Computational-Designed Enzyme for β-Tyrosine Production in Lignin Valorization
Abstract
:1. Introduction
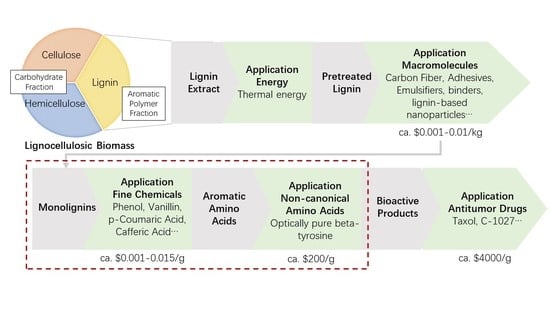
1.1. Lignin Valorization
1.2. β-Amino Acids and Anticancer Drugs
2. Results and Discussion
2.1. Computational Design of TchPAM for the Synthesis of β-tyrosine
2.2. Experimental Screening of the Designed TchPAM Mutants
2.3. Kinetic Analyses of Designed Enzymes
2.4. Enantioselectivity of Designed Enzymes
3. Conclusions and Perspectives
4. Materials and Methods
4.1. Computational Enzyme Design
4.2. Construction of TchPAM Mutants
4.3. Expression, Purification, and Concentration of TchPAM
4.4. TchPAM Activity Assays for Ammonia Elimination Reactions
4.5. Kinetics Measurements
4.6. HPLC Analysis in the Ammonia Addition Reaction
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Islam, M.R.; Hossain, M.E. Chapter 1—Introduction. In Drilling Engineering; Islam, M.R., Hossain, M.E., Eds.; Sustainable Oil and Gas Development Series; Gulf Professional Publishing: Houston, TX, USA, 2021; pp. 1–16. ISBN 978-0-12-820193-0. [Google Scholar]
- Ge, X.; Chang, C.; Zhang, L.; Cui, S.; Luo, X.; Hu, S.; Qin, Y.; Li, Y. Chapter Five—Conversion of Lignocellulosic Biomass Into Platform Chemicals for Biobased Polyurethane Application. In Advances in Bioenergy; Li, Y., Ge, X., Eds.; Elsevier: Amsterdam, The Netherlands, 2018; Volume 3, pp. 161–213. [Google Scholar]
- Yousuf, A.; Pirozzi, D.; Sannino, F. Chapter 1—Fundamentals of lignocellulosic biomass. In Lignocellulosic Biomass to Liquid Biofuels; Yousuf, A., Pirozzi, D., Sannino, F., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 1–15. ISBN 978-0-12-815936-1. [Google Scholar]
- Gutiérrez-Antonio, C.; Romero-Izquierdo, A.G.; Gómez-Castro, F.I.; Hernández, S. 5—Production processes from lignocellulosic feedstock. In Production Processes of Renewable Aviation Fuel; Gutiérrez-Antonio, C., Romero-Izquierdo, A.G., Gómez-Castro, F.I., Hernández, S., Eds.; Elsevier: Amsterdam, The Netherlands, 2021; pp. 129–169. ISBN 978-0-12-819719-6. [Google Scholar]
- Abdel-Hamid, A.M.; Solbiati, J.O.; Cann, I.K. Insights into lignin degradation and its potential industrial applications. Adv. Appl. Microbiol. 2013, 82, 1–28. [Google Scholar]
- Thomas, B.; Murphy, D.J.; Murray, B.G. Encyclopedia of Applied Plant Sciences; Academic Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Demuner, I.F.; Colodette, J.L.; Demuner, A.J.; Jardim, C.M. Biorefinery review: Wide-reaching products through kraft lignin. BioResources 2019, 14, 7543–7581. [Google Scholar] [CrossRef]
- Mahmood, N.; Yuan, Z.; Schmidt, J.; Xu, C. (Charles) Depolymerization of lignins and their applications for the preparation of polyols and rigid polyurethane foams: A review. Renew. Sustain. Energy Rev. 2016, 60, 317–329. [Google Scholar] [CrossRef]
- Bengtsson, A.; Bengtsson, J.; Sedin, M.; Sjöholm, E. Carbon Fibers from Lignin-Cellulose Precursors: Effect of Stabilization Conditions. ACS Sustain. Chem. Eng. 2019, 7, 8440–8448. [Google Scholar] [CrossRef] [Green Version]
- Ghaffar, S.H.; Fan, M. Lignin in straw and its applications as an adhesive. Int. J. Adhes. Adhes. 2014, 48, 92–101. [Google Scholar] [CrossRef]
- Yuliestyan, A.; García-Morales, M.; Moreno, E.; Carrera, V.; Partal, P. Assessment of modified lignin cationic emulsifier for bitumen emulsions used in road paving. Mater. Des. 2017, 131, 242–251. [Google Scholar] [CrossRef]
- Zhou, M.; Wang, D.; Peng, R.; Yang, D.; Qiu, X.; Qian, Y. Synthesis of a Hindered Amine-Grafted Lignin-Based Emulsifier and Its Application in a Green Emulsifiable Concentrate. J. Agric. Food Chem. 2019, 67, 11129–11136. [Google Scholar] [CrossRef]
- Gravitis, J.; Aboliņš, J.; Tupčiauskas, R.; Veveris, A. Lignin from steam-exploded wood as binder in wood composites. J. Environ. Eng. Landsc. Manag. 2010, 18, 75–84. [Google Scholar] [CrossRef]
- Xie, S.; Li, Q.; Karki, P.; Zhou, F.; Yuan, J.S. Lignin as Renewable and Superior Asphalt Binder Modifier. ACS Sustain. Chem. Eng. 2017, 5, 2817–2823. [Google Scholar] [CrossRef]
- Chauhan, P.S. Lignin nanoparticles: Eco-friendly and versatile tool for new era. Bioresour. Technol. Rep. 2020, 9, 100374. [Google Scholar] [CrossRef]
- Takada, M.; Rabemanolontsoa, H.; Minami, E.; Saka, S. Characterization of lignin-derived products from various lignocellulosics as treated by semi-flow hot-compressed water. J. Wood Sci. 2018, 64, 802–809. [Google Scholar] [CrossRef] [Green Version]
- Plesu Popescu, A.E.; Torralba, J.; Bonet, J.; Llorens, J. Vanillin production from lignin: Rigorous process simulation results for ethyl acetate versus aliphatic-alcohol-specific process designs. Clean. Eng. Technol. 2021, 4, 100133. [Google Scholar] [CrossRef]
- Beckham, G.T. Lignin Valorization: Emerging Approaches; Royal Society of Chemistry: London, UK, 2018. [Google Scholar]
- Becker, J.; Wittmann, C. A field of dreams: Lignin valorization into chemicals, materials, fuels, and health-care products. Biotechnol. Adv. 2019, 37, 107360. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Li, Y.; Zhang, W.; Li, S.; Gao, Y.; Ai, X.; Zhang, D.; Liu, B.; Li, Q. Metabolomics analysis reveals that elevated atmospheric CO2 alleviates drought stress in cucumber seedling leaves. Anal. Biochem. 2018, 559, 71–85. [Google Scholar] [CrossRef] [PubMed]
- Sarker, U.; Oba, S. Augmentation of leaf color parameters, pigments, vitamins, phenolic acids, flavonoids and antioxidant activity in selected Amaranthus tricolor under salinity stress. Sci. Rep. 2018, 8, 12349. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Yuan, B.; Huang, B. Differential heat-induced changes in phenolic acids associated with genotypic variations in heat tolerance for hard fescue. Crop Sci. 2019, 59, 667–674. [Google Scholar] [CrossRef]
- Baleroni, C.R.S.; Ferrarese, M.L.L.; Souza, N.E.; Ferrarese-Filho, O. Lipid accumulation during canola seed germination in response to cinnamic acid derivatives. Biol. Plant. 2000, 43, 313–316. [Google Scholar] [CrossRef]
- Timokhin, V.I.; Regner, M.; Motagamwala, A.H.; Sener, C.; Karlen, S.D.; Dumesic, J.A.; Ralph, J. Production of p-Coumaric Acid from Corn GVL-Lignin. ACS Sustain. Chem. Eng. 2020, 8, 17427–17438. [Google Scholar] [CrossRef]
- Marques, G.; Gutiérrez, A.; Del Río, J.C. Chemical characterization of lignin and lipophilic fractions from leaf fibers of curaua (Ananas erectifolius). J. Agric. Food Chem. 2007, 55, 1327–1336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- WHO. 2020. Available online: https://www.who.int/news-room/fact-sheets/detail/cancer (accessed on 30 September 2021).
- Christianson, C.V.; Montavon, T.J.; Van Lanen, S.G.; Shen, B.; Bruner, S.D. The structure of L-tyrosine 2,3-aminomutase from the C-1027 enediyne antitumor antibiotic biosynthetic pathway. Biochemistry 2007, 46, 7205–7214. [Google Scholar] [CrossRef]
- Malik, S.; Cusidó, R.M.; Mirjalili, M.H.; Moyano, E.; Palazón, J.; Bonfill, M. Production of the anticancer drug taxol in Taxus baccata suspension cultures: A review. Process Biochem. 2011, 46, 23–34. [Google Scholar] [CrossRef]
- Gond, S.K.; Kharwar, R.N.; White, J.F. Will fungi be the new source of the blockbuster drug taxol? Fungal Biol. Rev. 2014, 28, 77–84. [Google Scholar] [CrossRef]
- Baedeker, M.; Schulz, G.E. Autocatalytic peptide cyclization during chain folding of histidine ammonia-lyase. Structure 2002, 10, 61–67. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Murcia, P.A.; Bueren-Calabuig, J.A.; Camacho-Artacho, M.; Cortés-Cabrera, Á.; Gago, F. Stepwise Simulation of 3,5-Dihydro-5-methylidene-4H-imidazol-4-one (MIO) Biogenesis in Histidine Ammonia-lyase. Biochemistry 2016, 55, 5854–5864. [Google Scholar] [CrossRef] [Green Version]
- Sharma, A.; Shahzad, B.; Rehman, A.; Bhardwaj, R.; Landi, M.; Zheng, B. Response of phenylpropanoid pathway and the role of polyphenols in plants under abiotic stress. Molecules 2019, 24, 2452. [Google Scholar] [CrossRef] [Green Version]
- Wu, B.; Szymański, W.; Wybenga, G.G.; Heberling, M.M.; Bartsch, S.; Dewildeman, S.; Poelarends, G.J.; Feringa, B.L.; Dijkstra, B.W.; Janssen, D.B. Mechanism-inspired engineering of phenylalanine aminomutase for enhanced β-regioselective asymmetric amination of cinnamates. Angew. Chem. Int. Ed. 2012, 51, 482–486. [Google Scholar] [CrossRef]
- Wybenga, G.G.; Szymanski, W.; Wu, B.; Feringa, B.L.; Janssen, D.B.; Dijkstra, B.W. Structural investigations into the stereochemistry and activity of a phenylalanine-2,3-aminomutase from Taxus chinensis. Biochemistry 2014, 53, 3187–3198. [Google Scholar] [CrossRef] [PubMed]
- Tinberg, C.E.; Khare, S.D.; Dou, J.; Doyle, L.; Nelson, J.W.; Schena, A.; Jankowski, W.; Kalodimos, C.G.; Johnsson, K.; Stoddard, B.L.; et al. Computational design of ligand-binding proteins with high affinity and selectivity. Nature 2013, 501, 212–216. [Google Scholar] [CrossRef] [Green Version]
- Wu, B.; Szymanski, W.; Wietzes, P.; de Wildeman, S.; Poelarends, G.J.; Feringa, B.L.; Janssen, D.B. Enzymatic synthesis of enantiopure α- and β-amino acids by phenylalanine aminomutase-catalysed amination of cinnamic acid derivatives. ChemBioChem 2009, 10, 338–344. [Google Scholar] [CrossRef]
- Watts, K.T.; Mijts, B.N.; Lee, P.C.; Manning, A.J.; Schmidt-Dannert, C. Discovery of a Substrate Selectivity Switch in Tyrosine Ammonia-Lyase, a Member of the Aromatic Amino Acid Lyase Family. Chem. Biol. 2006, 13, 1317–1326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, Q.; Bourgeas, R.; Pucci, F.; Rooman, M. Computational analysis of the amino acid interactions that promote or decrease protein solubility. Sci. Rep. 2018, 8, 14661. [Google Scholar] [CrossRef]
- Burley, S.K.; Petsko, G.A. Amino-aromatic interactions in proteins. FEBS Lett. 1986, 203, 139–143. [Google Scholar]
- Lin, F.Y.; MacKerell, A.D. Improved Modeling of Cation-π and Anion-Ring Interactions Using the Drude Polarizable Empirical Force Field for Proteins. J. Comput. Chem. 2020, 41, 439–448. [Google Scholar] [CrossRef] [PubMed]
- Biot, C.; Buisine, E.; Kwasigroch, J.M.; Wintjens, R.; Rooman, M. Probing the energetic and structural role of amino acid/nucleobase cation-π interactions in protein-ligand complexes. J. Biol. Chem. 2002, 277, 40816–40822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Philip, V.; Harris, J.; Adams, R.; Nguyen, D.; Spiers, J.; Baudry, J.; Howell, E.E.; Hinde, R.J. A Survey of Aspartate À Phenylalanine and Glutamate À Phenylalanine. Biochemistry 2011, 50, 2939–2950. [Google Scholar] [CrossRef]
- McGaughey, G.B.; Gagné, M.; Rappé, A.K. π-Stacking interactions. Alive and well in proteins. J. Biol. Chem. 1998, 273, 15458–15463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janiak, C. A critical account on n-n stacking in metal complexes with aromatic nitrogen-containing ligands. J. Chem. Soc. Dalt. Trans. 2000, 3885–3896. [Google Scholar] [CrossRef]
- Feng, L.; Wanninayake, U.; Strom, S.; Geiger, J.; Walker, K.D. Mechanistic, mutational, and structural evaluation of a taxus phenylalanine aminomutase. Biochemistry 2011, 50, 2919–2930. [Google Scholar] [CrossRef]
- Tork, S.D.; Nagy, E.Z.A.; Cserepes, L.; Bordea, D.M.; Nagy, B.; Toşa, M.I.; Paizs, C.; Bencze, L.C. The production of l- and d-phenylalanines using engineered phenylalanine ammonia lyases from Petroselinum crispum. Sci. Rep. 2019, 9, 20123. [Google Scholar] [CrossRef]
- Rachid, S.; Krug, D.; Weissman, K.J.; Müller, R. Biosynthesis of (R)-β-tyrosine and its incorporation into the highly cytotoxic chondramides produced by Chondromyces crocatus. J. Biol. Chem. 2007, 282, 21810–21817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krug, D.; Müller, R. Discovery of additional members of the tyrosine aminomutase enzyme family and the mutational analysis of CmdF. ChemBioChem 2009, 10, 741–750. [Google Scholar] [CrossRef]
- Yan, J.; Aboshi, T.; Teraishi, M.; Strickler, S.R.; Spindel, J.E.; Tung, C.-W.; Takata, R.; Matsumoto, F.; Maesaka, Y.; McCouch, S.R.; et al. The Tyrosine Aminomutase TAM1 Is Required for β-Tyrosine Biosynthesis in Rice. Plant Cell 2015, 27, 1265–1278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deruiter, J. Carboxylic Acid Structure and Chemistry: Part 1. Princ. Drug Action 2005, 1, 1–11. [Google Scholar]
- Carretero-Molina, D.; Ortiz-Lopez, F.J.; Martín, J.; González, I.; Sánchez-Hidalgo, M.; Román-Hurtado, F.; Díaz, C.; de la Cruz, M.; Genilloud, O.; Reyes, F. Pentaminomycins F and G, first non-ribosomal peptides containing 2-pyridylalanine. J. Nat. Prod. 2021, 84, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Underlin, E.N.; Frommhagen, M.; Dilokpimol, A.; van Erven, G.; de Vries, R.P.; Kabel, M.A. Feruloyl Esterases for Biorefineries: Subfamily Classified Specificity for Natural Substrates. Front. Bioeng. Biotechnol. 2020, 8, 332. [Google Scholar] [CrossRef] [PubMed]
- Chaudhury, S.; Lyskov, S.; Gray, J.J. PyRosetta: A script-based interface for implementing molecular modeling algorithms using Rosetta. Bioinformatics 2010, 26, 689–691. [Google Scholar] [CrossRef]
- Boyle, N.M.O.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef] [Green Version]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 09, Revision A.02; Gaussian, Inc: Wallingford, CT, USA, 2016. [Google Scholar]
- BCL. Available online: http://www.meilerlab.org/index.php/servers/bcl-academic-license (accessed on 30 September 2021).
- Schrödinger, LLC. The {PyMOL} Molecular Graphics System, Version~1.8; Schrödinger, LLC: Portland, OR, USA, 2015. [Google Scholar]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Brückner, H.; Wittner, R.; Godel, H. Automated enantioseparation of amino acids by derivatization with o-phthaldialdehyde and n-acylated cysteines. J. Chromatogr. 1989, 476, 73–82. [Google Scholar] [CrossRef]







| (S)-α-Phenylalanine | |||
|---|---|---|---|
| TchPAM | KM (mM) | kcat (s−1) | kcat/KM (mM−1 s−1) |
| wt | 0.032 ± 0.001 | 0.02 ± 0.001 | 0.625 ± 0.02 |
| Y424C | 0.048 ± 0.006 | 0.024 ± 0.002 | 0.506 ± 0.041 |
| Y424D | n.d. | n.d. | n.d. |
| Y424N | 0.075 ± 0.001 | 0.03 ± 0.002 | 0.407 ± 0.019 |
| Y424Q | 0.006 1 | 0.003 1 | 0.608 ± 0.095 |
| (R)-β-Phenylalanine | |||
| TchPAM | KM (mM) | kcat (s−1) | kcat/KM (mM−1 s−1) |
| wt | 0.062 ± 0.012 | 0.026 ± 0.002 | 0.427 ± 0.078 |
| Y424C | 0.110 ± 0.017 | 0.035 ± 0.004 | 0.320 ± 0.044 |
| Y424D | 0.458 ± 0.091 | 0.024 ± 0.006 | 0.053 ± 0.006 |
| Y424N | 1.266 ± 0.108 | 0.138 ± 0.002 | 0.109 ± 0.01 |
| Y424Q | 0.422 ± 0.073 | 0.022 ± 0.005 | 0.053 ± 0.006 |
| (S)-α-Tyrosine | |||
| TchPAM | KM (mM) | kcat (s−1) | kcat/KM (mM−1 s−1) |
| wt | 2.435 ± 0.160 | 0.029 ± 0.005 | 0.011 ± 0.002 |
| Y424C | 0.347 ± 0.065 | 0.01 ± 0.002 | 0.03 ± 0.004 |
| Y424D | n.d. | n.d. | n.d. |
| Y424N | 0.330 ± 0.024 | 0.007 1 | 0.021 ± 0.001 |
| Y424Q | n.d. | n.d. | n.d. |
| (R)-β-Tyrosine | |||
| TchPAM | KM (mM) | kcat (s−1) | kcat/KM (mM−1 s−1) |
| wt | 0.465 ± 0.008 | 0.06 ± 0.001 | 0.130 ± 0.01 |
| Y424C | 0.299 ± 0.005 | 0.084 ± 0.007 | 0.281 ± 0.023 |
| Y424D | n.d. | n.d. | n.d. |
| Y424N | 0.480 ± 0.015 | 0.112 ± 0.003 | 0.234 ± 0.013 |
| Y424Q | 0.142 ± 0.001 | 0.014 ± 0.001 | 0.098 ± 0.008 |
| Enzyme | Specific Activity | eeβR | Reference | |||
|---|---|---|---|---|---|---|
| (S)-α- Tyrosine | (R)-α- Tyrosine | (S)-β- Tyrosine | (R)-β- Tyrosine | |||
| Wt | n.d. | n.d. | n.d. | n.d. | - | [1] |
| Y424N | n.d. | n.d. | n.d. | 0.104 ± 0.002 | >99% | This work |
| Y424C | n.d. | n.d. | n.d. | 0.141 ± 0.001 | >99% | This work |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, F.; Aliyu, H.; Delavault, A.; Engel, U.; Rudat, J. Computational-Designed Enzyme for β-Tyrosine Production in Lignin Valorization. Catalysts 2021, 11, 1310. https://doi.org/10.3390/catal11111310
Peng F, Aliyu H, Delavault A, Engel U, Rudat J. Computational-Designed Enzyme for β-Tyrosine Production in Lignin Valorization. Catalysts. 2021; 11(11):1310. https://doi.org/10.3390/catal11111310
Chicago/Turabian StylePeng, Fei, Habibu Aliyu, André Delavault, Ulrike Engel, and Jens Rudat. 2021. "Computational-Designed Enzyme for β-Tyrosine Production in Lignin Valorization" Catalysts 11, no. 11: 1310. https://doi.org/10.3390/catal11111310
APA StylePeng, F., Aliyu, H., Delavault, A., Engel, U., & Rudat, J. (2021). Computational-Designed Enzyme for β-Tyrosine Production in Lignin Valorization. Catalysts, 11(11), 1310. https://doi.org/10.3390/catal11111310