Structure Prediction of a Thermostable SR74 α-Amylase from Geobacillus stearothermophilus Expressed in CTG-Clade Yeast Meyerozyma guilliermondii Strain SO
Abstract
1. Introduction
2. Results and Discussion
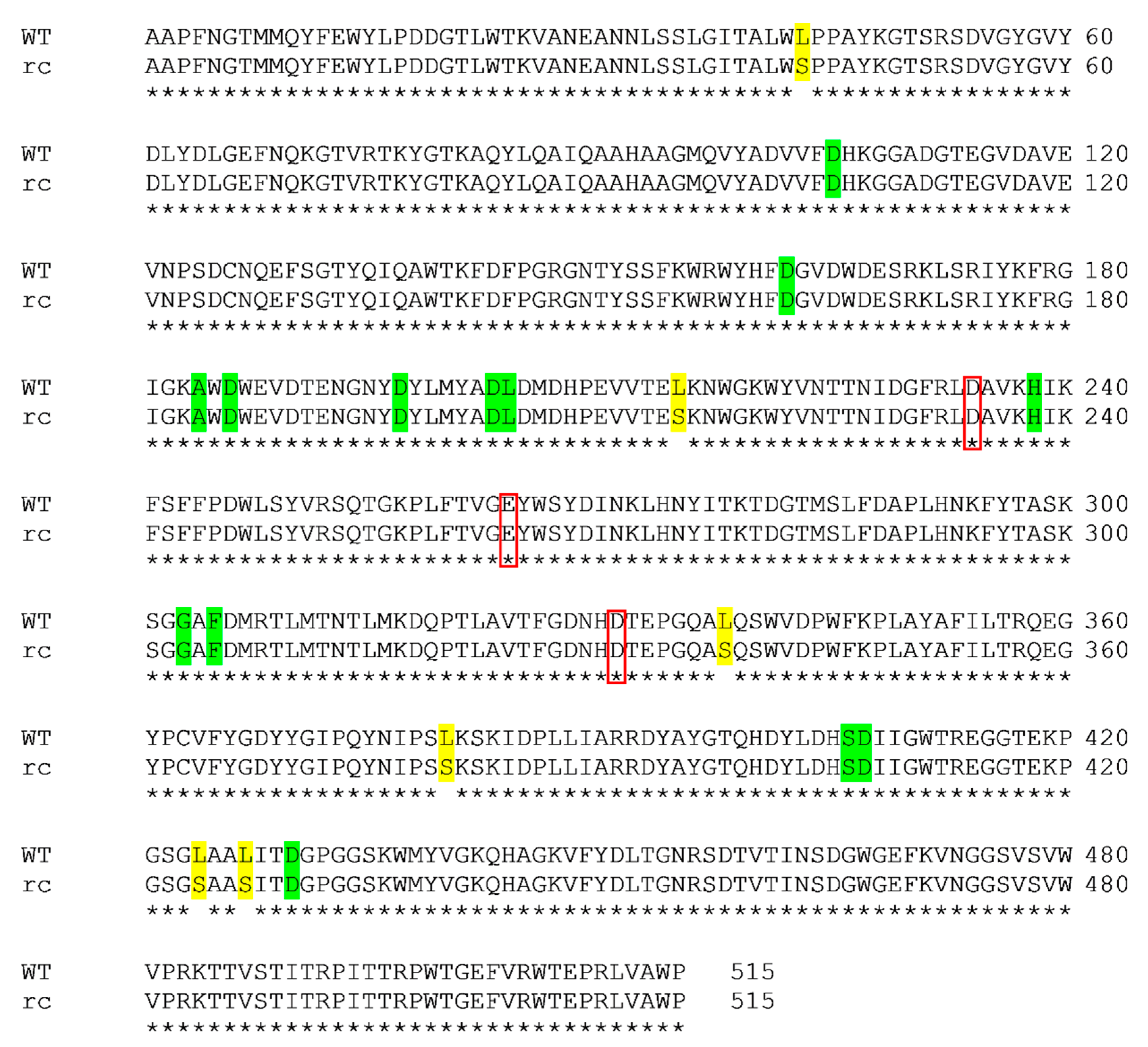
2.1. Structure and Sequence Analysis
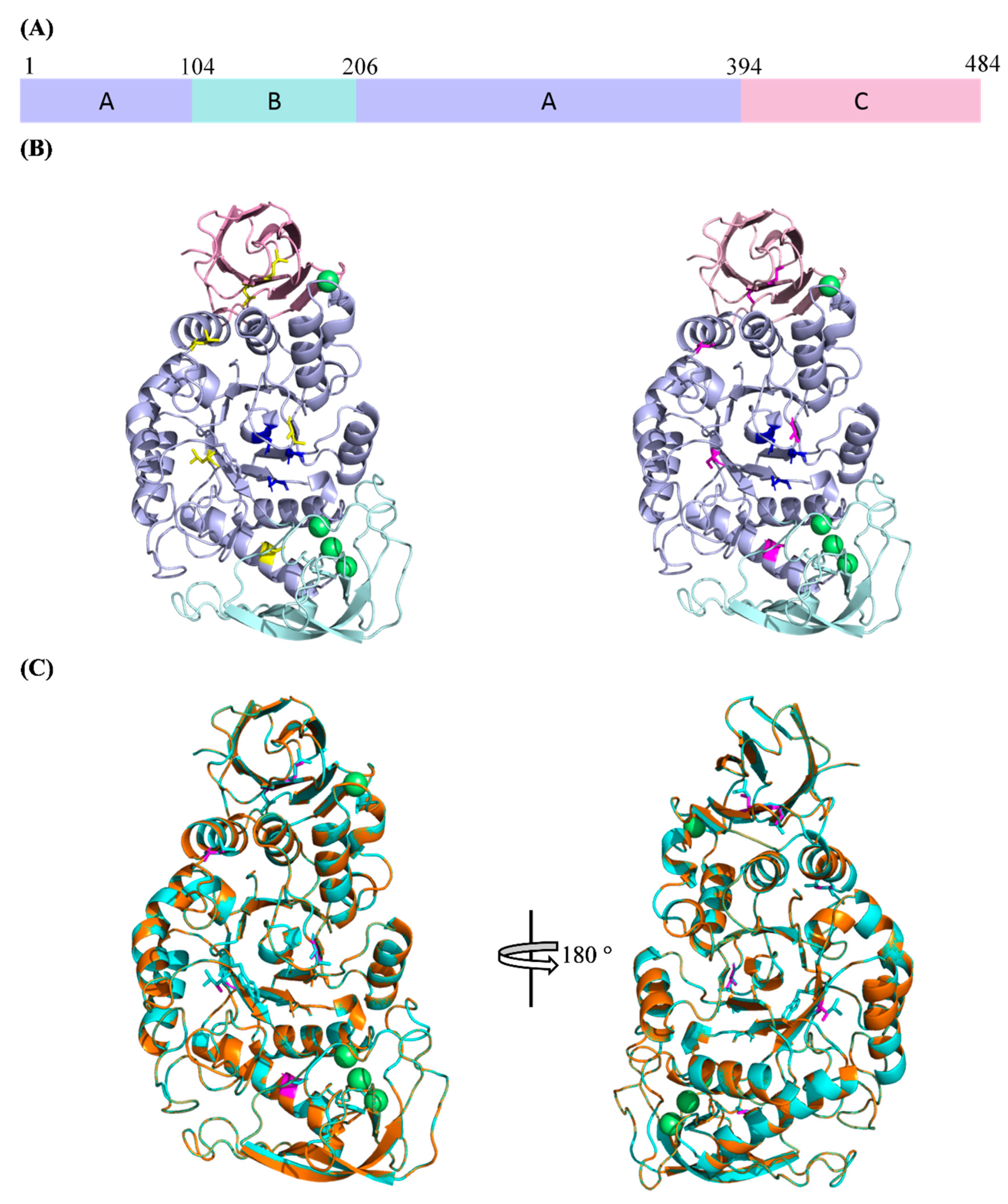
2.2. Homology Modelling and Structure Validation
2.3. Superimposition of WT and rc SR74 α-Amylases
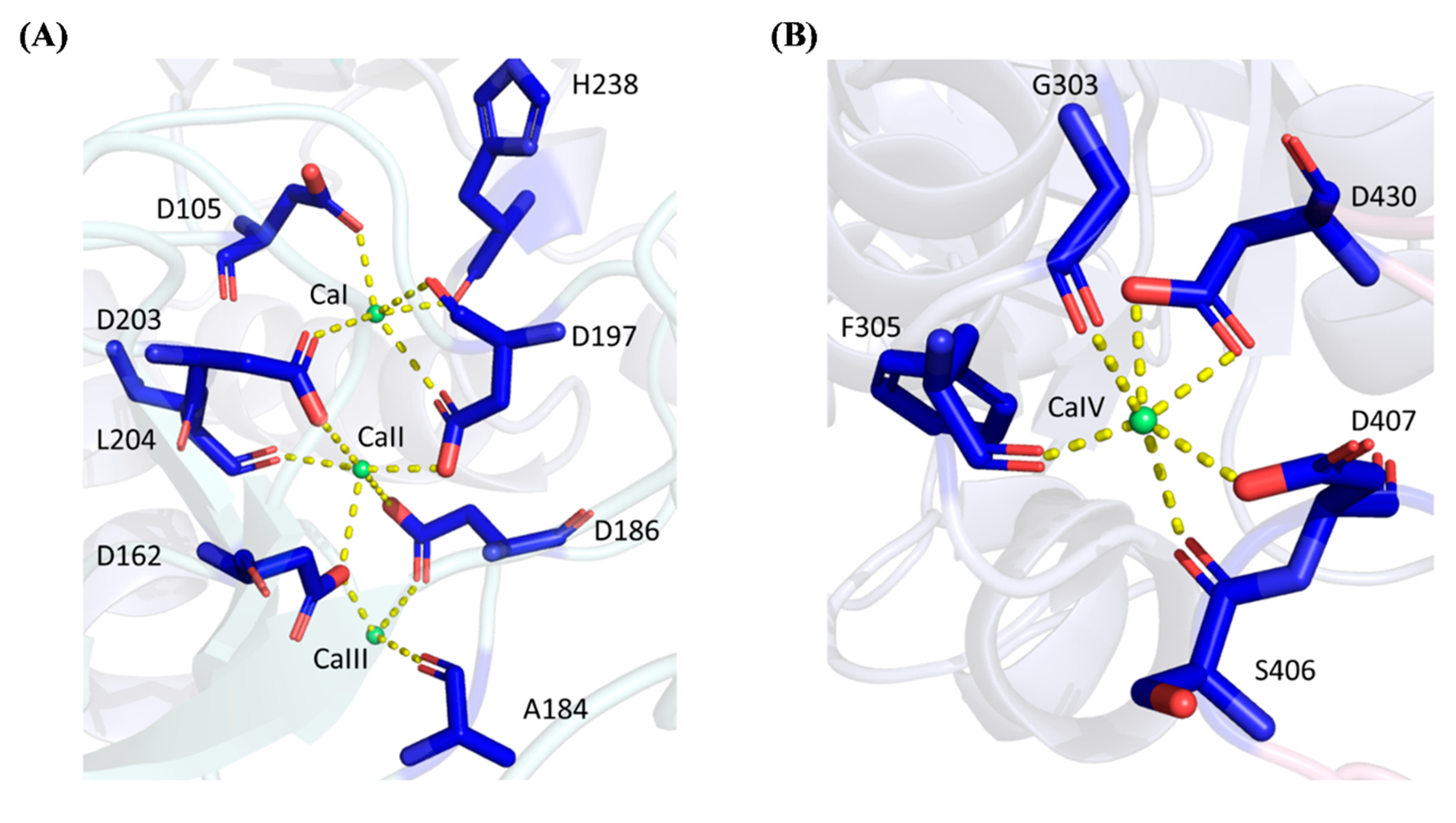
2.4. Protein–Ligand Interactions
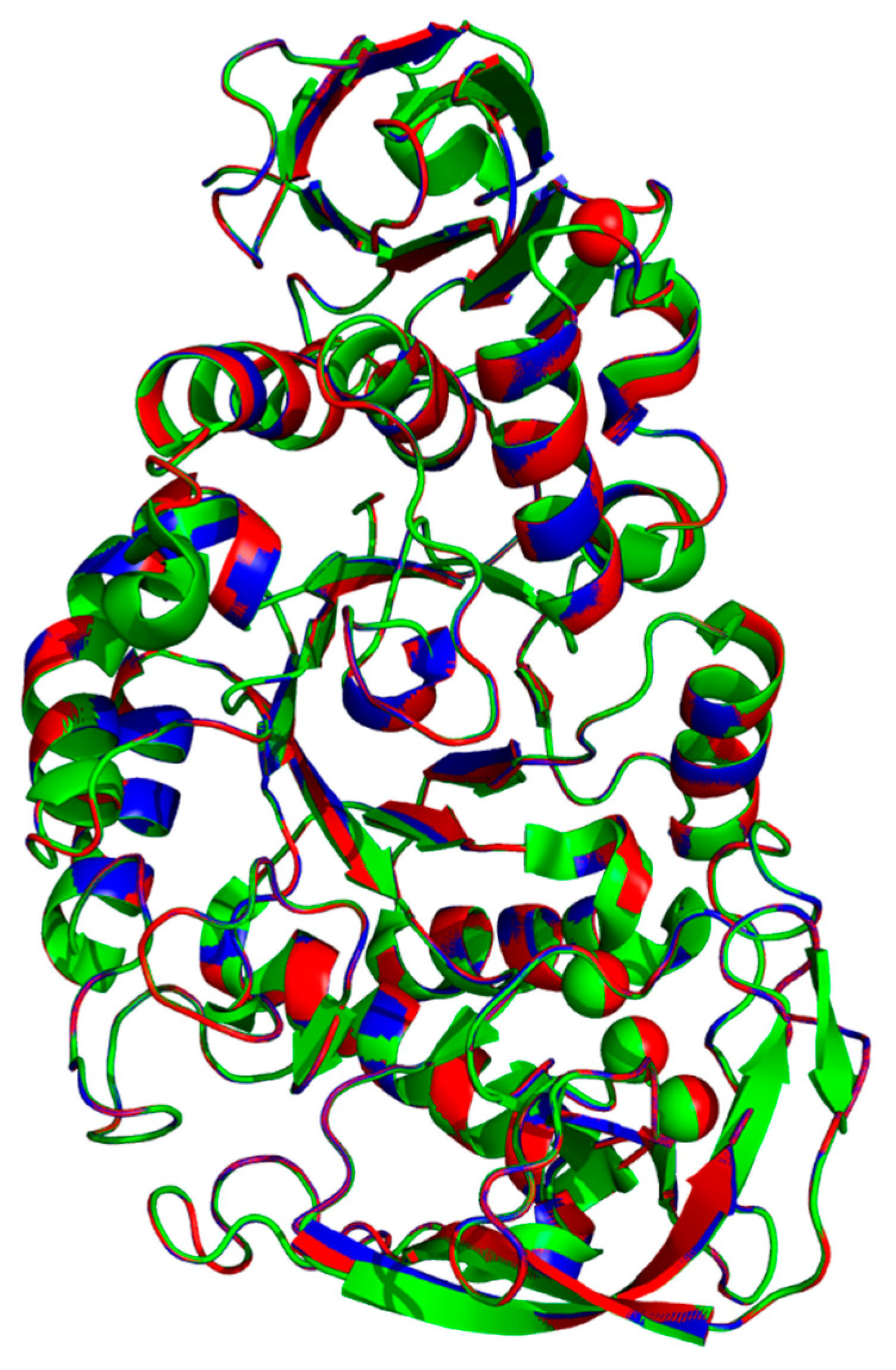
2.5. Superimposition of 6ag0.1.A, WT and rc SR74 α-Amylases
3. Materials and Methods
3.1. Acquisition of Nucleotide and Amino Acid Sequences of SR74 α-Amylases
3.2. Sequence and Structural Analysis of SR74 α-Amylases
3.3. In Silico 3D Structure Prediction of SR74 α-Amylases
3.4. Superimposition of SR74 α-Amylases with Template 6ag0.1.A
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Lim, S.J.; Hazwani-Oslan, S.N.; Oslan, S.N. Purification and Characterisation of Thermostable α-Amylases from Microbial Sources. BioResources 2020, 15, 2005–2029. [Google Scholar]
- Lombard, V.; Ramulu, H.G.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef]
- Janeček, Š.; Svensson, B.; MacGregor, E.A. α-Amylase: An enzyme specificity found in various families of glycoside hydrolases. Cell. Mol. Life Sci. 2014, 71, 1149–1170. [Google Scholar] [CrossRef]
- Du, R.; Song, Q.; Zhang, Q.; Zhao, F.; Kim, R.C.; Zhou, Z.; Han, Y. Purification and characterization of novel thermostable and Ca-independent α-amylase produced by Bacillus amyloliquefaciens BH072. Int. J. Biol. Macromol. 2018, 115, 1151–1156. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Ban, X.; Gu, Z.; Li, C.; Hong, Y.; Cheng, L.; Li, Z. Structure-Based Engineering of a Maltooligosaccharide-Forming Amylase to Enhance Product Specificity. J. Agric. Food Chem. 2020, 68, 838–844. [Google Scholar] [CrossRef] [PubMed]
- Karim, K.M.R.; Husaini, A.; Sing, N.N.; Sinang, F.M.; Roslan, H.A.; Hussain, H. Purification of an alpha amylase from Aspergillus flavus NSH9 and molecular characterization of its nucleotide gene sequence. 3 Biotech 2018, 8, 204. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, S.; Salleh, A.B.; Rahman, R.N.Z.R.A.; Chor Leow, T.; Oslan, S.N. Expression and characterization of geobacillus stearothermophilus sr74 recombinant α -amylase in pichia pastoris. Biomed. Res. Int. 2015, 2015, 529059. [Google Scholar] [CrossRef]
- Chai, K.P.; Othman, N.F.B.; Teh, A.H.; Ho, K.L.; Chan, K.G.; Shamsir, M.S.; Goh, K.M.; Ng, C.L. Crystal structure of Anoxybacillus α-amylase provides insights into maltose binding of a new glycosyl hydrolase subclass. Sci. Rep. 2016, 6, 23126. [Google Scholar] [CrossRef]
- Mehta, D.; Satyanarayana, T. Structural elements of thermostability in the maltogenic amylase of Geobacillus thermoleovorans. Int. J. Biol. Macromol. 2015, 79, 570–576. [Google Scholar] [CrossRef]
- Mehta, D.; Satyanarayana, T. Bacterial and archaeal α-amylases: Diversity and amelioration of the desirable characteristics for industrial applications. Front. Microbiol. 2016, 7, 1129. [Google Scholar] [CrossRef]
- Liao, S.-M.; Liang, G.; Zhu, J.; Lu, B.; Peng, L.-X.; Wang, Q.-Y.; Wei, Y.-T.; Zhou, G.-P.; Huang, R.-B. Influence of Calcium Ions on the Thermal Characteristics of α-amylase from Thermophilic Anoxybacillus sp. GXS-BL. Protein Pept. Lett. 2019, 26, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Li, Y.; Ban, X.; Zhang, Z.; Gu, Z.; Li, C.; Hong, Y.; Cheng, L.; Jin, T.; Li, Z. Crystal structure of a maltooligosaccharide-forming amylase from Bacillus stearothermophilus STB04. Int. J. Biol. Macromol. 2019, 138, 394–402. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Ban, X.; Gu, Z.; Li, C.; Hong, Y.; Cheng, L.; Li, Z. Insights into the thermostability and product specificity of a maltooligosaccharide-forming amylase from Bacillus stearothermophilus STB04. Biotechnol. Lett. 2020, 42, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Kassaye, E.K. Molecular Cloning and Expression of a Thermostable α-Amylase From Geobacillus sp. Master’s Thesis, Universiti Putra Malaysia, Seri Kembangan, Malaysia, 2009. [Google Scholar]
- Nasir, N.S.M.; Leow, C.T.; Oslan, S.N.H.; Salleh, A.B.; Oslan, S.N. Molecular expression of a recombinant thermostable bacterial amylase from Geobacillus stearothermophilus SR74 using methanol-free Meyerozyma guilliermondii strain SO yeast system. BioResources 2020, 15, 3161–3172. [Google Scholar]
- Oslan, S.N.; Salleh, A.B.; Rahman, R.R.A.; Basri, M.; Leow, T.C. Locally isolated yeasts from Malaysia: Identification, phylogenetic study and characterization. Acta Bochim. Pol. 2012, 59, 225–229. [Google Scholar] [CrossRef]
- Oslan, S.N.; Salleh, A.B.; Rahman, R.N.Z.R.A.; Leow, T.C.; Sukamat, H.; Basri, M. A newly isolated yeast as an expression host for recombinant lipase. Cell. Mol. Biol. Lett. 2015, 20, 279–293. [Google Scholar] [CrossRef]
- Periyasamy, N.A. Purification and Characterization of Recombinant Thermostable T1 Lipase Expressed from Pichia pastoris. Bachelor’s Thesis, Universiti Putra Malaysia, Seri Kembangan, Malaysia, 2015. [Google Scholar]
- De Marco, L.; Epis, S.; Capone, A.; Martin, E.; Bozic, J.; Crotti, E.; Ricci, I.; Sassera, D. The genomes of four Meyerozyma caribbica isolates and novel insights into the Meyerozyma guilliermondii species complex. G3 Genes Genomes Genet. 2018, 8, 755–759. [Google Scholar] [CrossRef]
- Krassowski, T.; Coughlan, A.Y.; Shen, X.X.; Zhou, X.; Kominek, J.; Opulente, D.A.; Riley, R.; Grigoriev, I.V.; Maheshwari, N.; Shields, D.C.; et al. Evolutionary instability of CUG-Leu in the genetic code of budding yeasts. Nat. Commun. 2018, 9, 1997. [Google Scholar] [CrossRef]
- Romi, W.; Keisam, S.; Ahmed, G.; Jeyaram, K. Reliable differentiation of Meyerozyma guilliermondii from Meyerozyma caribbica by internal transcribed spacer restriction fingerprinting. BMC Microbiol. 2014, 14, 52. [Google Scholar] [CrossRef]
- Gomes, A.C.; Miranda, I.; Silva, R.M.; Moura, G.R.; Thomas, B.; Akoulitchev, A.; Santos, M.A.S. A genetic code alteration generates a proteome of high diversity in the human pathogen Candida albicans. Genome Biol. 2007, 8, R206. [Google Scholar] [CrossRef]
- Ueda, T.; Suzuki, T.; Tokogawa, T.; Nishikawa, K.; Watanabe, K. Unique structure of new serine tRNAs responsible for decoding leucine codon CUG in various Candida species and their putative ancestral tRNA genes. Biochimie 1994, 76, 1217–1222. [Google Scholar] [CrossRef]
- Massey, S.E.; Moura, G.; Beltrão, P.; Almeida, R.; Garey, J.R.; Tuite, M.F.; Santos, M.A.S. Comparative evolutionary genomics unveils the molecular mechanism of reassignment of the CTG condon in Candida spp. Genome Res. 2003, 13, 544–557. [Google Scholar] [CrossRef] [PubMed]
- Bezerra, A.R.; Simões, J.; Lee, W.; Rung, J.; Weil, T.; Gut, I.G.; Gut, M.; Bayés, M.; Rizzetto, L.; Cavalieri, D.; et al. Reversion of a fungal genetic code alteration links proteome instability with genomic and phenotypic diversification. Proc. Natl. Acad. Sci. USA 2013, 110, 11079–11084. [Google Scholar] [CrossRef] [PubMed]
- Fraga, J.S.; Sárkány, Z.; Silva, A.; Correia, I.; Pereira, P.J.B.; Macedo-Ribeiro, S. Genetic code ambiguity modulates the activity of a C. albicans MAP kinase linked to cell wall remodeling. Biochim. Biophys. Acta Proteins Proteom. 2019, 1867, 654–661. [Google Scholar] [CrossRef] [PubMed]
- Simões, J.; Bezerra, A.R.; Moura, G.R.; Araújo, H.; Gut, I.; Bayes, M.; Santos, M.A.S. The fungus Candida albicans tolerates ambiguity at multiple codons. Front. Microbiol. 2016, 7, 401. [Google Scholar] [CrossRef] [PubMed]
- Dujon, B. Yeast evolutionary genomics. Nat. Rev. Genet. 2010, 11, 512–524. [Google Scholar] [CrossRef] [PubMed]
- Mehta, D.; Satyanarayana, T. Domain C of thermostable α-amylase of Geobacillus thermoleovorans mediates raw starch adsorption. Appl. Microbiol. Biotechnol. 2014, 98, 4503–4519. [Google Scholar] [CrossRef]
- Pan, S.; Gu, Z.; Ding, N.; Zhang, Z.; Chen, D.; Li, C.; Hong, Y.; Cheng, L.; Li, Z. Calcium and sodium ions synergistically enhance the thermostability of a maltooligosaccharide-forming amylase from Bacillus stearothermophilus STB04. Food Chem. 2019, 15, 170–176. [Google Scholar] [CrossRef]
- Koshland, D.E. Stereochemistry and the mechanism of enzymatic reactions. Biol. Rev. 1953, 28, 416–436. [Google Scholar] [CrossRef]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Cassarino, T.G.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014, 42, 252–258. [Google Scholar] [CrossRef]
- Kleywegt, G.J.; Jones, T.A. Phi/Psi-chology: Ramachandran revisited. Structure 1996, 4, 1395–1400. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Thornton, J.M. PROCHECK: Validation of protein-structure coordinates. In International Tables for Crystallography; Arnold, E., Himmel, D.M., Rossmann, M.G., Eds.; Wiley: Hoboken, NJ, USA, 2012; pp. 684–687. [Google Scholar]
- Rueda, M.; Orozco, M.; Totrov, M.; Abagyan, R. BioSuper: A web tool for the superimposition of biomolecules and assemblies with rotational symmetry. BMC Struct. Biol. 2013, 13, 32. [Google Scholar] [CrossRef] [PubMed]
- Kufareva, I.; Abagyan, R. Methods of protein structure comparison. Methods Mol. Biol. 2012, 857, 231–257. [Google Scholar] [PubMed]
- Chen, Y.H.; Chuang, L.Y.; Lo, H.F.; Hu, H.Y.; Wu, T.J.; Lin, L.L.; Chi, M.C. Mutational analysis of the proposed calcium-binding aspartates of a truncated a-amylase from Bacillus sp. strain TS-23. Ann. Microbiol. 2010, 60, 307–315. [Google Scholar] [CrossRef]
- Priyadharshini, R.; Gunasekaran, P. Site-directed mutagenesis of the calcium-binding site of α-amylase of Bacillus licheniformis. Biotechnol. Lett. 2007, 29, 1493–1499. [Google Scholar] [CrossRef]
- Offen, W.A.; Viksoe-Nielsen, A.; Borchert, T.V.; Wilson, K.S.; Davies, G.J. Three-dimensional structure of a variant “Termamyl-like” Geobacillus stearothermophilus α-amylase at 1.9Å resolution. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2015, 71, 66–70. [Google Scholar] [CrossRef]
- Arnold, K.; Bordoli, L.; Kopp, J.; Schwede, T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 2006, 22, 195–201. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, 296–303. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Remmert, M.; Biegert, A.; Hauser, A.; Söding, J. HHblits: Lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat. Methods 2012, 9, 173–175. [Google Scholar] [CrossRef]
- Bowie, J.U.; Lüthy, R.; Eisenberg, D. A method to identify protein sequences that fold into a known three-dimensional structure. Science 1991, 253, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Lüthy, R.; Bowie, J.U.; Eisenberg, D. Assessment of protein models with three-dimensional profiles. Nature 1992, 356, 83–85. [Google Scholar] [CrossRef] [PubMed]
- Colovos, C.; Yeates, T.O. Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Sci. 1993, 2, 1511–1519. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Rullmann, J.A.C.; MacArthur, M.W.; Kaptein, R.; Thornton, J.M. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 1996, 8, 477–486. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lim, S.J.; Muhd Noor, N.D.; Salleh, A.B.; Oslan, S.N. Structure Prediction of a Thermostable SR74 α-Amylase from Geobacillus stearothermophilus Expressed in CTG-Clade Yeast Meyerozyma guilliermondii Strain SO. Catalysts 2020, 10, 1059. https://doi.org/10.3390/catal10091059
Lim SJ, Muhd Noor ND, Salleh AB, Oslan SN. Structure Prediction of a Thermostable SR74 α-Amylase from Geobacillus stearothermophilus Expressed in CTG-Clade Yeast Meyerozyma guilliermondii Strain SO. Catalysts. 2020; 10(9):1059. https://doi.org/10.3390/catal10091059
Chicago/Turabian StyleLim, Si Jie, Noor Dina Muhd Noor, Abu Bakar Salleh, and Siti Nurbaya Oslan. 2020. "Structure Prediction of a Thermostable SR74 α-Amylase from Geobacillus stearothermophilus Expressed in CTG-Clade Yeast Meyerozyma guilliermondii Strain SO" Catalysts 10, no. 9: 1059. https://doi.org/10.3390/catal10091059
APA StyleLim, S. J., Muhd Noor, N. D., Salleh, A. B., & Oslan, S. N. (2020). Structure Prediction of a Thermostable SR74 α-Amylase from Geobacillus stearothermophilus Expressed in CTG-Clade Yeast Meyerozyma guilliermondii Strain SO. Catalysts, 10(9), 1059. https://doi.org/10.3390/catal10091059





