LPMOs (from AA9, AA10, or AA16) binary synergism must be established first via differential % mass protein loading or molar ratio. Therefore, the best CBH–CES and LPMO–CES can be mixed using different enzyme molar ratios to formulate a quaternary CES. Similarly, the binary or ternary synergism between EGs that are not processive from GH family 5, 6, 7, 9, 12, 44, 45, 51, and 74 must be formulated. We caution against the use of the processive EGs in cellulolytic enzyme cocktails intended to produce a high glucose yield because they compete with non-processive EGs for the cleavage sites on cellulose, resulting in “traffic jamming”. The best EG combinations for binary or ternary synergism must be used to formulate a quaternary or quinary synergy combination with LPMOs (C1- or/and C2-oxidizing enzymes). Finally, the CBH and LPMO–CES, and the EG and LPMOs can be used to formulate a cellulolytic enzyme cocktail that is effective, based on the theoretic concept that the cocktail will contain most of the enzyme required for effective cellulose hydrolysis. Expansin or SWO addition to the proposed cellulolytic enzyme cocktail must be at 1% and 10% of the total concentration because expansins, in particular, inhibit the current commercial cellulase cocktails at a higher concentrations.
Author Contributions
Conceptualization, M.S.M.; methodology, M.S.M., S.M. and B.I.P.; software, M.S.M. and S.M.; validation, M.S.M., S.M. and B.I.P.; writing—original draft preparation, M.S.M.; writing—review and editing, M.S.M., S.M. and B.I.P.; visualization, M.S.M., S.M. and B.I.P.; project administration, M.S.M.; funding acquisition, M.S.M. All authors have read and agreed to the published version of the manuscript.
Funding
MM received financial support from the Central Research Fund initiative (CRF) of the Natural and Agricultural Science Deans Office at University of the Free State (entity no 1-114-A5534). University of Pretoria and Rhodes University are gratefully acknowledged.
Institutional Review Board Statement
The current study did not use humans or animals.
Informed Consent Statement
The current study did not use humans or animals.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to reasons of privacy.
Acknowledgments
Any opinions, findings and conclusions or recommendations expressed in this material are those of the authors.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this review paper.
References
- McFarlane, H.E.; Doring, A.; Persson, S. The cell biology of cellulose synthesis. Annu. Rev. Plant Biol. 2014, 65, 69–94. [Google Scholar] [CrossRef] [PubMed]
- Coseri, S. Cellulose: To depolymerize… or not to? Biotechnol. Adv. 2017, 35, 251–266. [Google Scholar] [CrossRef] [PubMed]
- Malgas, S.; Rose, S.H.; van Zyl, W.H.; Pletschke, B.I. Enzymatic hydrolysis of softwood derived paper sludge by an in vitro recombinant cellulase cocktail for the production of fermentable sugars. Catalysts 2020, 10, 775. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Loosening of plant cell walls by expansins. Nature 2000, 407, 321–326. [Google Scholar] [CrossRef]
- Zhang, X.-Z.; Zhang, Y.-H.P. Cellulases: Characteristics, sources, production, and applications. In Bioprocessing Technologies in Biorefinery for Sustainable Production of Fuels, Chemicals, and Polymers; Yang, S.-T., El-Enshasy, H.A., Thongchul, N., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2013. [Google Scholar] [CrossRef]
- Jiang, F.; Ma, L.; Cai, R.; Ma, Q.; Guo, G.; Du, L.; Xiao, D. Efficient crude multi-enzyme produced by Trichoderma reesei using corncob for hydrolysis of lignocellulose. 3 Biotech. 2017, 7, 339. [Google Scholar] [CrossRef]
- Kuhad, R.C.; Deswal, D.; Sharma, S.; Bhattacharya, A.; Jain, K.K.; Kaur, A.; Pletschke, B.I.; Singh, A.; Karp, M. Revisiting cellulase production and redefining current strategies based on major challenges. Renew. Sustain. Energy Rev. 2016, 55, 249–272. [Google Scholar] [CrossRef]
- Thoresen, M.; Malgas, S.; Gandla, M.L.; Jönsson, L.J.; Sithole, B.; Pletschke, B.I. The effects of chemical and structural factors on the enzymatic saccharification of Eucalyptus sp. samples pre-treated by various technologies. Ind. Crop. Prod. 2021, 166, 113449. [Google Scholar] [CrossRef]
- Mafa, M.S.; Malgas, S.; Bhattacharya, A.; Rashamuse, K.; Pletschke, B.I. The effects of alkaline pretreatment on agricultural biomasses (corn cob and sweet sorghum bagasse) and their hydrolysis by a termite-derived enzyme cocktail. Agronomy 2020, 10, 1211. [Google Scholar] [CrossRef]
- Kostylev, M.; Wilson, D. Synergistic interactions in cellulose hydrolysis. Biofuels 2012, 3, 61–70. [Google Scholar] [CrossRef]
- Van Dyk, J.S.; Pletschke, B.I. A review of lignocellulose bioconversion using enzymatic hydrolysis and synergistic cooperation between enzymes-Factors affecting enzymes, conversion and synergy. Biotechnol. Adv. 2012, 30, 1458–1480. [Google Scholar] [CrossRef]
- Thoresen, M.; Malgas, S.; Mafa, M.S.; Pletschke, B.I. Revisiting the phenomenon of cellulase action: Not all endo-and exo-cellulase interactions are synergistic. Catalysts 2021, 11, 170. [Google Scholar] [CrossRef]
- Pletschke, B.; Malgas, S.; Bhattacharya, A.; Bhattacharya-Shrivastava, A.; Clarke, M.D.; Mafa, M.S.; Morake, S.; Thoresen, M. Enzyme synergism: A powerful tool for decreasing enzyme loading for efficient biomass conversion. In Proceedings of the 24th European Biomass Conference and Exhibition, Amsterdam, The Netherlands, 6–9 June 2016. [Google Scholar] [CrossRef]
- Pellegrini, V.O.; Bernardes, A.; Rezende, C.A.; Polikarpov, I. Cellulose fiber size defines efficiency of enzymatic hydrolysis and impacts degree of synergy between endo and exoglucanases. Cellulose 2018, 25, 1865–1881. [Google Scholar] [CrossRef]
- Mafa, M.S.; Malgas, S.; Rashamuse, K.; Pletschke, B.I. Delineating functional properties of a cello-oligosaccharide and β-glucan specific cellobiohydrolase (GH5_38): Its synergism with Cel6A and Cel7A for β-(1,3)-(1,4)-glucan degradation. Carbohydr. Res. 2020, 495, 108081. [Google Scholar] [CrossRef]
- Jalak, J.; Kurasin, M.; Teugjas, H.; Valjamae, P. Endo-exo synergism in cellulose hydrolysis revisited. J. Biol. Chem. 2012, 287, 28802–28815. [Google Scholar] [CrossRef]
- Kang, K.; Wang, S.; Lai, G.; Liu, G.; Xing, M. Characterization of a novel swollenin from Penicillium oxalicum in facilitating enzymatic saccharification of cellulose. BMC Biotech. 2013, 13, 42. [Google Scholar] [CrossRef] [PubMed]
- Bunterngsook, B.; Eurwilaichitr, L.; Thamchaipenet, A.; Champreda, V. Binding characteristics and synergistic effects of bacterial expansins on cellulosic and hemicellulosic substrates. Bioresour. Technol. 2015, 176, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Bunterngsook, B.; Mhuantong, W.; Champreda, V.; Thamchaipenet, A.; Eurwilaichitr, L. Identification of novel bacterial expansins and their synergistic actions on cellulose degradation. Bioresour. Technol. 2014, 159, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Badino, S.F.; Christensen, S.J.; Kari, J.; Windahl, M.S.; Hvidt, S.; Borch, K.; Westh, P. Exo-exo synergy between Cel6A and Cel7A from Hypocrea jecorina: Role of carbohydrate binding module and the endolytic character of the enzymes. Biotechnol. Bioeng. 2017, 114, 1639–1647. [Google Scholar] [CrossRef] [PubMed]
- Ogunyewo, O.A.; Randhawa, A.; Gupta, M.; Kaladhar, V.C.; Verma, P.K.; Yazdania, S.S. Synergistic Action of a Lytic Polysaccharide Monooxygenase and a Cellobiohydrolase from Penicillium funiculosum in Cellulose Saccharification under High-Level Substrate Loading. Appl. Environ. Microbiol. 2020, 86, e01769-20. [Google Scholar] [CrossRef]
- Uchiyama, T.; Uchihashi, T.; Nakamura, A.; Watanabe, H.; Kaneko, S.; Samejima, M.; Igarashi, K. Convergent evolution of processivity in bacterial and fungal cellulases. Proc. Natl. Acad. Sci. USA 2020, 117, 19896–19903. [Google Scholar] [CrossRef]
- Forsberg, Z.; Vaaje-Kolstad, G.; Westereng, B.; Bunæs, A.C.; Stenstrøm, Y.; MacKenzie, A.; Sørlie, M.; Horn, S.J.; Eijsink, V.G.H. Cleavage of cellulose by a CBM33 protein. Protein Sci. 2011, 20, 1479–1483. [Google Scholar] [CrossRef]
- Kadowaki, M.A.S.; Magri, S.; de Godoy, M.O.; Monclaro, A.V.; Zarattini, M.; Cannella, D. A fast and easy strategy for lytic polysaccharide monooxygenase-cleavable His6-Tag cloning, expression, and purification. Enzym. Microb. Technol. 2021, 143, 109704. [Google Scholar] [CrossRef] [PubMed]
- Filiatrault-Chastel, C.; Navarro, D.; Haon, M.; Grisel, S.; Herpoël-Gimbert, I.; Chevret, D.; Fanuel, M.; Henrissat, B.; Heiss-Blanquet, S.; Margeot, A.; et al. AA16, a new lytic polysaccharide monooxygenase family identified in fungal secretomes. Biotechnol. Biofuels 2019, 12, 55. [Google Scholar] [CrossRef]
- Tuveng, T.R.; Jensen, M.S.; Fredriksen, L.; Vaaje-Kolstad, G.; Eijsink, V.G.H.; Forsberg, Z. A thermostable bacterial lytic polysaccharide monooxygenase with high operational stability in a wide temperature range. Biotechnol. Biofuels 2020, 13, 194. [Google Scholar] [CrossRef] [PubMed]
- Isaksen, T.; Westereng, B.; Aachmann, F.L.; Agger, J.W.; Kracher, D.; Kittl, R.; Ludwig, R.; Haltrich, D.; Eijsink, V.G.H.; Horn, S.J. A c4-oxidizing lytic polysaccharide monooxygenase cleaving both cellulose and cello-oligosaccharides. J. Biol. Chem. 2014, 289, 2632–2642. [Google Scholar] [CrossRef] [PubMed]
- Vermaas, J.V.; Crowley, M.F.; Beckham, G.T.; Payne, C.M. Effects of lytic polysaccharide monooxygenase oxidation on cellulose structure and binding of oxidized cellulose oligomers to cellulases. J. Phys. Chem. B 2015, 119, 6129–6143. [Google Scholar] [CrossRef]
- Keller, N.B.; Badino, F.S.; Røjel, N.; Sørensen, T.H.; Kari, J.; McBrayer, B.; Borch, K.; Blossom, B.M.; Westh, P. A comparative biochemical investigation of the impeding effect of C1-oxidizing LPMOs on cellobiohydrolases. J. Biol. Chem. 2021, 296, 100504. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Lee, S.; Ko, H.; Kim, H.K.; Choi, I. An expansin-like protein from Hahella chejuensis binds cellulose and enhances cellulase activity. Mol. Cells 2010, 29, 379–385. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Su, R.; Duan, Y.; Cui, M.; Huang, R.; Qi, W.; He, Z.; Thielemans, W. Synergy between endo/exo-glucanases and expansin enhances enzyme adsorption and cellulose conversion. Carbohydr. Polym. 2021, 253, 117287. [Google Scholar] [CrossRef]
- Kim, I.J.; Lee, H.J.; Choi, I.; Kim, K.H. Synergistic proteins for the enhanced enzymatic hydrolysis of cellulose by cellulase. Appl. Microbiol. Biotechnol. 2014, 98, 8469–8480. [Google Scholar] [CrossRef] [PubMed]
- Saloheimo, M.; Paloheimo, M.; Hakola, S.; Pere, J.; Swanson, B.; Nyyssonen, E.; Bhatia, A.; Ward, M.; Penttila, M. Swollenin, a Trichoderma reesei protein with sequence similarity to the plant expansins, exhibits disruption activity on cellulosic materials. Eur. J. Biochem. 2002, 269, 4202–4211. [Google Scholar] [CrossRef] [PubMed]
- Malgas, S.; Thoresen, M.; van Dyk, J.S.; Pletschke, B.I. Time dependence of enzyme synergism during the degradation of model and natural lignocellulosic substrates. Enzym. Microb. Technol. 2017, 103, 1–11. [Google Scholar] [CrossRef]
- Zhou, Q.; Lv, X.; Zhang, X.; Meng, X.; Chen, G.; Liu, W. Evaluation of swollenin from Trichoderma pseudokoningii as a potential synergistic factor in the enzymatic hydrolysis of cellulose with low cellulase loadings. World J. Microbiol. Biotechnol. 2011, 27, 905–1910. [Google Scholar] [CrossRef]
- Malgas, S.; van Dyk, S.J.; Pletschke, B.I. β-Mannanase (Man26A) and α-galactosidase (Aga27A) synergism–a key factor for the hydrolysis of galactomannan substrates. Enzym. Microb. Technol. 2015, 70, 1–8. [Google Scholar] [CrossRef]
- Tokin, R.; Ipsen, J.O.; Westh, P.; Johansen, K.S. The synergy between LPMOs and cellulases in enzymatic saccharification of cellulose is both enzyme- and substrate-dependent. Biotechnol. Lett. 2020, 42, 1975–1984. [Google Scholar] [CrossRef]
- Andersen, N.; Johansen, K.S.; Michelsen, M.; Stenby, E.H.; Krogh, K.B.R.M.; Olsson, L. Hydrolysis of cellulose using mono-component enzymes shows synergy during hydrolysis of phosphoric acid swollen cellulose (PASC), but competition on Avicel. Enzym. Microb. Technol. 2008, 42, 362–370. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, Y.; Brunecky, R.; Yao, B.; Xie, X.; Zheng, F.; Luo, H. A swollenin from Talaromyces leycettanus JCM12802 enhances cellulase hydrolysis toward various substrates. Front. Microbiol. 2021, 12, 658096. [Google Scholar] [CrossRef]
- Liu, X.; Ma, T.; Zhang, M. Research advances in expansins and expansion-like proteins involved in lignocellulose degradation. Biotechnol. Lett. 2015, 37, 1541–1551. [Google Scholar] [CrossRef] [PubMed]
- Pech-Cervantes, A.A.; Ogunade, I.M.; Jiang, Y.; Muhammad Irfan, M.; Arriola, K.G.; Amaro, F.X.; Gonzalez, C.F.; Nicolas DiLorenzo, N.D.; Bromfield, J.J.; Vyas, D.; et al. An expansin-like protein expands forage cell walls and synergistically increases hydrolysis, digestibility and fermentation of livestock feeds by fibrolytic enzymes. PLoS ONE 2019, 14, e0224381. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.S.; Lee, H.J.; Bang, W.; Choi, I.; Kim, K.H. Functional characterization of a bacterial expansin from bacillus subtilis for enhanced enzymatic hydrolysis of cellulose. Biotechnol. Bioeng. 2009, 102, 1342–1353. [Google Scholar] [CrossRef]
- Santos, C.A.; Ferreira-Filho, J.A.; O’Donovan, A.; Gupta, V.K.; Maria, G.; Tuohy, M.G.; Souza, A.P. Production of a recombinant swollenin from Trichoderma harzianum in Escherichia coli and its potential synergistic role in biomass degradation. Microb. Cell Fact. 2017, 16, 83. [Google Scholar] [CrossRef]
- Boisset, C.; Fraschini, C.; Schulein, M.; Henrissat, B.; Chanzy, H. Imaging the enzymatic digestion of bacterial cellulose ribbons reveals the endo character of the cellobiohydrolase Cel6A from Humicola insolens and its mode of synergy with cellobiohydrolase Cel7A. Appl. Environ. Microbiol. 2000, 66, 1444–1452. [Google Scholar] [CrossRef] [PubMed]
- Boisset, C.; Petrequin, C.; Chanzy, H.; Henrissat, B.; Schulein, M. Optimized mixtures of recombinant Humicola insolens cellulases for the biodegradation of crystalline cellulose. Biotechnol. Bioeng. 2001, 72, 3. [Google Scholar] [CrossRef]
- Junior, A.T.; Dolce, L.G.; Neto, M.O.; Polikarpov, I. Xanthomonas campestris expansin-like X domain is a structurally disordered beta-sheet macromolecule capable of synergistically enhancing enzymatic efficiency of cellulose hydrolysis. Biotechnol. Lett. 2015, 37, 2419–2426. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-Z.; Sathitsuksanoh, N.; Zhang, Y.-H.P. Glycoside hydrolase family 9 processive endoglucanase from Clostridium phytofermentans: Heterologous expression, characterization, and synergy with family 48 cellobiohydrolase. Bioresour. Technol. 2010, 101, 5534–5538. [Google Scholar] [CrossRef]
- Tang, Z.; Li, P.; Sun, R.; Liu, M.; Jin, W.; Gou, L.; Chen, H.; Wu, Q.; Bu, T.; Li, C. Optimum mixture and synergy analysis of three main cellulases. Int. J. Agric. Biol. 2018, 20, 1814–9596. [Google Scholar]
- Yang, M.; Zhang, K.; Zhang, P.; Zhou, X.; Ma, X.; Li, F. Synergistic cellulose hydrolysis dominated by a multi-modular processive endo glucanase from Clostridium cellulosi. Front. Microbiol. 2016, 7, 932. [Google Scholar] [CrossRef]
- Zhou, S.; Ingram, L.O. Synergistic hydrolysis of carboxymethyl cellulose and acid-swollen cellulose by two endoglucanases (CelZ and CelY) from Erwinia chrysanthemi. J. Bacterial. 2000, 182, 5676–5682. [Google Scholar] [CrossRef] [PubMed]
- Cairo, J.P.L.F.; Leandro, C.; Oliveira, L.C.; Uchima, C.A.; Alvarez, T.M.; Citadini, A.P.S.; Leonardo, J.C.F.C.; Costa-Leonardo, A.M.; Carazzolle, M.F.; Costa, F.F.; et al. Deciphering the synergism of endogenous glycoside hydrolase families 1 and 9 from Coptotermes gestroi. Insect Biochem. Mol. Biol. 2013, 43, 970–981. [Google Scholar] [CrossRef]
- Liu, Y.; Nemmaru, B.; Chundawat, P.S. Thermobifida fusca cellulases exhibit increased endo−exo synergistic activity, but lower exocellulase activity, on cellulose-III. ACS Sustain. Chem. Eng. 2020, 8, 5028–5039. [Google Scholar] [CrossRef]
- Woodward, J.; Lima, M.; Lee, N.E. The role of cellulase concentration in determining the degree of synergism in the hydrolysis of microcrystalline cellulose. Biochem. J. 1988, 255, 895–899. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Wu, D.; Zhang, L.; Simmons, B.A.; Singh, S.; Yang, B.; Wyman, C.E. Dynamic changes of substrate reactivity and enzyme adsorption on partially hydrolyzed cellulose. Biotechnol. Bioeng. 2017, 114, 503–515. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, T.; Karlsson, J.; Tjerneld, F. A model explaining declining rate in hydrolysis of lignocellulose substrates with cellobiohydrolase I (Cel7A) and endoglucanase I (Cel7B) of Trichoderma rees. Appl. Biochem. Biotechnol. 2002, 101, 42–58. [Google Scholar] [CrossRef]
- Zhou, H.; Li, T.; Yua, Z.; Jua, J.; Zhanga, H.; Tan, H.; Li, K.; Yin, H. A lytic polysaccharide monooxygenase from Myceliophthora thermophila and its synergism with cellobiohydrolases in cellulose hydrolysis. Int. J. Biol. Macromol. 2019, 139, 570–576. [Google Scholar] [CrossRef] [PubMed]
- Karnaouri, A.; Muraleedharan, M.N.; Dimarogona, M.; Topakas, E.; Rova, U.; Sandgren, M.; Christakopoulos, P. Recombinant expression of thermostable processive MtEG5 endoglucanase and its synergism with MtLPMO from Myceliophthora thermophila during the hydrolysis of lignocellulosic substrates. Biotechnol. Biofuels 2017, 10, 126. [Google Scholar] [CrossRef] [PubMed]
- Zverlov, V.; Mah, S.; Riedel, K.; Bronnenmeier, K. Properties and gene structure of a bifunctional cellulolytic enzyme (CelA) from the extreme thermophile ‘Anaerocellum thermophilum’ with separate glycosyl hydrolase family 9 and 48 catalytic domains. Microbiology 1998, 144, 457–465. [Google Scholar] [CrossRef]
- Irwin, D.C.; Zhang, S.; Wilson, D.B. Cloning, expression and characterization of a Family 48 exocellulase, Cel48A, from Thermobifida fusca. Eur. J. Biochem. 2000, 267, 4988–4997. [Google Scholar] [CrossRef]
- Zhang, K.-D.; Li, W.; Wang, Y.-F.; Zheng, Y.-L.; Tan, F.-C.; Ma, X.-Q.; Yao, L.-S.; Bayer, E.A.; Wang, L.-S.; Li, F.-L.; et al. Processive degradation of crystalline cellulose by a multimodular endoglucanase via a wirewalking mode. Biomacromolecules 2018, 19, 1686–1696. [Google Scholar] [CrossRef]
- Foreman, P.K.; Brown, D.; Dankmeyer, L.; Dean, R.; Stephen Diener, S.; Dunn-Coleman, N.S.; Goedegebuur, F.; Houfek, T.D.; England, G.J.; Kelley, A.S.; et al. Transcriptional regulation of biomass-degrading enzymes in the filamentous fungus Trichoderma reesei. J. Biol. Chem. 2003, 278, 31988–31997. [Google Scholar] [CrossRef]
- Wood, T.M.; Mccrae, S.I.; Bhat, K.M. The mechanism of fungal cellulase action. Biochem. J. 1989, 260, 37–43. [Google Scholar] [CrossRef]
- Bhat, K.M.; Hay, A.J.; Claeyssens, M.; Wood, T.M. Study of the mode of action and site-specificity of the endo-(1 -4)-β-D-glucanases of the fungus Penicillium pinophilum with normal, 1-3H-labelled, reduced and chromogenic cello-oligosaccharides. Biochem. J. 1990, 266, 371–378. [Google Scholar] [CrossRef]
- Bhat, M.K.; Bhat, S. Cellulose degrading enzymes and their potential industrial applications. Biotechnol. Adv. 1997, 15, 583–620. [Google Scholar] [CrossRef]
- Segato, F.; Dias, B.; Bertoa, G.L.; de Oliveira, D.M.; De Souza, F.H.M.; Citadini, A.P.; Murakami, M.T.; Damásio, A.R.L.; Squina, F.M.; Polikarpova, I. Cloning, heterologous expression and biochemical characterization of a non-specific endoglucanase family 12 from Aspergillus terreus NIH2624. Biochim. Biophys. Acta 2017, 1865, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Takizawa, S.; Asano, R.; Fukuda, Y.; Feng, M.; Baba, Y.; Abe, K.; Tada, C.; Nakai, Y. Change of endoglucanase activity and rumen microbial community during biodegradation of cellulose using rumen microbiota. Front. Microbiol. 2020, 11, 603818. [Google Scholar] [CrossRef] [PubMed]
- Scapin, S.M.N.; Souza, F.H.M.; Zanphorlin, L.M.; de Almeida, T.S.; Sade, Y.B.; Cardoso, A.M.; Pinheiro, G.L.; Murakami, M.T. Structure, and function of a novel GH8 endoglucanase from the bacterial cellulose synthase complex of Raoultella ornithinolytica. PLoS ONE 2017, 12, e0176550. [Google Scholar] [CrossRef] [PubMed]
- Hakamada, Y.; Endo, K.; Takizawa, S.; Kobayashi, T.; Shirai, T.; Yamane, T.; Ito, S. Enzymatic properties, crystallization, and deduced amino acid sequence of an alkaline endoglucanase from Bacillus circulans. Biochim. Biophys. Acta 2002, 1570, 174–180. [Google Scholar] [CrossRef]
- Zainudin, M.H.M.; Mustapha, N.A.; Hassan, M.A.; Bahrin, E.K.; Tokura, M.; Yasueda, H.; Shirai, Y. A highly thermostable crude endoglucanase produced by a newly isolated Thermobifida fusca strain UPMC 901. Sci. Rep. 2019, 9, 13526. [Google Scholar] [CrossRef] [PubMed]
- Sadhu, S.; Maiti, T.K. Cellulase production by bacteria: A Review. Br. Microbiol. Res. J. 2013, 3, 235–258. [Google Scholar] [CrossRef]
- Carli, S.; Carneiro, A.L.C.B.; Ward, R.J.; Meleiro, L.P. Immobilization of a β-glucosidase and an endoglucanase in ferromagnetic nanoparticles: A study of synergistic effects. Protein Expr. Purif. 2019, 160, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, R.S.S.; da Silvaa, A.S.; Kime, J.-H.J.; Ishikawad, K.; Endod, T.; Leec, S.-H.; Bon, E.P.S. Combining biomass wet disk milling and endoglucanase/β-glucosidase hydrolysis for the production of cellulose nanocrystals. Carbohydr. Polym. 2015, 128, 75–81. [Google Scholar] [CrossRef]
- Jeon, E.; Hyeon, J.; Eun, L.S.; Park, B.-S.; Kim, S.W.; Lee, J.; Han, S.O. Cellulosic alcoholic fermentation using recombinant Saccharomyces cerevisiae engineered for the production of Clostridium cellulovorans endoglucanase and Saccharomycopsis fibuligera β-glucosidase. FEMS Microbiol. Lett. 2009, 301, 130–136. [Google Scholar] [CrossRef]
- Liu, M.; Gu, J.; Xie, W.; Yu, H. Directed co-evolution of an endoglucanase and a β-glucosidase in Escherichia coli by a novel high-throughput screening method. Chem. Commun. 2013, 49, 7219–7221. [Google Scholar] [CrossRef]
- Rosales-Calderon, O.; Trajano, H.L.; Duff, S.J.B. Stability of commercial glucanase and β-glucosidase preparations under hydrolysis conditions. PeerJ 2014, 2, e402. [Google Scholar] [CrossRef]
- Lutzen, N.W.; Nielsen, M.K.; Oxenboell, K.M.; Schtlein, M.; Stentebjerg-olesen, B. Cellulases and their application in the conversion of lignocellulose to fermentable sugars. Phil. Trans. R. Soc. Lond. B 1983, 300, 283–291. [Google Scholar]
- Oh, C.H.; Park, C.S.; Lee, Y.G.; Song, Y.; Bae, H.-J. Characterization of acidic endoglucanase Cel12A from Gloeophyllum trabeum and its synergistic effects on hydrogen peroxide–acetic acid (HPAC)-pretreated lignocellulose. J. Wood Sci. 2019, 65, 24. [Google Scholar] [CrossRef]
- Samejima, M.; Sugiyama, J.; Igarashi, K.; Eriksson, K.L. Enzymatic hydrolysis of bacterial cellulose. Carbohydr. Res. 1998, 305, 281–288. [Google Scholar] [CrossRef]
- Forsberg, Z.; Mackenziea, A.K.; Sørlie, M.; Røh, A.K.; Helland, R.; Arvai, A.S.; Gustav Vaaje-Kolstad, G.; Vincent, G.H.; Eijsink, V.G.H. Structural and functional characterization of a conserved pair of bacterial cellulose-oxidizing lytic polysaccharide monooxygenases. Proc. Natl. Acad. Sci. USA 2014, 111, 23. [Google Scholar] [CrossRef]
- Shang, B.Z.; Chu, J. Kinetic modeling at single-molecule resolution elucidates the mechanisms of cellulase synergy. ACS Catal. 2014, 4, 2216–2225. [Google Scholar] [CrossRef][Green Version]
- Buchert, J.; Heikinheimo, L. New cellulase processes for the textile industry. Carbohydr. Eur. 1998, 22, 32–34. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).