Abstract
Cutinases (EC 3.1.1.74) are serin esterases that belong to the / hydrolases superfamily and present in the Ser-His-Asp catalytic triad. They show characteristics between esterases and lipases. These enzymes hydrolyze esters and triacylglycerols and catalyze esterification and transesterification reactions. Cutinases are synthesize by plant pathogenic fungi, but some bacteria and plants have been found to produce cutinases as well. In nature they facilitate a pathogen’s invasion by hydrolyzing the cuticle that protects plants, but can be also used for saprophytic fungi as a way to nourish themselves. Cutinases can hydrolyze a wide range of substrates like esters, polyesters, triacylglycerols and waxes and that makes this enzyme very attractive for industrial purposes. This work discusses techniques of industrial interest such as immobilization and purification, as well as some of the most important uses of cutinases in industries.
1. Introduction
Aerial plant organs are protected by a cuticle composed of an insoluble polymeric structural compound, cutin, which is a polyester composed of hydroxy and hydroxyepoxy fatty acids. Cutinases are enzymes that catalyze the breakdown of those polyesters that form the cuticle that protects plants. These enzymes are synthesized by pathogenic fungi of plants, as well as by some bacteria and plants [1]. In addition to participating in the invasion of the host plant [2,3], cutinases also show other functions in fungal biology such as recognition of the surface where the spores land [4], the morphological development of fungi [5,6] or nutrition in saprophytic fungi [7,8].
In addition to natural polyesters, cutinases are able to hydrolyze synthetic esters, triacylglycerols and waxes, and to catalyze esterification and transesterification reactions [9]. This variety of substrates presented by cutinases makes them suitable for various applications by the textile, food and detergent industries, among others. This paper resumes the applications of cutinase in these industries, as well as several methods to improve the performance of cutinases at the industrial level.
2. Classification and Structure
Cutinases are serine esterases belonging to the superfamily of / hydrolases. The subfamily of cutinases currently consists of about 20 members, based on their amino acid sequence [10]. They are formed by simple polypeptides with a molecular weight of less than 40 kDa and their tertiary structure is formed by five chains and four helices (Figure 1).

Figure 1.
Diagram representing the folding of the cutinases. Five chains (enumerated from 3 to 7) and four helices (A, B, C, F) are shown. Near the C-terminal end is the active site formed by the catalytic triad Ser, Asp and His.
As a characteristic sign of the / hydrolases, the cutinases present the catalytic triad Ser-His-Asp, where the serine is exposed to the solvent and performs the hydrolysis of the substrate [9]. According to the International Union of Biochemistry and Molecular Biology they receive the EC number 3.1.1.74, which defines them as hydrolases that act on the cutin’s sour bonds. Brenda [11] classifies them as EC 3.1.1.74 (cutin hydrolases), as well as EC 3.1.1.3 (triacylglycerol lipases). Cutinases are often considered as intermediaries between lipases and esterases, since they show several characteristics belonging to both enzymes. Unlike lipases, which have a secondary structure or hydrophobic “cap” that covers the active site, cutinases do not have their catalytic esterase protected. Although a cutinase has recently been found with its active site protected by a “cap” [12]. In addition, high-resolution studies show that the active site of cutinases exhibits a certain degree of flexibility [13] and also speak of a “mini-cap” on the active site. These two characteristics can lead to a possible adaptation of the active site to different substrates and to large substrates such as cutin. Finally, the oxyanion hole is already formed in cutinases while in lipases it is formed once the substrate is bound [13,14]; this, together with the absence of a hydrophobic coverage in its active site, could be the reason why the cutinases do not demonstrate interfacial activation, i.e., being able to hydrolyze their dissolved substrate or form aggregates in an emulsion. Cutinases are able to hydrolyze a wide variety of substrates such as soluble low molecular weight esters, short and long chain triacylglycerols, natural and synthetic polyesters and waxes. It also catalyses esterification and transesterification reactions [9]. For an extensive structural analysis, we recommend the manuscript prepared by Nikolaivits et al. [15].
3. Evolutionary History
Regarding the study of the cutinase gene, several advances have recently been made in that direction. Skamnioti et al. [16] have performed a taxonomy of different genes for cutinases in five species of Ascomycete. Through the prediction of genes, 147 cutinase genes have been studied that show an evolutionary history of duplications and losses in the family of this gene, indicating a clear division between two ancestral families. Skamnioti et al. [16] associate this divergence to the duplication of an ancestral gene before the divergence between Basidiomycetes and Ascomycetes, since cutinases are found in almost all fungal subfamilies. In addition, the positions of the introns in the sequence are not shared through the most ancestral divergences and many sequences contain introns that are not found in others. These facts support the theory that these genes diverged very early and have diverged widely since then. Interestingly no cutinases have been found in any of the Saccharomycotina yeast species studied, this may indicate that evolution has not kept this protein in fungi non-parasites [16]. Another point to take into account is the transversal transference of the lateral gene transfer (LGT) of these sequences. All cutinase genes studied have a higher GC content compared to the genome that hosts them. This amount of GC can be an LGT test between pathogenic bacteria and fungi. However, the same authors affirm that the lateral transfer has not been a relevant event in the evolution of the gene family of the cutinase, since phylogenetic analyses place the bacterial cutinase in a different group from its fungal homologs. Although that same study shows that two of the fungal cutinases analyzed are more related to the cutinases of Mycobacteria than to those of their own group. On the other hand, Belbahri et al. [17] consider the LGT as a key event in the distribution of cutinases in different phylogenetic groups. Belbahri et al. [17] show that the Phytophthora cutinase sequence, an Oomycete, has a high homology with the sequence of the bacterial cutinase of Mycobacterium. In this way, they suggest the acquisition of this sequence via LGT and that this mechanism has played an important role in the evolution of this species, allowing them to colonize their host plants. In the evolutionary study of the cutinase, they performed bioinformatic and phylogenetic analyses on the known cutinase homologs. These homologs were found in Mycobacteria, fungi and Oomycetes. While the Mycobacteria, which belong to the Actinobacteria, are prokaryotes, fungi and Oomycetes are eukaryotes, the first grouping among the Opistokonta and the last among the Heterokonta. In addition, no homologs have been found between the Archaeas, between other groups belonging to the Opistokonta, nor in groups belonging to the Chromalveolata—the super-group where the Heterokonta are included. For Belbahri et al. [17] this indicates that the appearance of the sequence in these groups is of recent origin. Thus, as the existence of a common ancestor seems quite improbable, it suggests the recent appearance of the gene followed by a distribution of the same LGT pathway.
4. Biology
The role and importance of cutinases in pathogenicity is a controversial issue. On the one hand, there are studies that show an important role of these enzymes in the virulence of the pathogen [2,3]. On the other hand, organisms such as Magnaporthe grisea, which penetrates the cuticle through mechanical processes, and “oxide fungi” that enter their hosts through stomata [4], seem to indicate that the role of cutinases is not so decisive. That cutinase is used by pathogenic fungi seems to be a well-contrasted fact, since a large number of them produce extracellular cutinase in vitro [18] and cutinase has been found at the site of penetration of the fungus [19], but no results have been found that demonstrate a definitive role of cutinase in the penetration of the cuticle. Studies with deficient mutants for the cutinase in Fusarium solani pisi and Colletotrichum gloeosporioides [2,3], both ascomycetes, show that the virulence of the pathogen decreases when the enzyme is not available, but that it is re-established by adding exogenous cutinase. It has also been found that by providing Mycosphaerella spp., a parasitic fungus that infects papaya fruits only if they present a wound, the cutinase gene of F. solani pisi allows it to parasitize its host even without wounds [20]. Studies of disruption of the cutinase gene in Pieris brassicae, which generates a germinative tube that penetrates directly into the host, show that, although the germinative tube is generated in the same way, it is unable to penetrate the cuticle, producing subcuticular growth of the fungus as if it occurs in the wild variety [21]. In addition, it has been possible to prevent the infection of both F. solani pisi in peas and C. gloeosporioides in papaya, using cutinase antibodies in the first case [19] and with the inhibitor diisopropylfluorophosphate (DFP) in the second [22].
On the other hand, there are studies of genetic modification carried out in F. solani [23], M. grisea [24] and Botrytis cinerea [25] that are not so enlightening. Virulence does not decrease when cutinase is no longer involved. These results suggest that cutinase exerts additional functions in this process, beyond the hydrolysis of the cutin itself.
The cutinase also seems to have a role in recognizing the substrate on which the spores land, as well as guaranteeing the adhesion of these. Deising et al. [4] show that the spores of Uromyces viciae-fabae excrete cutinases and esterases that help in the formation of an adhesion-pad that allows the attachment of the spore to the substrate. They verified the importance of these enzymes when observing that autoclaved spores were fixed in smaller numbers and that this adhesion was recovered when washing with cutinases and esterases [4]. The studies carried out suggest that the cutinase excreted by the spores acts to recognize the surface in which they are found. The cutinases released by the spores release cutin monomers that induce the production of more quantity of enzyme, thus initiating the penetration process, since the cutin monomers act as inducers of the transcription of the cutinase itself, and glucose inhibits this transcription. The inhibition of this process, either by mutation of the gene, specific inhibition of the cutinase, or presumably, because the spore does not meet a valid objective, interrupts the penetration [26]. Cutinases also play a crucial role in the development of fungi. Erysiphe graminis conidia have been observed to excrete a protein mixture, including cutinase. Cutinase releases cutin monomers from the surface of host leaves. The importance of cutinase in the differentiation of these organs is evident when it is found that its formation in subjects treated with ebelactones A and B, inhibitors of the cutinase, is reduced by up to 50% [5]. In M. grisea, the cutinase is involved both in the formation of conidia and in that of the appressorial organ. Skamnioti and Gurr [6] showed that M. grisea mutants for the CUT2 gene have reduced their conidial formation by 52% and that they also produce appressoria in smaller numbers and with aberrant morphologies.
The number of genes that code for cutinase also provides us with valuable information about the biology of the fungus. Up to 13 cutinase genes have been identified in Curvularia lunata, which could indicate that this fungus is capable of degrading several types of cuticles, thus explaining the great variety of hosts that can parasitize, including are rice, strawberry and spinach, among others [27]. The same is found in M. grisea that hosts 17 putative genes that could be due to the organism’s need to colonize its hosts [16].
In other cases, it has been proposed that the function exerted by the cutinase is due to the ecological niche occupied by the fungus [28]. Therefore, fungi whose nutrition is fundamentally saprophytic, would need cutinase to take advantage of the cutin of the plant remains. This is shown by studies of gene disruption where the pathogenicity of the fungus is not altered by not having cutinase, but its growth is strongly inhibited when growing in media where cutin is its only carbon source since cutin is no longer accepted as carbon source [7,8]. It is important to say that fungi seem to make use of one type or another of cutinase according to their needs or their availability of nutrients. Alternaria brassicicola seems to alternate between two cutinases with different functions. In the first instance, the gene is induced by cutin monomers, induction that stops until at least 24 h, thus recognizing the cutin as a barrier to penetrate. If the pathogen manages to penetrate into its host the carbohydrates that this provides, they repress the transcription of more cutinase. On the other hand, if this repression does not occur within 24 h, the cutin monomers will stimulate the synthesis of more cutinase, thus recognizing the cutin as a carbon source [28].
5. Use of Biocatalysts in Industrial Production
Biocatalysts have been used as an alternative to chemical catalysts in the industry due to their enzymatic characteristics: a high reaction rate, catalytic activity in both directions, a good degree of selectivity against the type of reaction, high substrate specificity and high activity at room temperature and atmospheric pressure. In addition, the use of biocatalysts against chemical catalysts reduces the environmental impact and consumes less energy, which makes them economically more profitable. Even with this, biocatalysts are not widely used in the industry, since the enzymes are not stable at the high temperatures used, they are not reusable, it is difficult to maintain precise control of the reaction and, even with their advantages, they remain economically less profitable than chemical catalysts. However, there are several methods to improve the characteristics of enzymes, either by modifying the enzyme itself or by techniques that improve its performance at the industrial level [29].
5.1. Immobilization
One of the techniques employed is the immobilization of the biocatalyst in which the enzyme is fixed or confined in a defined region while maintaining its catalytic activity [30]. Several supports for the immobilization of cutinases have been studied, such as zeolites and supports of silica, alumina and polyamide. Among them, the NaY zeolite is the one that gives more stability to the cutinase [31,32,33,34]. Zeolites are microporous aluminosilicate minerals in which the aluminum is in a tetrahedral arrangement and contains one cation for each aluminum atom. These cations can be easily changed and have a great impact on the properties of zeolites [32]. Su et al. [30] used three well-characterized cutinases (Aspergillus oryzae Cutinase (AoC), Humicola insolens Cutinase (HiC) and Thielavia terrestris Cutinase (TtC)) using Lewatit VP OC 1600 as the macroporous support, describing immobilization yields of >98% (Table 1). The effect of the Si:Al ratio on the composition of the zeolite was also studied. The aluminum content determines the total load and the number of cations that the support will possess, and defines the affinity to water and the acidity or basicity of the structure. Zeolites with a Si:Al ratio have low enzymatic activities since zeolites with a high silica content contain less associated cations and are considered hydrophobic and not very basic. On the contrary, zeolites with a high content of alumina are considered as hydrophilic and basic. On the other hand, the Si:Al ratio does not seem to affect water activity. Apparently, there is a Si:Al ratio that is the optimum for the enzymatic activity, which corresponds approximately to that of the NaY zeolite (2.7), which, coincidentally, turns out to be one of the cheapest and available zeolites (Table 1) [32]. The effectiveness of the support for the reaction of alcoholysis of butyl acetate with hexanol to isoctane catalyzed by the cutinase of F. solani pisi was studied. The NaY zeolite obtained the highest catalytic activity compared to the other supports and maintains a large part of the activity of the cutinase at 150 C while in other supports it is deactivated [33]. Furthermore, it does not induce any conformational change in the enzyme and the pH value has less impact on the catalytic activity than in the other supports, although the optimum pH value is in any case about 7.2–8, a value close to the isoelectric point of cutinase, 7.9 [34]. The Fe-O magnetic nanoparticles have emerged as good solid support for the immobilization of enzymes owing to their desirable physico-chemical properties and several advantages viz. biocompatibility and high ratio of surface area:volume [35,36]. This results in high enzyme loading capacity with improved catalytic efficiency after immobilization, ease of surface modification, outstanding stability, improved mass transfer by the reduction in barriers to substrate diffusion, high dispersion, minor steric hindrance space and higher substrate affinity, and efficient separation of the magnetite loaded enzymes from the reaction mixtures simply by applying an external magnetic field and without any mechanical process like filtration or centrifugation [37].

Table 1.
Enzymatic activity, amount of water and water activity for zeolites A according to their associated cation. Higher enzymatic activity in bold letters.
5.2. Modification of Cutinases
The heterologous transformation can be used to make post-translational modifications that increase the stability and activity of the enzyme. By cloning the cutinase AnCUT2 from Aspergillus niger and expressing it in Pichia pastoris Al-Tammar et al. [38] verified that the glycosylation of cutinase by P. pastoris increased its molecular mass and modified the value of its isoelectric point allowing the enzyme to be stable over a wide range of pH.
Mutations have also been made in the cutinase gene of F. solani pisi to adapt the enzyme to what purpose. Araújo et al. [39] used site-directed mutagenesis to increase the size of the active site of the enzyme and thus accommodate larger substrates. Brissos et al. [40] searched from among a large number of mutated cutinases, generated by saturated mutagenesis, a variant that presented greater stability to the dioctyl sulfosuccinate sodium surfactant (DOSS) than the wild variety.
5.3. Industrial Applications of Cutinases
The generally recognized applications of cutinases are in the textile industry, in laundry detergents, in the processing of biomass and food, in biocatalysis and in detoxification of environmental pollutants [9,41,42].
5.3.1. Textile Industry
Cotton Fibers
Cotton fabric is the most popular textile product in the world. The cotton is washed in the industry for better wettability and thus facilitates a uniform stain and a better finish of the product. This wash, which removes the hydrophobic cuticle that covers the cotton, has been done by alkaline hydrolysis at high temperatures (100 C) using sodium hydroxide (NaOH), a method that requires large amounts of water and energy as well as damaging the fibers and generates large amounts of alkaline waste. Therefore, the use of biocatalysts, such as cutinases, cellulases and pectinases, has been proposed in order to reduce the environmental impact [9,43].
The hydrophobic cotton cuticle is composed of cutin, wax, pectin and protein. Efforts in research to achieve a cotton wash using biocatalysts or biowashing have focused on the use of enzymes, or sets of enzymes, that hydrolyze the cuticle components, avoiding the use of high temperatures and the waste generated in chemical washing [44,45]. Cellulases are the only enzymes that give good results when applied alone, although they reduce the strength of the fibers and therefore also their quality. On the other hand, the combined use of cellulases and proteases or pectinases, or lipases and pectinases give the best results in washing the cuticle. Even so, the use of boiling water is required to achieve an improvement in the wettability of the fibers [43].
For a while now, the value of cutinases in the washing of cotton has been studied, since they are capable of hydrolyzing both the cutin of cotton fibers and wax. Unfortunately, the wax can not be completely removed at low temperatures and high temperatures (80–90 C) are required in which most cutinases are not stable [9]. Although it has been possible to increase the stability of the cutinase in high temperatures by immobilizing the enzyme in zeolite NY [31,33], this material interferes between the cutinase and the wax of the fibers, so that immobilization is not a viable technique in this industry [9].
Washing with a cutinase of bacterial origin (Pseudomonas mandocino) has been achieved, with results similar to that of other enzymes (with a moistening time of 20–30 s) and without the use of boiling water [43]. The action of the cutinase in this process improves when adding detergents. When carrying out the process with a mixture of cutinase and pectinase they show a synergic effect, in which the cutinase would enlarge the pores of the cutin, and open new ones, allowing a rapid action of the pectinase, achieving a wetting time of 5 s. Studies have also been carried out to check the value of this enzyme in the washing of cotton. Fungal cutinases can eliminate the waxes of the cuticle of the cotton at low temperature(Agrawal et al. [44]). The degradation of the wax at 30 C was achieved with an incubation time of only 15–30 min, a much shorter time than that needed by Degani et al. [43] (10–20 h) (Figure 2). This reduction in incubation time could be explained by the different origins of the cutinase. To improve the action of pectinase in the process, it must penetrate the fibers and access the pectin. The surfactants and the elimination of the waxes fulfill this objective. For an optimal result, compatibility between the surfactant and the cutinase must be given. While the anionic surfactants negatively affect the action of the cutinase, the non-ionic surfactants do not interfere with the action of the enzymes. As the increase in the hydrophilicity of the fibers decreased the action of the cutinase, it was necessary to find a balance in which the increasing hydrophilicity of the fibers did not interfere with an optimum activity of the cutinase. Up to 70% of the original cutinase activity was retained for Triton X-100 values greater than 0.3 g L.

Figure 2.
Wetting times in seconds after treatment of cotton garments. DDW, control without enzymes; PL, pectinase alone; CUT, cutinase alone; CUT + PL, mixture of pectinase and cutinase. Adapted from [43].
When carrying out the process with cutinase, Triton X-100 and pectinase (also applying mechanical force with a wedge) the cotton was washed at 30 C in 15 min, so it can be affirmed that the cutinase can be used in the process of bio-washed by being able to remove cotton waxes at low-temperature [44]. The same author has managed to reduce the washing time using both mechanical strength [45] and ultrasound [46]. In the second case, the ultrasound intensifies the mass transfer and the kinetics of the enzymes, accelerating the washing process. Using a mixture of cutinase, pectinase, and Triton X-100 and applying ultrasound at a frequency of 30 kHz, the washing process was achieved in a cotton fabric of 280 g m in just 2–5 min.
Synthetic Fibers
Synthetic fibers are widely used in the textile industry (50% of the world market) due to their strength and elasticity. Textile products made with synthetic fibers shrink less, dry faster and can be mass-produced in a short time at a low cost. However, they have low permeability and evaporation of sweat due to their hydrophobicity, which also hinders the process of staining and finishing of fabric, making them uncomfortable to dress and difficult to wash [47,48]. Therefore, the treatment of this type of material is focused on reducing its hydrophobicity to improve its wettability, facilitating its staining and finishing. Traditionally this process has been carried out using strong chemical treatments with alkaline agents, which not only requires a large amount of energy and diminish the strength of the fibers, but also have a great environmental impact. The aim of the treatment of this type of fibers with enzymes is to modify the surface of the tissue without affecting its interior so as not to damage the tissue. On the other hand, cutinases are able to hydrolyze the surfaces of polyethylene terephthalate (PET), polyamide (PA) and acrylonitrile fibers or acrylic fibers [49].
Polyester Fabrics
PET is a polymer composed of terephthalic acid (TPA) and ethylene glycol. Cutinases from various organisms, such as F. solani pisi [47], Thermobifida halotoleran [50], T. fusca [51], Fusarium oxysporum [48] and Thermobifida cellulosilytica [52], can hydrolyze the ester bonds found on the surface of PET fibers, thus improving their hydrophobicity without compromising the integrity of the material [49]. As a result of hydrolysis, molecules of terephthalic acid, mono (2-hydroxyethyl)-terephthalate and bis (2-hydroxyethyl)-terephthalate are released [47].
Unlike the harsh conditions necessary for the chemical hydrolysis of PET, cutinases can perform this function in milder conditions. At 40 C the maximum activity is observed on the fiber esters, decaying at temperatures higher than 45 C due to the low stability of the enzyme. Regarding pH, the highest release of TPA, and therefore of hydrolysis of PET, was found at pH 8 (Figure 3).

Figure 3.
Effect of temperature and pH on the hydrolysis of PET by the cutinase of Fusarium oxysporum. Image extracted from [48].
If pH values are greater than 8, there is decreased activity of the enzyme. Alkaline pHs can increase the chemical hydrolysis of PET fibers producing the same tissue damage as chemical hydrolysis [48]. The disposition of the PET molecules also affects the effectiveness of the enzymatic activity on this, in such a way that the enzymes have a lower ability to hydrolyze crystallized substrates [53]. This phenomenon also happens for cutinases, which have greater catalytic activity on amorphous polyesters than on highly crystallized ones [47].
An attempt has also been made to increase the efficiency of cutinases in the hydrolysis of PET fibers. By means of site-directed mutagenesis, it was possible to increase the size of the active site of the cutinase of F. solani pisi, which improves the accommodation and stability of the large chains of PET esters, thus increasing the activity of the enzyme with respect to the native enzyme. Of the five mutant enzymes tested, the cutinase L182A was the one that obtained the best results, presenting a catalytic activity against the PET four times greater than the native enzyme and maintaining a 95% adsorption to the substrate [39].
It has also been possible to improve the hydrolysis of PET by the cutinase using hydrophobins [52]. The hydrolysis of the PET was measured (measuring the release of TPA and HET) in three different assays, cutinases and hydrophobins combined. An increase in the hydrolysis of the PET 16 times greater than the control (only cutinase) was obtained using the cutinases fused with hydrophobins. The other two trials also increased the efficacy of cutinase, although in a much more modest way. The results varied among the three hydrophobins used for each of the trials. The specific mechanism by which hydrophobins increase the activity of cutinases on PET is not known, but it may be due to its interaction with the substrate.
Polyamide Fabrics
Synthetic polyamide fabrics (nylon) offer abrasion resistance, low abrasion coefficient, good resistance to chemicals and firmness at high and low temperatures [54]. They are composed of hexanodioic acid and hexamethylenediamine bound by amide bonds. As with the fabrics seen above, the hydrophilicity of polyamide fabrics can be improved by alkaline treatments, which also damage this type of fibers and harms the environment [42]. Although the cutinase is an esterase, it is able to hydrolyze the polyamide, because it has a structure similar to cutin. Even so, it does so at a low rate, possibly due to the high crystallization of the polyamide structure [47,49,54]. However, it is more active against polyamides than against PET, releasing hexamethylenediamine molecules and amino groups during the process [49]. The cutinase has lower catalytic activity and adsorption against polyamide fabrics than against polyester fabrics. The cutinase of F. solani pisi mutant (L182A) of Araújo et al. [39] only increased the catalytic activity by 19% on the polyamide and decreased the adsorption by 5% with respect to the native enzyme. Silva et al. [54] also studied the effect of mechanical agitation on the hydrolysis of polyamides using the cutinase of F. solani pisi native and the mutant form L182A.
Wool Fabrics
The efficiency of the use of biocatalysts in the treatments to fabrics made from wool has also be investigated. The wool is covered by a cuticle composed of superimposed cells, in which the lipids connect cysteine residues by ester or thioester bonds, or form a network of isopeptides joined by amide bonds. This structure of scales and its composition gives a great hydrophobicity to wool, so it presents the same problems seen with the previous fabrics, in addition to a tendency to felting and shrinking, especially in aqueous medium and subject to mechanical forces as it happens in the washing processes [55].
The commercial method chlorination-Hercosett is used to avoid shrinkage of the fabric. Chlorinated chemicals are used to degrade cuticle scales, and then synthetic resins are applied to coat the fibers, resulting in washable fabrics. As a disadvantage, the process consumes large amounts of water, produces toxic residues and damages the fibers, making them difficult to stain and negatively affecting the touch [55]. The wool can also be pre-treated with alkaline substances, oxidation or chlorination to facilitate the penetration of proteases to the fibers and improve the hydrophilicity of the fabric. However, some of these methods cause excessive damage to the fibers and produce uneven treatment on the tissue surface [56].
Wang et al. [56] compared the effect of cutinases and hydrogen peroxide as pretreatment on the wettability of wool fibers. The use of cutinases managed to decrease the wettability time, from more than 30 min of the untreated control tissue to 7.9 min, reaching 1.3 min when proteases were subsequently applied. In contrast, hydrogen peroxide decreased this time to 21.2 min, 11.5 when applying proteases. To check the action of the proteases, and therefore the efficiency of the treatment to facilitate its penetration, the weight loss of the tissue is measured. Cutinases achieved a 6.3% weight loss, while hydrogen peroxide achieved 6.9%, approximately 3% more than the untreated control. This weight loss is usually accompanied by damage to the fibers, which can be checked by measuring the breaking stress (Figure 4).

Figure 4.
Loss of weight, breaking stress and shrinkage area for wool fabrics before (white bars) and after (gray bars). (a) control; (b) pretreated with cutinase; (c) pretreated with hydrogen peroxide. Adapted from [56].
For the fibers treated only with cutinase, no tensile stress values different to those obtained in the control were obtained, while the fibers treated with hydrogen peroxide showed a slightly lower value. The subsequent treatment with proteases did have an obvious effect on the fibers. The fibers pretreated with cutinases presented breaking stress slightly lower than the control fibers, while the loss of strength in those pretreated with hydrogen peroxide was more evident. This effect of the treatment of cutinases may be due to the fact that these increase the adsorption of the proteases to the tissue, resulting in a greater enzymatic action of these which would result in damage to the fibers. The treatment with cutinases shows promising results to avoid the shrinkage of wool tissues. The area of shrinkage of the tissue treated with cutinases was similar to that of the control tissue (about 10%), while that treated with hydrogen peroxide increased by 12%. However, after treatment with proteases, the area of shrinkage of the tissue pretreated with cutinases decreased up to 4%, below 6% of the control tissue, and of the sample pretreated with hydrogen peroxide.
5.3.2. Detergents
Enzymes have become a common addition to detergents, but they must be able to perform their catalytic activity under normal washing conditions. Therefore, the enzymes used as detergent components must be active in the alkaline pH range of the detergents, between 7 and 11, and at the temperatures at which the washings are carried out, from 4 to 60 C. They must also withstand washing times and detergent compositions, such as surfactants, bleaches and other enzymes, both during washing and while they are stored. In addition, the enzymes used must have sufficient substrate specificity to be able to act on the dirt in the multiple ways in which it can occur [57].
Hydrolases are added to the detergents to solubilize the stains and make it easier to remove them from the tissue. Proteases degrade protein spots in small fragments that are removed by the other components of the detergent, so they are often used to remove blood and grass spots. Amylases are applied to food stains such as potatos or chocolate since they degrade starch. Lipases are used to remove grease stains from clothes. Cellulases are used as a softener since their enzymatic activity degrades tissue microfibers that harden clothes without affecting it, and help to release stains trapped in the fabric and to revive the colors of the garments [57,58].
Cutinases are effective in the elimination of fat spots, since they are a type of lipase, and they are active both in solution and in water-oil interfaces [40]. Lipolase is a commercial lipase created by genetic engineering used as a detergent. Lipolase is calcium-dependent, which is a disadvantage since calcium chelators are added to the wash cycle because calcium produces stains that are difficult to remove from the garments (Figure 5).
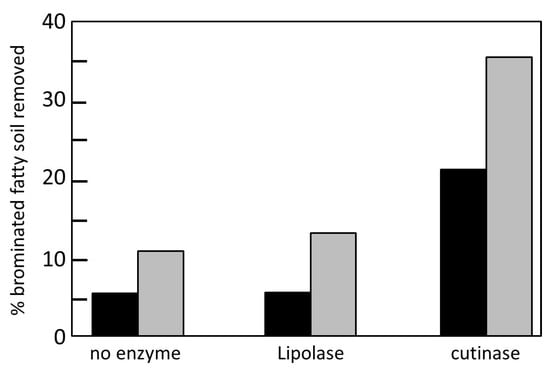
Figure 5.
Percentage of fatty spots eliminated by Lipolase and the cutinase of F. solani pisi using the sequential washing system. Black column: incubation 1; gray column: incubation 2. Adapted from [59].
Flipsen et al. [59] compared the effectiveness of the cutinase of F. solani pisi with that of Lipolase in the removal of olive oil stains on cotton and polyester garments. A sequential washing system was used, in which the enzymes were first incubated for 30 min and then a second incubation of another 30 min in the presence of detergent was carried out.
For Lipolase, no increase in the elimination of fats was observed with respect to the untreated control, due to the fact that the absence of calcium prevents the enzyme from carrying out its function. Although in the second phase the detergent containing calcium is added, the elimination of fats only increased by 2.7%. Flipsen et al. [59] managed to increase the elimination of these fats by performing an additional washing step.
5.3.3. Food Industry
Fruits and Vegetables
Fruits and vegetables are protected by a waterproof membrane that prevents evaporation of the liquids they contain. This membrane is composed of cutin, cellulose, pectin, wax and triacylglycerides, among other biomolecules. For the commercialization of these products it is necessary to increase their permeability to water to be able to dry them. Through chemical products such as methanol, chloromethanes, alkali hydroxides and others, this purpose can be achieved. However, the use of these products can leave traces in the food and its intake can have harmful effects, as well as being harmful to workers exposed to them. On the other hand, the treatment of waste generated by these chemicals is expensive [60].
Therefore, the use of enzymes to increase the permeability of fruits and vegetables is desirable, which also allows the classification of these products as natural by the food authorities [61]. The cutinases can be used to hydrolyze the materials that make up the cuticle of fruits and vegetables and, thus, improve their permeability to water. In this way, cutinases can be used to dehydrate these products for their preservation. In addition, drying some fruits and vegetables, such as raisins, plums, apricots and tomatoes, modifies their flavor and texture. Finally, the increase in permeability can be used for the administration of substances such as sweeteners, flavor enhancers or preservatives [60].
Aromas and Flavors
Short-chain esters (less than 10 carbon atoms) are very important compounds in the food industry because they have a pleasant fruit aroma and are used to give artificial flavors and aromas to foods and snacks. For example: ethyl butyrate is used as an artificial flavoring of strawberry, pineapple, apple and banana, and is added to candies, juices and pastries; Hexyl acetate is present in the aromas of fruits such as apple, apricot, pear and plum, and can be used as a fruit flavor enhancer in various soft drinks, candies and pastries. Furthermore, compounds such as ethyl acetate, hexanoate, butyl butyrate and octanoatealso are used to achieve fruity, herbal and butter aromas [62,63].
These aliphatic esters can be synthesized chemically by catalyzing the reaction between an alcohol and a carboxylic acid. This method is fast and cheap but, due to the strong acids and bases that are used as catalysts together with the high temperatures and pressures required, the resulting compounds cannot be classified as natural [62,63]. They can also be obtained naturally by extracting them directly from plants. However, in this way a complex mixture of compounds is obtained where the desired compound is in very low concentration and which changes during the ripening of the fruits, besides being subject to climatic conditions [62,63].
Therefore, the use of biocatalysts is very attractive, since more moderate conditions can be used, and the selectivity of the enzymes ensures the purity of the compounds. In this area lipases and esterases have been used for the synthesis of a wide variety of esters, although they show greater affinity towards long-chain (>C8) and short (C1 and C2) substrates than towards substrates of 3 to 8 atoms of carbon that are the most desirable [63].
Cutinases are able to catalyze the formation of esters from alcohols and fatty acids. De Barros et al. [61] studied the synthesis of ethyl hexanoate using the cutinase of F. solani pisi as a catalyst.
To study the effect of the concentration of the enzyme in the reaction, tests were carried out at 30 C with enzyme concentrations of 0.7 to 5.6 mg mL, at two different alcohol:acid ratios (1 and 2) with a constant alcohol concentration of 0.2 M. The highest production values were always given for the ratio 2, and the highest production, 97%, was given for an enzyme concentration of 2.8 mg mL. Using a temperature range of 25 to 50 C, the highest production of esters was obtained, 70%, at 30 C after an incubation of 10 h. The activity of the cutinase was also increased by increasing the value of the water activity (A), reaching a maximum between 0.68 and 0.75.
The concentrations of the substrates, acid and alcohol, also have an effect on the reaction, since the equilibrium of the reaction towards the synthesis of esters can be decanted. For equimolar concentrations of the substrates the highest production of esters was given, 77%, at a concentration 0.2 M. Concentrations greater than these resulted in a decrease in production. To study the effect of the acid concentration, the alcohol concentration was set at 0.2 M and acid concentrations of 0.025 to 0.25 M were tested. The highest production of esters, 88%, was given for an acid concentration of 0.1 M (alcohol: acid ratio 2), and the highest initial reaction rate (2.58 mol min mg) at 0.2 M (alcohol ratio:acid 2). Higher concentrations have inhibitory effects. The same method was used to check the effect of alcohol concentration. The highest initial velocity (2.91 mol min mg) was found at an ethanol concentration of 0.15 M, and an inhibitory effect of this velocity was 0.25M. No large changes were observed in the concentration of converted esters, which remained between 50 and 70%, between the 0.025 and 0.125 M ethanol concentrations (alcohol: acid ratio of 1.6 to 8). From 0.125 M ethanol production decreases to 48%.
Finally, the effect of the length of the acid chain was studied. Acids of chain length increasing from C2 to C18 were used (Table 2). It was observed that cutinase shows a great preference towards short chain acids, mainly towards butyric (C4) and valeric (C5) acids, since initial reaction rates of 1.15 and 1.06 min mg, respectively, were obtained. In contrast, for the acids of 8, 10 and 18 carbon atoms in length the reaction did not start until after 10, 20 and 40 min, respectively. In addition, in 6 h of incubation the highest production of esters was obtained, 96.5%, for valeric acid, while for butyric and caproic acids (C6) 84 and 62% were obtained, respectively. In contrast, with oleic acid (C18) 43% of production was obtained in 8 h of incubation [61].

Table 2.
Influence of the acid length on the initial rate of the esterification reaction. Note the delay time (lag) for the longest chain acids (C8, C10 and C18).
Recently Su et al. [63] studied the efficiency of T. fusca cutinase in the synthesis of ethyl hexanoate. They obtained a production of 99% using an enzyme concentration of 50 U mL and above. The highest initial velocity (0.14 M h) was obtained for an enzyme concentration of 300 U mL. A similar ester production was observed for temperatures between 30 and 50 C, with a peak of 99.2% at 40 C. At higher temperatures the production decreases. For acid and alcohol concentrations, the highest production value was obtained, 99.2%, for a 0.1 M acid concentration (ratio 2), and for a 0.1 M alcohol concentration (ratio 0.5) a maximum production of 98.3% was recorded.
The T. fusca cutinase has a greater affinity for short chain acids, as does the cutinase of F. solani pisi [61], although a greater selectivity of the substrate towards acids of longer chains is observed. The production was greater than 98% for acids with lengths between C3–C8, and the lowest productions were given for capric (C10) and lauric (C12) acids with 90.9 and 84.2% respectively. Regarding the length of the alcohols, values above 95% of production were obtained for alcohols with lengths between C1–C6. For alcohols greater than C6, the production decreased as the alcohol length increased (Figure 6).

Figure 6.
Effect of acid length on the synthesis of esters by the cutinase of F. solani pisi. White bars: alcohol: acid 2 ratio; black bars: ratio alcohol: acid 4. Image extracted from [61].
In addition to esters, milk fats are also of great importance in the food industry. The partially hydrolyzed milk fats are used to flavor cheese in a wide range of products such as cereals, sweets, dairy products, pastries, etc. [64,65]. The hydrolysis of these fatty acids can be done by the action of the enzymes of the native microbiome, as in the cheesemaking process, or by the addition of exogenous enzymes. Fats show different degrees of flavor depending on the degree and specificity of the hydrolysis, such as butter, cream or cheese flavor. Short-chain acids, such as butyric and caproic acids, are volatile and, in addition to imparting aroma, give pleasant flavors. In contrast, longer chain acids, between C10 and C18, give soapy and bitter flavors [64,65].
Normally animal lipases are suitable for the release of short chain fatty acids, as they are very specific for carbon atoms in positions 1 and 3 of fatty acids, unlike lipases of microbial origin that have a range of activity and a wider specificity. As the production of animal enzymes is very expensive, it is very desirable to use lipases of microbial origin that release short-chain fatty acids, since their production is simpler (Horri et al., 2010).
Regado et al. [64] compared the hydrolysis of fats from cow, goat and sheep milk, using 10 microbial lipases and the cutinase of F. solani pisi. The results of the hydrolysis using cutinase differed with respect to those of the other lipases. The proportion of short chain (C4–C8) and medium (C10–C14) acids released was significantly higher for cutinase than for the rest of lipases (Table 3). The levels of free butyric acid were between 16–17% and those of caproic acid between 13–14%. Furthermore, the release of C16 chain acids did not occur, and only a very low level of C18 chain acid was found, which agrees with the specificity of the cutinase studied by De Barros et al. [61]. Horri et al. [65] used the cutinase of Aspergillus oryzae, in its secreted form and on the cell surface of Saccharomyces cerevisiae, and verified its efficiency by hydrolyzing butter fats. They also compared the results of this cutinase with those of pancreatin, a lipase of commercial animal origin. After 96 h of incubation, the amount of fatty acids released by the secreted cutinase was 5.0%, 1.7 times higher than that obtained by pancreatin. In addition, the proportion of butyric acid in total fatty acids was 16.8%, a value comparable to that obtained with pancreatin. The value of fatty acids released using the cutinase expressed on the surface of S. cerevisiae was 4.2%, a value comparable to that obtained with the secreted cutinase. In contrast, a 45.5% proportion of butyric acid was obtained, 2.7 times higher than that of the secreted cutinase.

Table 3.
Proportions of fatty acids according to the length of their chain in lipolyzed milk fats of cow, sheep and goat. The results are shown as short chain (C4–C8), medium chain (C10–C14) and long chain (C16–C18).
Plastics
In recent years, important advances have been made in the study of the hydrolytic action of cutinases applied to the degradation of plastics [66]. It has been found that cutinase can biodegrade plastic and biomodify synthetic fibers. Cutinase is the most important enzyme in solving plastic pollution. Polyethylene terephthalate (PET) has a long half-life. This factor makes it a huge problem, and new technologies must be implemented to enable its treatment as urban solid waste. These enzymes have been successfully applied for the biodegradation of plastics, as well as for the delicate superficial hydrolysis of polymeric materials prior to their functionalization, as has been reported by Nikolaivits et al. [15]. Gomes et al. [67] characterized and improved the stability of the enzyme to optimize the degradation process by enzymatic hydrolysis of PET by recombinant cutinases. The objectives were met using the cutinase from Fusarium solani pisi and its C-terminal fusion with the cellulose-binding domain N1 of Cellulomonas fim, produced by genetically modified Escherichia coli. The maximum activity of cutinases produced in lactose broth in the presence of ampicillin and isopropyl -D-1-thiogalactopyranoside (IPTG) was 1.4 IU/mL. Both cutinases had an optimum pH around 7.0 and were stable between 30 and 50 C for 90 min.
6. Production of Cutinases
The production of cutinases has been achieved in bacteria, fungi and plants, the latter being the most complicated to work with since the cultivation and isolation of the enzyme on a large scale is easier to carry out with microorganisms (Table 4). Presumably, a long list of microorganisms can be added to this list, as indicated by the study by Belbahri et al. [17]. Fett et al. [68,69] have studied various cutin-producing microbial strains and the effect of several products as inductors or repressors of production. Among more than 38 bacterial strains studied Thermobifida fusca and Thermobifida vulgaris exhibited the highest production levels, with an esterase activity greater than 500 nmol min mL during 7 days of incubation, cultured in the liquid TYE enriched with a 0.4% (w/v) of apple cutin at pH 8.0. They verified that the skin, suberin and apple skin, tomato skin, potato suberin and commercial cork show an inducer effect in production, with the apple derivatives and the tomato skin being more efficient inducers. Most of these inducers are agricultural byproducts, so they have great potential as low cost inductors. In contrast, glucose suppresses the production of cutinase and olive oil and 16-hydroxypalmitic acid inhibits the bacterial growth of the producing strain as well as fiber and corn bran. Cutin hydrolyzate does not induce cutinase production.

Table 4.
Cutinases producing organisms.
Currently, cutinase is produced by Novozymes and Genencor companies. DE19706023 A1 (1998, BAYER AG; NOVOZYMES AS) discloses the use of a cutinase from Humicola insolens and lipases from Aspergillus niger, Mucor miehei(Lipozyme 20,000 L) and Candida antarctica (lipase component B) to degrade substrate polymers that are aliphatic polyesters, aromatic poly(ester amide)s, or partially aromatic poly(ester urethane)s [77,78]. WO9733001 A1 (1997, University of California; GENENCOR INT) and US6254645 B1 (2001, GENENCOR INT) describe the use of lipases or polyesterases to modify polyester fibers to enhance wettability and absorbency of textiles [79,80].
6.1. Purification of Cutinases
6.1.1. Purification by Ion Exchange Chromatography
It has been possible to purify cutinases using ion exchange chromatographies. Chen et al. [75] purified cutinases of Colletotrichum kahawaeand C. gloeosporioides by submitting them to ion exchange chromatography in two phases. The proteins were separated from the other components by ammonium sulfate and then subjected to precipitation by acetic acid. The set of proteins was then subjected to chromatography using two stationary phases in two steps. First a diethylaminoethyl matrix (DEAE), which functions as an anion exchanger, at pH 8.5, was used to separate the cutinases other 40-kDa carboxylases, since these bind to the matrix. Then another chromatography, using an SP (sulfopropyl) matrix, at pH 7.6, as a cation exchanger, manages to purify the cutinases of the remaining proteins. With these two chromatographies the purified cutinases should have a pI between 7.5 and 8.5, since they do not bind to any of the two matrices being at pHs 8.5 and 7.6, respectively.
6.1.2. Purification by Affinity Chromatography
Affinity chromatography is a very useful method for the purification of enzymes. In these chromatography the ligand forming the stationary phase is required to have a high affinity for the desired enzyme and which in turn forms a weak bond with it to be easily separated [81]. Although cutinases can be purified by this method, the results are not as good as one might wish [76].
Theorizing that, since 1,1,1-Trifluoro-3-((4-mercaptobutyl) thio)-2-propanone (MBTFP) is used for the purification of hormones esterases, which act as serine esterases and are members of the superfamily / hydrolase as well as cutinases, Wang G. et al. [76] purified Monilinia fructicola cutinases by affinity chromatography using MBTFP. Using the MBTFP as a stationary phase, they were able to bind 90.6% of the esterase activity present, thus highlighting the potential of this compound for the purification of cutinases.
For the washing of the column, two inhibitors of 1,1,1-trifluoro-2-propane (TFK), 3-n-octylthiio-1,1,1-trifluoro-2-propanone (OTFP) and 3-n-pentthio-1,1,1-trifluoro-2-propanone (PTFP) were considered. These compounds were used because TFK is a covalent inhibitor of serine esterases, and proteins bound to affinity matrices can be eluted using one of their inhibitors. When testing the ability of these compounds to inhibit the cutinase bound to the MBTFP, the OTFP was chosen to perform the column washes, due to the small differences between the I50 of PTFP and MBTFP for the cutinase of M. fructicola. The result of this chromatography is not a pure solution of cutinases, so the use of an electrophoresis with SDS-PAGE is required for the correct purification of the cutinase [76].
6.1.3. Purification by Means of Two Aqueous Phase Systems
An alternative to the use of chromatographies in the purification of proteins are the systems of two aqueous phases or aqueous two phase systems (ATPS). ATPS are formed by mixing two polymers or a polymer and a salt in a solution with a high water content. When adding proteins to the system, these will move towards one or another phase, and can easily separate them. This separation is influenced by the characteristics of the system (pH, temperature, type and concentration of salts and polymers), as well as by the characteristics of the proteins themselves (molecular weight, load and hydrophobicity) [82,83]. This system is useful for the separation of biological material due to its high water content and does not require specialized equipment for its operation, thus lowering costs [82]. Even so, ATPS are not used in the industry due to the scarce knowledge about how the separation process works and because its use is almost totally restricted to laboratory work scale and not to large-scale production [83].
Almeida et al. [82] studied the purification of cutinases using ATPS systems with polyethylene glycol (PEG) and purified hydroxypropyl starch (Reppal PES 100) and unrefined (HPS). They investigated how they affect the pH, type and concentration of the polymer and salts in these systems. In the PEG-hydroxypropyl systems in the absence of salts, the purification of cutinases was not satisfactory. The variations in the cutinase partition coefficient (cutinase activity ratio per milliliter between the two phases, K) were studied by modifying the pH and molecular mass values for the two system types (PEG-Reppal PES 100 and PEG-HPS). of the polymer. Although it was found that the mass of the polymer slightly affects the process and that pH values (8.0) close to the pI of the protein (7.8) increase the distribution of this to the upper phase, the results were not satisfactory. The distribution coefficients in each of the systems studied are between 0.5 and 1, values that show the low efficiency of these systems in the purification of cutinases, since the values should be greater than 1 to consider them satisfactory. The addition of salts to the system significantly improved the partition coefficient of the cutinase. The effect of the addition of sodium chloride (NaCl), ammonium sulfate ((NH)SO) and sodium sulfate (NaSO) was studied. For the PEG-Reppal system, the highest coefficient (K = 3.7) was obtained in the presence of ammonium sulfate (0.5 M) and pH 4.0, while for PEG-HPS the highest value (K = 12) was obtained with chloride. sodium (1 M) also at pH 4.0. These differences between the two systems are explained by Almeida et al. [82] because of the impurities presented by the unrefined HPS, because these could change the ionic strength thus altering the electrical potential between the phases. There were large differences in the distribution coefficients between the systems with and without salts at extreme pH values (4.0 and 9.0), but not at pH 8.0 since, being the cutinase pI of 7.8, it is less affected by the loads of salts.
7. Conclusions
Cutinases have characteristics that make them attractive for use in various industries. It has been seen that these enzymes can be useful in the textile industry. They can degrade cotton cuticle wax at low temperatures and in short times, making them viable for washing cotton. They can also be used for the hydrolysis of synthetic fiber fabrics, such as polyester and polyamide fabrics, thus improving their hydrophilicity and contributing to a better finish in this type of garments. Regarding the treatment of woolen fabrics, the cutinase achieves a wettability time less than that observed when treating these tissues with hydrogen peroxide, in addition it damages the fibers less and reduces the amount of tissue that shrinks a very small area. These results mean that cutinases should be considered as an interesting option in the textile area. On the other hand, the property of cutinases to be active in water-oil interfaces is very attractive for use as a detergent and is also active in the absence of calcium, which is an advantage in the current washing systems and make them more efficient than other enzymes already commercial. Regarding the food industry, the preference of cutinases towards short-chain alcohols and acids, as well as short chain triacylglycerols, make it more efficient than other enzymes in the production of aliphatic esters and partially hydrolyzed fats used in this sector.
The use of biocatalysts in the industry allows the use of smoother conditions and reduces the environmental impact produced by the chemicals normally used, which should be of economic benefit to the companies, since they have to manage the waste they generate and use large quantities of energy to achieve the conditions necessary for its production. Even so, this does not occur because the enzymes have several drawbacks, such as their instability at high temperatures, their low level of production and low reuse, among others, which make them unviable at industrial production levels. Therefore, it is necessary to research enzymes that present attractive characteristics for industrial production processes, as well as the development of techniques that improve the productivity of biocatalysts at this level. It is also important to continue investigating methods of immobilization that improve the performance of enzymes, as well as production methods that generate sufficient amounts of enzyme for these processes. The study of the structure of the cutinases, as well as studies of mutagenesis, can lead to modifications in the structure of this enzyme that improve its properties and make them more viable in the industry. Thus, it has been possible to find cutinase variants that are more stable in the presence of surfactants or with their wider active site, which makes them more attractive in their use as a detergent or in the treatment of polyester fibers, respectively.
Author Contributions
A.M. and S.M. wrote the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| LGT | Lateral gene transfer |
| DOSS | Dioctyl sulfosuccinate sodium surfactant |
| DEAE | Diethylaminoethyl |
| MBTFP | 1,1,1-Trifluoro-3-((4-mercaptobutyl) thio) -2-propanone |
| TFK | 1,1,1-Trifluoro-2-propane |
| OTFP | 3-n-Octylthiio-1,1,1-trifluoro-2-propanone |
| PTFP | 3-n-Pentthio-1,1,1-trifluoro-2-propanone |
| SDS-PAGE | Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis |
| ATPS | Aqueous two phase systems |
| PET | Polyethylene386terephthalate |
| PA | Polyamide |
| PEG | Polyethylene glycol |
References
- Dutta, K.; Sen, S.; Veeranki, V.D. Production characterization and applications of microbial cutinases. Process. Biochem. 2009, 44, 127–134. [Google Scholar] [CrossRef]
- Dantzig, A.H.; Zuckerman, S.H.; Andonov-Roland, M.M. Isolation of a Fusarium solani mutant reduced in cutinase activity and virulence. J. Bacteriol. 1986, 2, 911–916. [Google Scholar] [CrossRef]
- Dickman, M.B.; Patil, S.S. Cutinase deficient mutants of Colletotrichum gloeosporioides are nonpathogenic to papaya fruit. Physiol. Mol. Plant Pathol. 1986, 28, 235–242. [Google Scholar] [CrossRef]
- Deising, H.; Nicholson, R.L.; Haug, M.; Howard, R.J.; Mendgen, K. Adhesion pad formation and the involvement of cutinase and esterases in the attachment of uredospores to the host cuticle. Plant Cell 1992, 4, 1101–1111. [Google Scholar] [CrossRef]
- Francis, S.A.; Dewey, F.M.; Gurr, S. The role of cutinase in germling development and infection by Erysiphe graminis f. sp. hordei. Physiol. Mol. Plant Pathol. 1996, 49, 201–211. [Google Scholar] [CrossRef]
- Skamnioti, P.; Gurr, S.J. Magnaporthe grisea cutinase mediates appressorium differentiation and host penetration and is required for full virulence. Plant Cell 2007, 19, 2674–2689. [Google Scholar] [CrossRef] [PubMed]
- Stahl, D.J.; Schafer, W. Cutinase is not required for fungal pathogenicity on pea. Plant Cell 1992, 4, 621–629. [Google Scholar] [PubMed]
- Yao, C.; Koller, W. Diversity of cutinases from plant pathogenic fungi: Different cutinases are expressed during saprophytic and pathogenic stages of Alternaria brassicicola. Mol. Plant Microbe Interact. 1995, 8, 122–130. [Google Scholar] [CrossRef]
- Chen, S.; Su, L.; Chen, J.; Wu, J. Cutinase: Characteristics preparation, and application. Biotechnol. Adv. 2013, 31, 1754–1767. [Google Scholar] [CrossRef]
- Egmond, M.R.; de Vlieg, J. Fusarium solani pisi cutinase. Biochimie 2000, 82, 1015–1021. [Google Scholar] [CrossRef]
- Chang, A.; Jeske, L.; Ulbrich, S.; Hofmann, J.; Koblitz, J.; Schomburg, I.; Neumann-Schaal, M.; Jahn, D.; Schomburg, D. BRENDA, the ELIXIR core data resource in 2021: New developments and updates. Nucleic Acids Res. 2020, 49, 498–508. [Google Scholar] [CrossRef]
- Roussel, A.; Amara, S.; Nyyssölä, A.; Mateos-Diaz, E.; Blangy, S.; Kontkanen, H.; Westerholm-Parvinen, A.; Carrière, F.; Cambillau, C. A cutinase from Trichoderma reesei with a lid-covered active site and kinetic properties of true lipases. J. Mol. Biol. 2014, 426, 3757–3772. [Google Scholar] [CrossRef]
- Longhi, S.; Czjzek, M.; Lamzin, V.; Nicolas, A.; Cambillau, C. Atomic resolution (1.0 A) crystal structure of Fusarium solani. Stereochemical analysis. J. Mol. Biol. 1997, 268, 779–799. [Google Scholar] [CrossRef] [PubMed]
- Bornscheuer, U.T. Microbial carboxyl esterases: Classification, properties and application in biocatalysis. Fems Microbiol. Rev. 2002, 26, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Nikolaivits, E.; Kanelli, M.; Dimarogona, M.; Topakas, E. A middle-aged enzyme still in its prime: Recent advances in the field of cutinases. Catalysts 2018, 8, 612. [Google Scholar] [CrossRef]
- Skamnioti, P.; Furlong, R.F.; Gurr, S.J. Evolutionary history of the ancient cutinase family in five filamentous Ascomycetes reveals differential gene duplications and losses and in Magnaporthe grisea shows evidence of sub-and neo-functionalization. New Phytol. 2008, 180, 711–721. [Google Scholar] [CrossRef]
- Belbahri, L.; Calmin, G.; Mauch, F.; Andersson, J.O. Evolution of the cutinase gene family: Evidence for lateral gene transfer of a candidate phytophthora virulence factor. Gene 2008, 408, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Davies, K.; De Lorono, I.; Foster, S.; Li, D.; Johnstone, K.; Ashby, A. Evidence for a role of cutinase in pathogenicity of Pyrenopeziza brassicae on brassicas. Physiol. Mol. Plant Pathol. 2000, 57, 63–75. [Google Scholar] [CrossRef]
- Shaykh, M.; Soliday, C.; Kolattukudy, P.E. Proof for the production of cutinase byFusarium solani f. pisi during penetration into its host Pisum sativum. Plant Physiol. 1977, 60, 170–172. [Google Scholar] [CrossRef]
- Dickman, M.; Podila, G.; Kolattukudy, P. Insertion of cutinase gene into a wound pathogen enables it to infect intact host. Nature 1989, 342, 446–448. [Google Scholar] [CrossRef]
- Li, D.; Ashby, A.M.; Johnstone, K. Molecular evidence that the extracellular cutinase Pbc1 is required for pathogenicity of Pyrenopeziza brassicae on oilseed rape. Mol.-Plant-Microbe Interact. 2003, 16, 545–552. [Google Scholar] [CrossRef]
- Dickman, M.B.; Patil, S.S.; Kolattukudy, P. Purification, characterization and role in infection of an extracellular cutinolytic enzyme from Colletotrichum gloeosporioides penz on Carica papaya L. Physiol. Plant Pathol. 1982, 20, 333–347. [Google Scholar] [CrossRef]
- Stahl, D.J.; Theuerkauf, A.; Heitefuss, R.; Schafer, W. Cutinase of Nectria haematococca (Fusarium solani f. sp. pisi) is not required for fungal virulence or organ specificity on pea. Mol. Plant Microbe Interact. 1994, 7, 713–725. [Google Scholar] [CrossRef]
- Sweigard, J.A.; Chumley, F.G.; Valent, B. Disruption of a Magnaporthe grisea cutinase gene. Mol. Gen. Genet. 1992, 232, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Van Kan, J.; van’t Klooster, J.W.; Wagemakers, C.A.; Dees, D.C.; Van der Vlugt-Bergmans, C. Cutinase A of Botrytis cinerea is expressed, but not essential, during penetration of gerbera and tomato. Mol.-Plant-Microbe Interact. 1997, 10, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Köller, W.; Parker, D.; Becker, C. Role of cutinase in the penetration of apple leaves by Venturia inaequalis. Phytopathology 1991, 81, 1375–1379. [Google Scholar] [CrossRef]
- Liu, T.; Hou, J.; Wang, Y.; Jin, Y.; Borth, W.; Zhao, F.; Liu, Z.; Hu, J.; Zuo, Y. Genome-wide identification, classification and expression analysis in fungal-plant interactions of cutinase gene family and functional analysis of a putative ClCUT in Curvularia lunata. Mol. Genet. Genom. 2016, 7, 1–11. [Google Scholar] [CrossRef]
- Fan, C.; Köller, W. Diversity of cutinases from plant pathogenic fungi: Differential and sequential expression of cutinolytic esterases by Alternaria brassicicola. Fems Microbiol. Lett. 1998, 158, 33–38. [Google Scholar] [CrossRef]
- Arroyo, M.I.d.e. Fundamentos, métodos y aplicaciones. Ars Pharm. 1998, 39, 23–39. [Google Scholar]
- Su, A.; Shirke, A.; Baik, J.; Zou, Y.; Gross, R. Immobilized cutinases: Preparation, solvent tolerance and thermal stability. Enzym. Microb Technol. 2018, 116, 33–40. [Google Scholar] [CrossRef]
- Gonçalves, A.; Lopes, J.; Lemos, F.; Ribeiro, F.R.; Prazeres, D.; Cabral, J.; Aires-Barros, M.R. Effect of the immobilization support on the hydrolytic activity of a cutinase from Fusarium solani pisi. Enzym. Microb. Technol. 1997, 20, 93–101. [Google Scholar] [CrossRef]
- Serralha, F.; Lopes, J.; Lemos, F.; Prazeres, D.; Aires-Barros, M.; Cabral, J.; Aires-Barros, M.R. Zeolites as supports for an enzymatic alcoholysis reaction. J. Mol. Catal. B Enzym. 1998, 4, 303–311. [Google Scholar] [CrossRef]
- Serralha, F.; Lopes, J.; Aires-Barros, M.; Prazeres, D.; Cabral, J.; Lemos, F.; Ramôa Ribeiro, F. Stability of a recombinant cutinase immobilized on zeolites. Enzym. Microb. Technol. 2002, 31, 29–34. [Google Scholar] [CrossRef]
- Serralha, F.; Lopes, J.; Lemos, F.; Ribeiro, F.R.; Prazeres, D.; Aires-Barros, M.; Cabral, J.M.S. Application of factorial design to the study of an alcoholysis transformation promoted by cutinase immobilized on NaY zeolite and accurel PA6. J. Mol. Catal. B Enzym. 2004, 27, 19–27. [Google Scholar] [CrossRef]
- Bedade, D.K.; Muley, A.B.S.R.S. Magnetic cross-linked enzyme aggregates of acrylamidase from Cupriavidus oxalaticus ICTDB921 for biodegradation of acrylamide from industrial waste water. Bioresour. Technol. 2019, 272, 137–145. [Google Scholar] [CrossRef]
- Lucena, G.N.; dos Santos, C.; Pinto, G.; da Rocha, C.; Brandt, J.; de Paula, A.; Junior, M.; Marques, R. Magnetic cross-linked enzyme aggregates (MCLEAs) applied to biomass conversion. J. Solid State Chem. 2019, 10, 58–70. [Google Scholar] [CrossRef]
- Muley, A.B.; Thorat, A.S.; Singhal, R.S.; Babu, K.H. A tri-enzyme co-immobilized magnetic complex: Process details, kinetics, thermodynamics and applications. Int. J. Biol. Macromol. 2018, 118, 1781–1795. [Google Scholar] [CrossRef] [PubMed]
- Al-Tammar, A.; Omar, K.; Murad, O.; Munir, A.; Bakar, A.; Diba, A. Expression and and characterization of a cutinase (AnCUT2) from Aspergillus niger. Open Life Sci. 2016, 11, 29–38. [Google Scholar] [CrossRef][Green Version]
- Araujo, R.; Silva, C.; O’Neill, A.; Micaelo, N.; Guebitz, G.; Soares, C.M.; Casal, M.; Cavaco-Paulo, A. Tailoring cutinase activity towards polyethylene terephthalate and polyamide 6, 6 fibers. J. Biotechnol. 2007, 128, 849–857. [Google Scholar] [CrossRef]
- Brissos, V.; Eggert, T.; Cabral, J.M.; Jaeger, K.E. Improving activity and stability of cutinase towards the anionic detergent AOT by complete saturation mutagenesis. Protein Eng. Des. Sel. 2008, 21, 387–393. [Google Scholar] [CrossRef]
- Pio, T.; Macedo, G. Chapter 4 Cutinases: Properties and Industrial Applications. Adv. Appl. Microbiol. 2009, 66, 77–95. [Google Scholar] [CrossRef] [PubMed]
- Nyyssola, A. Which properties of cutinases are important for applications? Appl. Microbiol. Biotechnol. 2015, 99, 4931–4942. [Google Scholar] [CrossRef] [PubMed]
- Degani, O.; Gepstein, S.; Dosoretz, C.G. Potential use of cutinase in enzymatic scouring of cotton fiber cuticle. Appl. Biochem. Biotechnol. 2002, 102, 277–289. [Google Scholar] [CrossRef]
- Agrawal, P.; Agrawal, P.; Nierstrasz, V.; Bouwhuis, G.; Warmoeskerken, M. Biocatalysis and Biotransformation. Cutinase Pectinase Cotton Bioscouring Innov. Fast Bioscouring Process. 2008, 26, 412–421. [Google Scholar]
- Agrawal, P.; Nierstrasz, V.; Warmoeskerken, M. Role of mechanical action in low-temperature cotton scouring with F. solani pisicutinase and pectate lyase. Enzym. Microb. Technol. 2008, 42, 473–482. [Google Scholar] [CrossRef]
- Agrawal, P.; Nierstrasz, V.A.; Warmoeskerken, M. Ultrasound-boosted enzymatic cotton scouring process with cutinase and pectate lyase. Biocatal. Biotransform. 2010, 28, 320–328. [Google Scholar] [CrossRef]
- Vertommen, M.; Nierstrasz, V.; van der Veer, D.; Warmoeskerken, M.M.C.G. Enzymatic surface modification of poly (ethylene terephthalate). J. Biotechnol. 2005, 120, 376–386. [Google Scholar] [CrossRef]
- Kanelli, M.; Vasilakos, S.; Nikolaivits, E.; Ladas, S.; Christakopoulos, P.; Topakas, E. Surface modification of poly (ethylene terephthalate) (PET) fibers by a cutinase from Fusarium oxysporum. Process. Biochem. 2015, 50, 1885–1892. [Google Scholar] [CrossRef]
- Silva, C.M.; Carneiro, F.; O’Neill, A.; Fonseca, L.P.; Cabral, J.S.; Guebitz, G.; Cavaco-Paulo, A. Cutinase-A new tool for biomodification of synthetic fibers. J. Polym. Sci. Part Polym. Chem. 2005, 43, 2448–2450. [Google Scholar] [CrossRef]
- Ribitsch, D.; Herrero Acero, E.; Greimel, K.; Dellacher, A.; Zitzenbacher, S.; Marold, A.; Rodriguez, R.D.; Steinkellner, G.; Gruber, K.; Schwab, H.; et al. A new esterase from Thermobifida halotolerans hydrolyses polyethylene terephthalate (PET) and polylactic acid (PLA). Polymers 2012, 4, 617–629. [Google Scholar] [CrossRef]
- Oeser, T.; Wei, R.; Baumgarten, T.; Billig, S.; Föllner, C.; Zimmermann, W. High level expression of a hydrophobic poly (ethylene terephthalate)-hydrolyzing carboxylesterase fromThermobifida fusca KW3 in Escherichia coli BL21 (DE3). J. Biotechnol. 2010, 146, 100–104. [Google Scholar] [CrossRef]
- Ribitsch, D.; Herrero-Acero, E.; Prylucka, A.; Zitzenbacher, S.; Marold, A.; Gamerith, A.C.; Tscheließnig, R.; Jungbauer, A.; Rennhofer, H.; Lichtenegger, H.; et al. Enhanced cutinase-catalyzed hydrolysis of polyethylene terephthalate by covalent fusion to hydrophobins. Appl. Environ. Microbiol. 2015, 81, 3586–3592. [Google Scholar] [CrossRef]
- Donelli, I.; Taddei, P.; Smet, P.F.; Poelman, D.; Nierstrasz, V.A.; Freddi, G. Enzymatic surface modification and functionalization of PET: A water contact angle, FTIR, and fluorescence spectroscopy study. Biotechnol. Bioeng. 2009, 103, 845–856. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.; Araujo, R.; Casal, M.; Gübitz, G.M.; Cavaco-Paulo, A. Influence of mechanical agitation on cutinases and protease activity towards polyamide substrates. Enzym. Microb. Technol. 2007, 40, 1678–1685. [Google Scholar] [CrossRef]
- García, S.; Olivera, N. Oportunidades de innovación biotecnológica en la industria textil lanera. In Proceedings of the VII Congreso De Ingeniería Industrial y Afines, Chubut, Argentina, 2014. [Google Scholar]
- Wang, P.; Wang, Q.; Fan, X.; Cui, L.; Yuan, J.; Chen, S.; Wu, J. Effects of cutinase on the enzymatic shrink-resist finishing of wool fabrics. Enzym. Microb. Technol. 2009, 44, 302–308. [Google Scholar] [CrossRef]
- Aehle, W. Enzymes in Household Detergents. In Enzymes in Industry: Products and Applications; Aehle, W., Ed.; John Wiley & Sons: Hoboken, NJ, USA, 2006; pp. 154–177. [Google Scholar]
- Silva, C.; Azoia, N.G.; Martins, M.; Matamá, T.; Wu, J.; Cavaco-Paulo, A. Molecular recognition of esterase plays a major role on the removal of fatty soils during detergency. J. Biotechnol. 2012, 161, 228–234. [Google Scholar] [CrossRef] [PubMed]
- Flipsen, J.; Appel, A.; Van der Hijden, H.; Verrips, C. Mechanism of removal of immobilized triacylglycerol by lipolytic enzymes in a sequential laundry wash process. Enzym. Microb. Technol. 1998, 23, 274–280. [Google Scholar] [CrossRef]
- Poulose, A.J.; Boston, M. Enzyme Assisted Degradation of Surface Membranes of Harvested Fruits and Vegetables. U.S. Patent 5298265, 5 August 1991. [Google Scholar]
- De Barros, D.P.; Fonseca, L.P.; Fernandes, P.; Cabral, J.M.; Mojovic, L. Biosynthesis of ethyl caproate and other short ethyl esters catalyzed by cutinase in organic solvent. J. Mol. Catal. Enzym. 2009, 60, 178–185. [Google Scholar] [CrossRef]
- De Barros, D.P.; Azevedo, A.M.; Cabral, J.; Fonseca, L.P. Optimization of flavor esters synthesis by Fusarium solani pisi cutinase. J. Food Biochem. 2012, 36, 275–284. [Google Scholar] [CrossRef]
- Su, L.; Hong, R.; Guo, X.; Wu, J.; Xia, Y. Short-chain aliphatic ester synthesis using Thermobifida fusca cutinase. Food Chem. 2016, 206, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Regado, M.A.; Cristóvao, B.M.; Moutinho, C.G.; Balcao, V.M.; Aires-Barros, R.; Ferreira, J.P.M.; Malcata, F.X. Flavour development via lipolysis of milkfats: Changes in free fatty acid pool. Int. J. Food Sci. Technol. 2007, 42, 961–968. [Google Scholar] [CrossRef]
- Horii, K.; Adachi, T.; Tanino, T.; Tanaka, T.; Kotaka, A.; Sahara, H.; Hashimoto, T.; Kuratani, N.; Shibasaki, S.; Ogino, C.; et al. Fatty acid production from butter using novel cutinase-displaying yeast. Enzym. Microb. Technol. 2010, 46, 194–199. [Google Scholar] [CrossRef]
- Xu, Z. Research Progress on bacterial cutinases for plastic pollution. IOP Conf. Ser. Earth Environ. Sci. 2020, 450, 012077. [Google Scholar] [CrossRef]
- Gomes, D.S.; Matamá, T.; Cavaco-Paulo, A.; Campos-Takaki, G.M.; Salgueiro, A.A. Production of heterologous cutinases by E. coli and improved enzyme formulation for application on plastic degradation. Electron. J. Biotechnol. 2013, 16, 3. [Google Scholar] [CrossRef]
- Fett, W.; Wijey, C.; Moreau, R.; Osman, S. Production of cutinase by Thermomonospora fusca ATCC 27730. J. Appl. Microbiol. 1999, 86, 561–568. [Google Scholar] [CrossRef]
- Fett, W.; Wijey, C.; Moreau, R.; Osman, S. Production of cutinolytic esterase by filamentous bacteria. Lett. Appl. Microbiol. 2000, 31, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.S.; Kolattukudy, P.E. Structural studies on cutinase, a glycoprotein containing novel amino acids and glucuronic acid amide at the N terminus. Eur. J. Biochem. 1980, 106, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Fett, W.F.; Gerard, H.C.; Moreau, R.A.; Osman, S.F.; Jones, L.E. Cutinase production by Streptomyces spp. Curr. Microbiol. 1992, 25, 165–171. [Google Scholar] [CrossRef]
- Sebastian, J.; Kolattukudy, P.E. Purification and and characterization of cutinase from a fluorescent Pseudomonas putida bacterial strain isolated from phyllosphere. Arch Biochem. Biophys. 1988, 263, 77–85. [Google Scholar] [CrossRef]
- Kim, Y.H.; Lee, J.; Moon, S.H. Uniqueness of microbial cutinases in hydrolysis of p-nitrophenyl esters. J. Microbiol. Biotechnol. 2003, 13, 57–63. [Google Scholar]
- Hijden, V.; Htjd, M.; Warr, J.F.; Klugkist, J.; Musters, W.; DHA, H. Enzyme-Containing Surfactants Compositions. Unilever Patent WO943578.94.2, 1994. [Google Scholar]
- Chen, Z.; Franco, C.F.; Baptista, R.P.; Cabral, J.M.; Coelho, A.V.; Rodrigues, C.J.; Melo, E.P. Purification and identification of cutinases from Colletotrichum kahawae and Colletotrichum gloeosporioides. Appl. Microbiol. Biotechnol. 2007, 73, 1306–1313. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Michailides, T.J.; Hammock, B.D.; Lee, Y.; Bostock, R.M. Affinity purification and characterization of a cutinase from the fungal plant pathogen Monilinia fructicola (Wint.) Honey. Arch. Biochem. Biophys. 2000, 382, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Villalba, M.; Verdasco-Martín, C.M.; dos Santos, J.C.; Fernandez-Lafuente, R.; Otero, C. Operational stabilities of different chemical derivatives of Novozym 435 in an alcoholysis reaction. Enzym. Microb. Technol. 2016, 90, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Feder, D.; Gross, R. Exploring chain length selectivity in HIC-catalyzed polycondensation reactions. Biomacromolecules 2010, 3, 690–697. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Lee, J.; Seung-Hyeon, M. Increasing Pharmacological Effect of Agricultural Chemicals. U.S. Patent 88-08945, 1988. [Google Scholar]
- Kolattukudy, P.; Poulose, A.J. Cutinase for Use in Detergent Composition Produced by Culturing Pseudomonas Putida. U.S. Patent 89-02922, 1989. [Google Scholar]
- Voet, D.P.; Voet, J.G.; Pratt, C.W. Purificación y análisis de proteínas. In Fundamentos de Bioquímica: La Vida a Nivel Molecular; Editorial Médica Panamericana: Madrid, Spain, 2007; pp. 97–107. [Google Scholar]
- Almeida, M.; Venancio, A.; Teixeira, J.; Aires-Barros, M. Cutinase purification on poly (ethylene glycol)-hydroxypropyl starch aqueous two-phase systems. J. Chromatogr. Biomed. Sci. Appl. 1998, 711, 151–159. [Google Scholar] [CrossRef]
- Rito-Palomares, M. Practical application of aqueous two-phase partition to process development for the recovery of biological products. J. Chromatogr. B 2004, 807, 3–11. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).