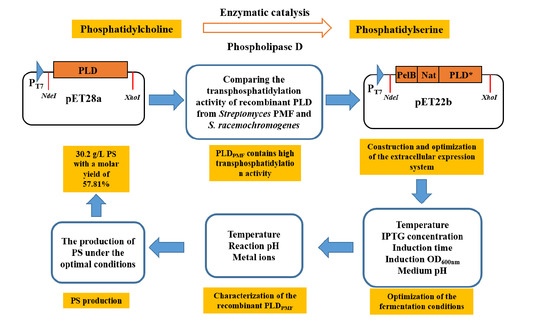
Highly Efficient Extracellular Production of Recombinant Streptomyces PMF Phospholipase D in Escherichia coli
Abstract
1. Introduction
2. Results
2.1. Intracellular Expression of PLD in E. coli
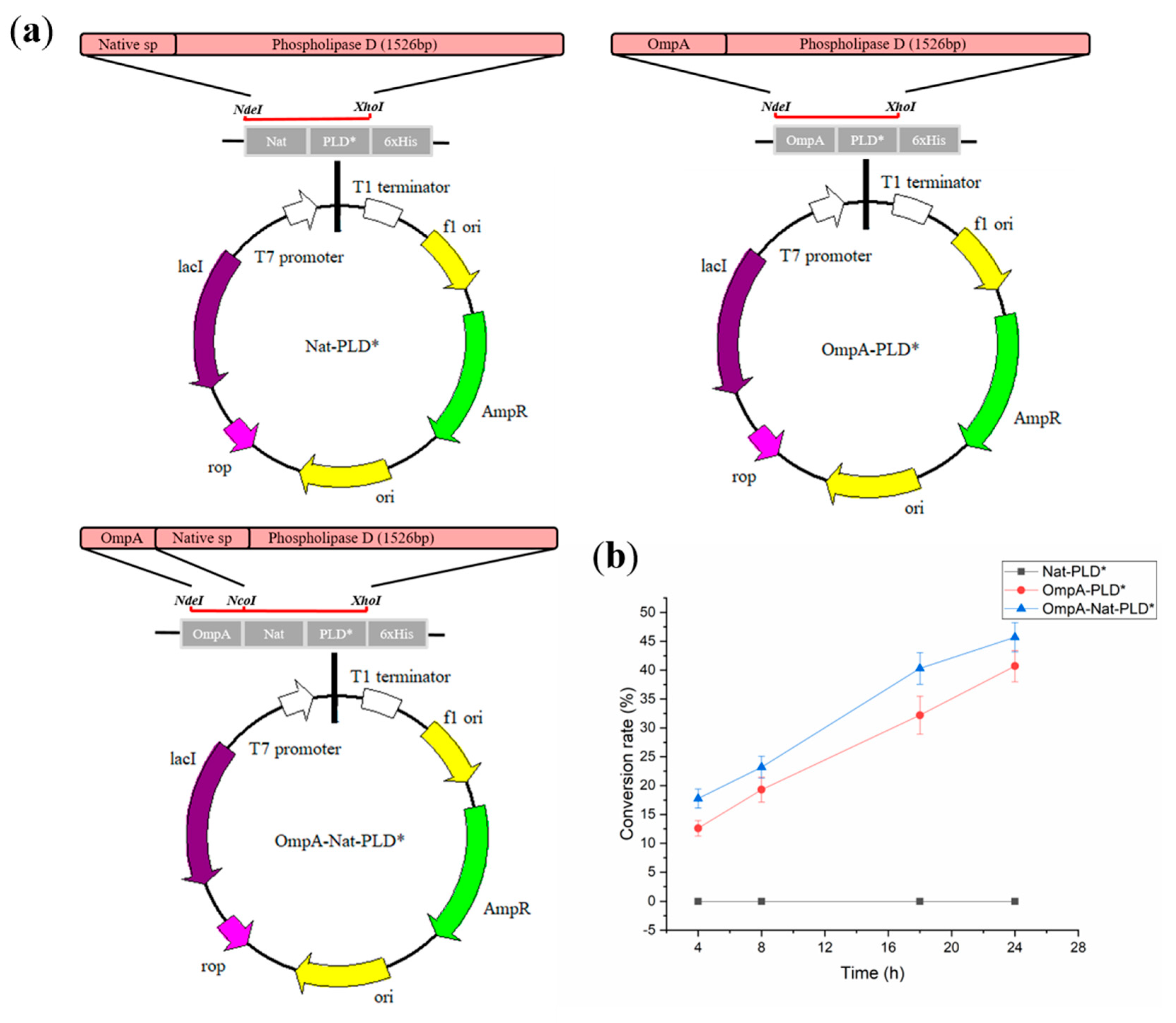
2.2. Secretory Expression of PLD in E. coli by Optimizing Signal Peptides
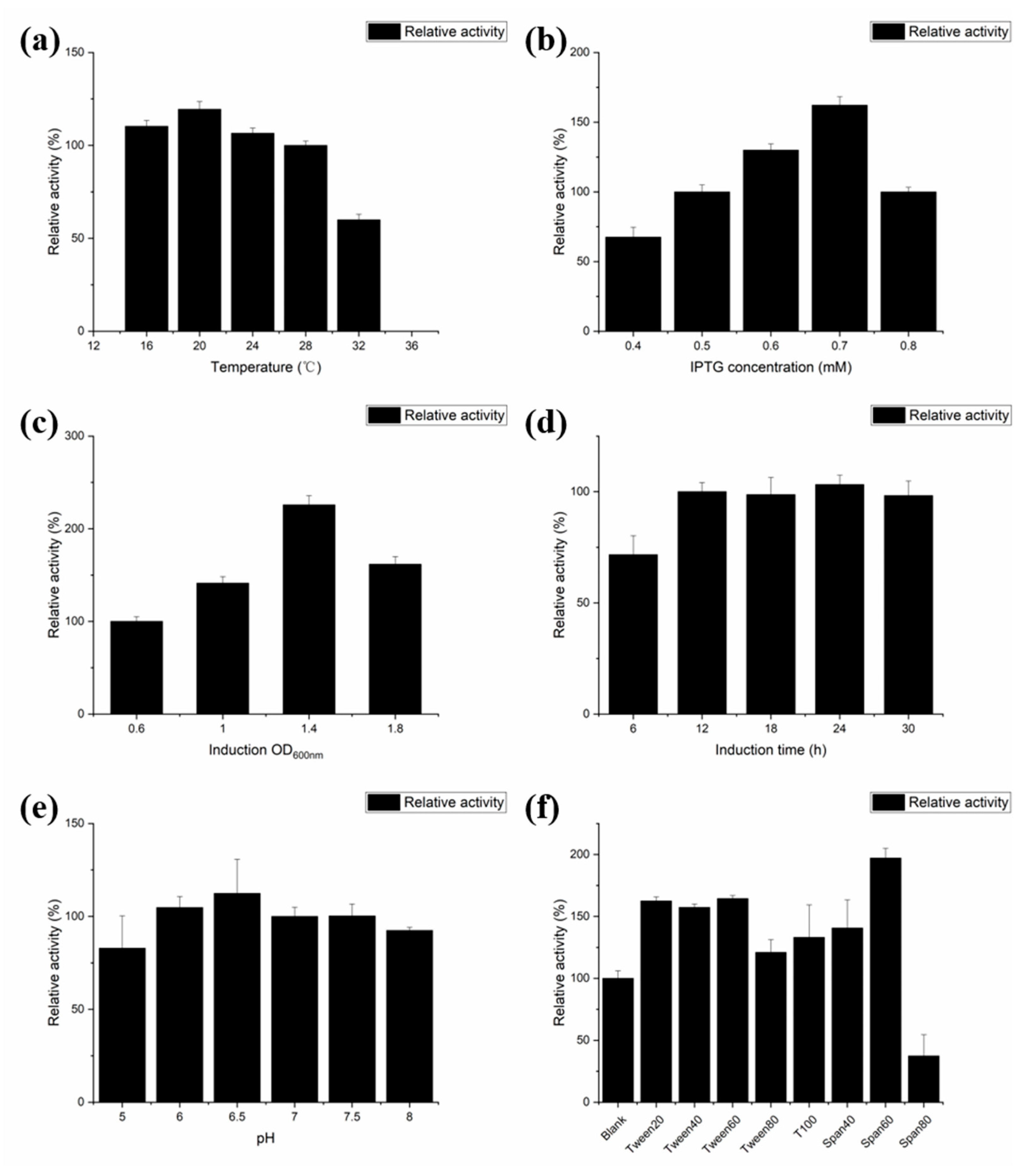
2.3. Effect of Fermentation Conditions on the Secretory Expression of PLD
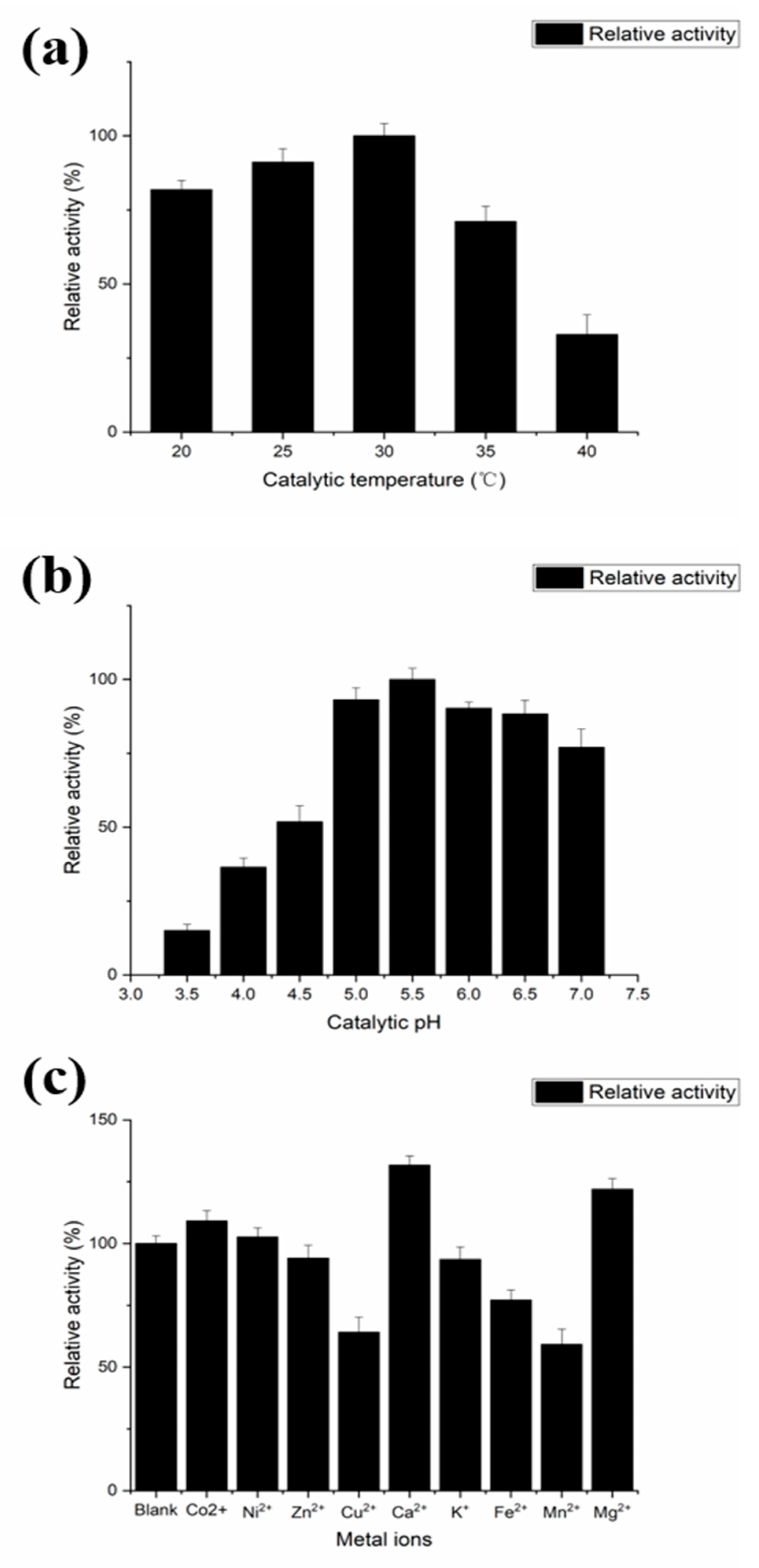
2.4. Characterization of the Recombinant PLD Activity
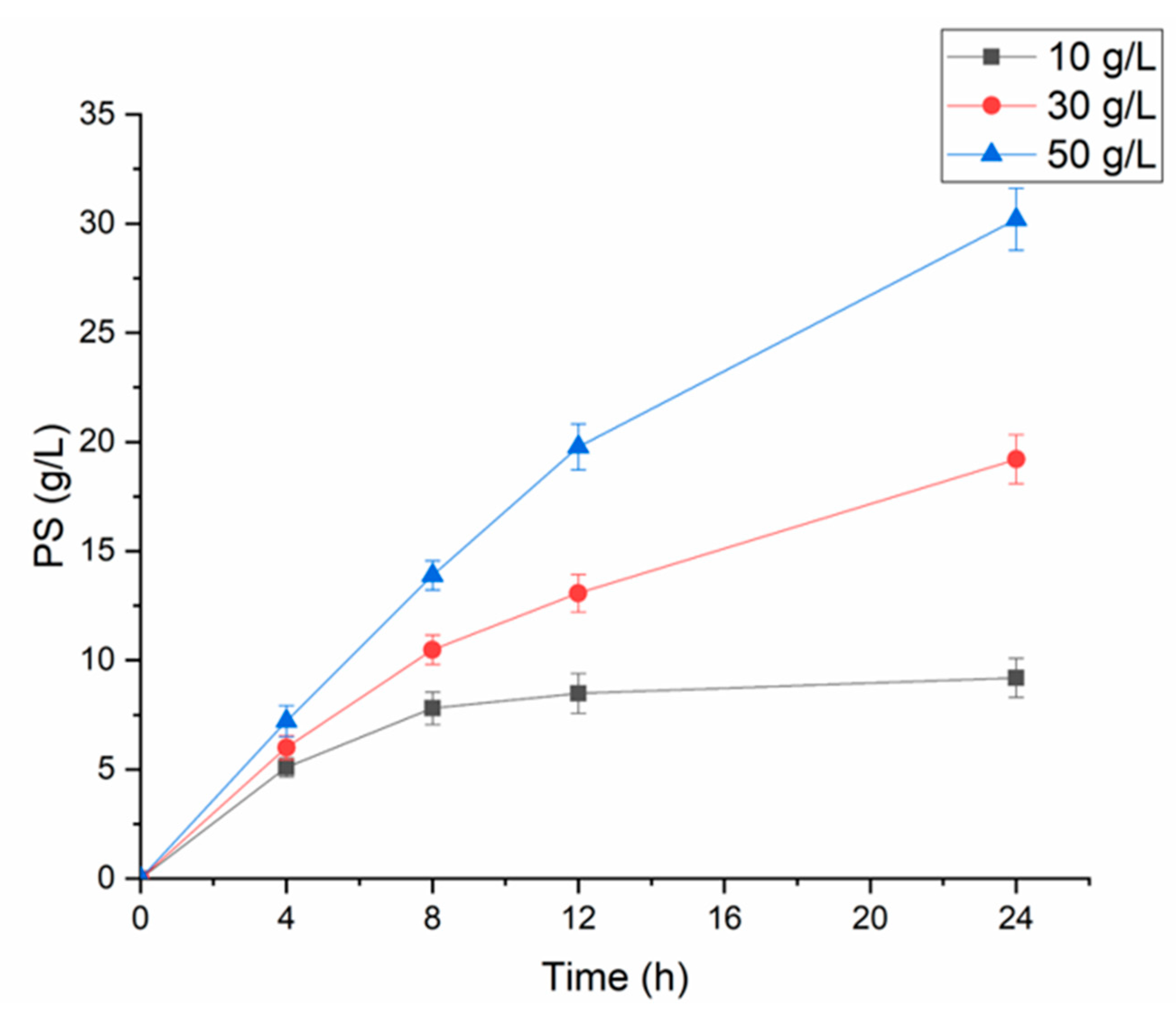
2.5. The Application of Recombinant PLDPMF for the Bioconversion of PC to PS
3. Discussion
4. Materials and Methods
4.1. Microorganisms and Media
4.2. Plasmid Construction
4.3. Protein Expression and Cell Fractionation
4.4. Enzyme Assay
4.5. Analytical Methods
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Nakazawa, Y.; Sagane, Y.; Sakurai, S.-I.; Uchino, M.; Sato, H.; Toeda, K.; Takano, K. Large-Scale Production of Phospholipase D from Streptomyces racemochromogenes and Its Application to Soybean Lecithin Modification. Appl. Biochem. Biotechnol. 2011, 165, 1494–1506. [Google Scholar] [CrossRef] [PubMed]
- Doig, S.D.; Diks, R.M.M. Toolbox for exchanging constituent fatty acids in lecithins. Eur. J. Lipid Sci. Technol. 2003, 105, 359–367. [Google Scholar] [CrossRef]
- Altshuller, Y.M. Human ADP-ribosylation Factor-activated Phosphatidylcholine-specific Phospholipase D Defines a New and Highly Conserved Gene Family. J. Boil. Chem. 1995, 270, 29640–29643. [Google Scholar] [CrossRef] [PubMed]
- Khatoon, H.; Mansfeld, J.; Schierhorn, A.; Ulbrich-Hofmann, R. Purification, sequencing and characterization of phospholipase D from Indian mustard seeds. Phytochemistry 2015, 117, 65–75. [Google Scholar] [CrossRef]
- Frohman, M.A. The phospholipase D superfamily as therapeutic targets. Trends Pharmacol. Sci. 2015, 36, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Hagishita, T.; Nishikawa, M.; Hatanaka, T. Isolation of phospholipase d producing microorganisms with high transphosphatidylation activity. Biotechnol. Lett. 2000, 22, 1587–1590. [Google Scholar] [CrossRef]
- Ramrakhiani, L.; Chand, S. Recent Progress on Phospholipases: Different Sources, Assay Methods, Industrial Potential and Pathogenicity. Appl. Biochem. Biotechnol. 2011, 164, 991–1022. [Google Scholar] [CrossRef]
- Carrea, G.; D’Arrigo, P.; Piergianni, V.; Roncaglio, S.; Secundo, F.; Servi, S. Purification and properties of two phospholipases D from Streptomyces sp. Biochim. Biophys. Acta (BBA) Lipids Lipid Metab. 1995, 1255, 273–279. [Google Scholar] [CrossRef]
- Ogino, C.; Kanemasu, M.; Fukumoto, M.; Kubo, T.; Yoshino, T.; Kondo, A.; Fukuda, H.; Shimizu, N. Continuous production of phospholipase D using immobilized recombinant Streptomyces lividans. Enzym. Microb. Technol. 2007, 41, 156–161. [Google Scholar] [CrossRef]
- Chen, S.; Xu, L.; Li, Y.; Hao, N.; Yan, M. Bioconversion of phosphatidylserine by phospholipase D from Streptomyces racemochromogenes in a microaqueous water-immiscible organic solvent. Biosci. Biotechnol. Biochem. 2013, 77, 130388. [Google Scholar] [CrossRef][Green Version]
- Lee, J.-S.; Bat-Ochir, M.; Demirev, A.V.; Nam, D.H. Molecular cloning of the phospholipase D gene from Streptomyces sp. YU100 and its expression in Escherichia coli. J. Microbiol. 2009, 47, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.-B.; Gong, J.-S.; Hou, H.-J.; Li, H.; Lu, Z.-M.; Xu, H.-Y.; Xu, Z.-H.; Shi, J.-S. Mining of a phospholipase D and its application in enzymatic preparation of phosphatidylserine. Bioengineered 2018, 9, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Choojit, S.; Bornscheuer, U.T.; Upaichit, A.; H-Kittikun, A. Efficient phosphatidylserine synthesis by a phospholipase D fromStreptomycessp. SC734 isolated from soil-contaminated palm oil. Eur. J. Lipid Sci. Technol. 2016, 118, 803–813. [Google Scholar] [CrossRef]
- Ogino, C.; Kanemasu, M.; Hayashi, Y.; Kondo, A.; Shimizu, N.; Tokuyama, S.; Tahara, Y.; Kuroda, S.; Tanizawa, K.; Fukuda, H. Over-expression system for secretory phospholipase D by Streptomyces lividans. Appl. Microbiol. Biotechnol. 2004, 64, 823–828. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, T.; Qiao, J.; Liu, X.; Bo, J.; Wang, J.; Lu, F. High-Yield Phosphatidylserine Production via Yeast Surface Display of Phospholipase D from Streptomyces chromofuscus on Pichia pastoris. J. Agric. Food Chem. 2014, 62, 5354–5360. [Google Scholar] [CrossRef]
- Zambonelli, C. Cloning and expression in Escherichia coli of the gene encoding Streptomyces PMF PLD, a phospholipase D with high transphosphatidylation activity. Enzym. Microb. Technol. 2003, 33, 676–688. [Google Scholar] [CrossRef]
- Xiong, W.; Zeng, X.; Ho, S.-H.; Ling, X.; Shen, L.; Yao, C.; Lu, Y. Strategies for achieving high-level and stable production of toxic Streptomyces phospholipase D in Escherichia coli. J. Chem. Technol. Biotechnol. 2019, 94, 1220–1229. [Google Scholar] [CrossRef]
- Hikita, C.; Mizushima, S. Effects of total hydrophobicity and length of the hydrophobic domain of a signal peptide on in vitro translocation efficiency. J. Boil. Chem. 1992, 267, 4882–4888. [Google Scholar]
- Li, Z.; Li, B.; Liu, Z.-G.; Wang, M.; Gu, Z.-B.; Du, G.-C.; Wu, J.; Chen, J. Calcium Leads to Further Increase in Glycine-Enhanced Extracellular Secretion of Recombinant α-Cyclodextrin Glycosyltransferase inEscherichia coli. J. Agric. Food Chem. 2009, 57, 6231–6237. [Google Scholar] [CrossRef]
- Shin, H.-D.; Chen, R. Extracellular recombinant protein production from anEscherichia coli lppdeletion mutant. Biotechnol. Bioeng. 2008, 101, 1288–1296. [Google Scholar] [CrossRef]
- Su, L.; Chen, S.; Yi, L.; Woodard, R.W.; Chen, J.; Wu, J. Extracellular overexpression of recombinant Thermobifida fusca cutinase by alpha-hemolysin secretion system in E. coli BL21(DE3). Microb. Cell Factories 2012, 11, 8. [Google Scholar] [CrossRef] [PubMed]
- Selvy, P.E.; Lavieri, R.R.; Lindsley, C.W.; Brown, H.A. Phospholipase D: Enzymology, Functionality, and Chemical Modulation. Chem. Rev. 2011, 111, 6064–6119. [Google Scholar] [CrossRef]
- Cerminati, S.; Paoletti, L.; Aguirre, A.; Peirú, S.; Menzella, H.G.; Castelli, M.E. Industrial uses of phospholipases: Current state and future applications. Appl. Microbiol. Biotechnol. 2019, 103, 2571–2582. [Google Scholar] [CrossRef] [PubMed]
- Jeucken, A.; Helms, J.B.; Brouwers, J.F. Cardiolipin synthases of Escherichia coli have phospholipid class specific phospholipase D activity dependent on endogenous and foreign phospholipids. Biochim. Biophys. Acta (BBA) Mol. Cell Boil. Lipids 2018, 1863, 1345–1353. [Google Scholar] [CrossRef]
- Mayr, J.A.; Kohlwein, S.D.; Paltauf, F. Identification of a novel, Ca2+-dependent phospholipase D with preference for phosphatidylserine and phosphatidylethanolamine inSaccharomyces cerevisiae. FEBS Lett. 1996, 393, 236–240. [Google Scholar] [CrossRef]
- Huang, T.; Lv, X.; Li, J.; Shin, H.-d.; Du, G.; Liu, L. Combinatorial Fine-Tuning of Phospholipase D Expression by Bacillus subtilis WB600 for the Production of Phosphatidylserine. J. Microbiol. Biotechnol. 2018, 28, 2046–2056. [Google Scholar] [CrossRef]
- Makino, T.; Skretas, G.; Georgiou, G. Strain engineering for improved expression of recombinant proteins in bacteria. Microb. Cell Factories 2011, 10, 32. [Google Scholar] [CrossRef]
- Pechsrichuang, P.; Songsiriritthigul, C.; Haltrich, D.; Roytrakul, S.; Namvijtr, P.; Bonaparte, N.; Yamabhai, M. OmpA signal peptide leads to heterogenous secretion of B. subtilis chitosanase enzyme from E. coli expression system. SpringerPlus 2016, 5, 1200. [Google Scholar] [CrossRef]
- Valent, Q.A.; Scotti, P.A.; High, S.; De Gier, J.L.; Von Heijne, G.; Lentzen, G.; Wintermeyer, W.; Oudega, B.; Luirink, J. The Escherichia coli SRP and SecB targeting pathways converge at the translocon. EMBO J. 1998, 17, 2504–2512. [Google Scholar] [CrossRef]
- Choi, J.H.; Lee, S.Y. Secretory and extracellular production of recombinant proteins using Escherichia coli. Appl. Microbiol. Biotechnol. 2004, 64, 625–635. [Google Scholar] [CrossRef]
- Chen, R. Permeability issues in whole-cell bioprocesses and cellular membrane engineering. Appl. Microbiol. Biotechnol. 2007, 74, 730–738. [Google Scholar] [CrossRef] [PubMed]
- Simkhada, J.R.; Lee, H.J.; Jang, S.Y.; Kim, J.H.; Lee, H.C.; Sohng, J.K.; Yoo, J.C. A novel low molecular weight phospholipase D from Streptomyces sp. CS684. Bioresour. Technol. 2009, 100, 1388–1393. [Google Scholar] [CrossRef] [PubMed]
- Zambonelli, C.; Roberts, M.F. An Iron-dependent Bacterial Phospholipase D Reminiscent of Purple Acid Phosphatases. J. Boil. Chem. 2003, 278, 13706–13711. [Google Scholar] [CrossRef] [PubMed]
- Duan, Z.-Q.; Hu, F. Efficient synthesis of phosphatidylserine in 2-methyltetrahydrofuran. J. Biotechnol. 2013, 163, 45–49. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Lu, F.-P.; Tian, L.; Li, Y.; Du, L. Cloning and expression of phospholipase D genepld fromStreptomyces chromofuscus. Ann. Microbiol. 2008, 58, 227–231. [Google Scholar] [CrossRef]
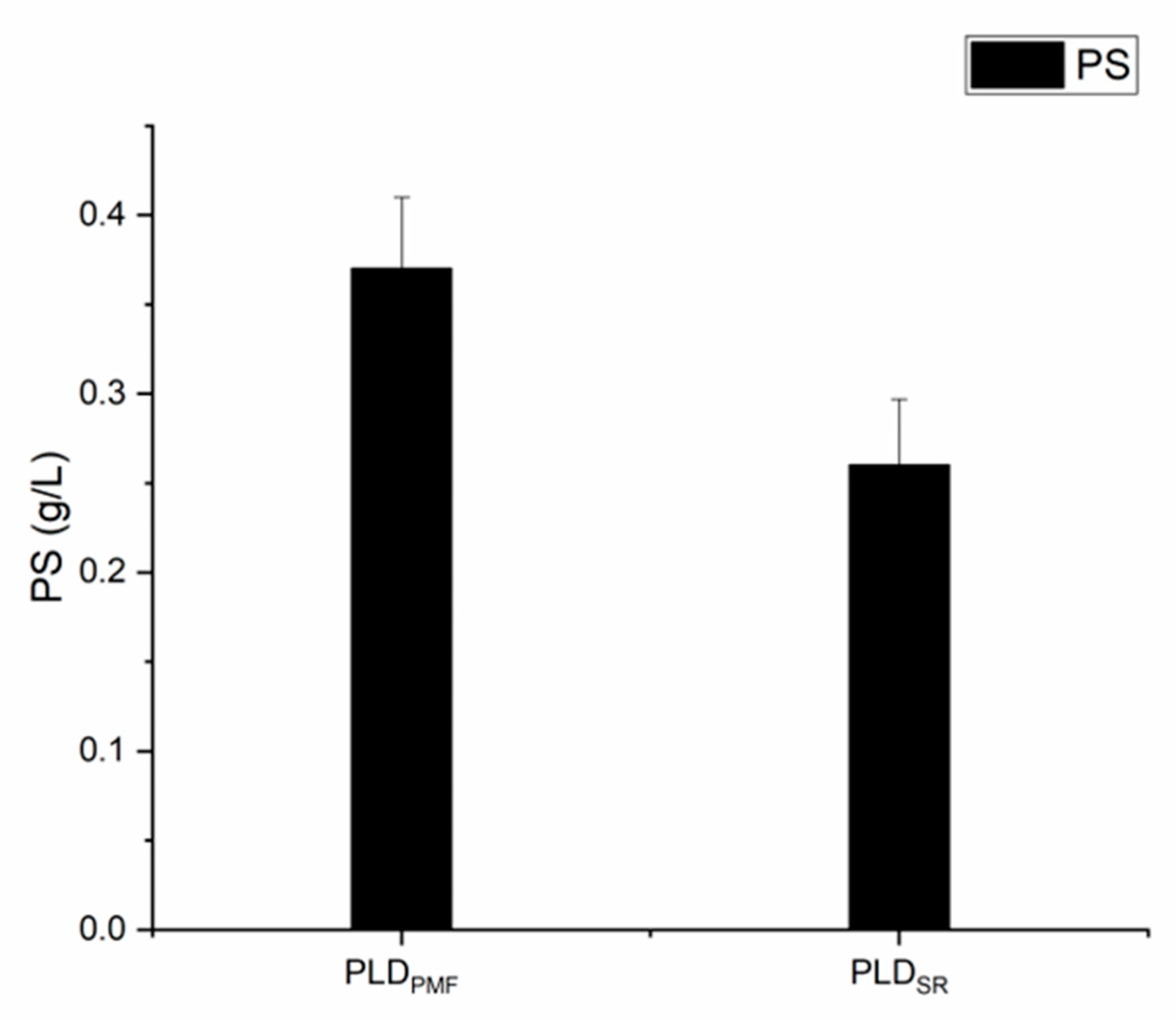


| PLD Origin | Expression Host | PS (g/L) | References |
|---|---|---|---|
| Streptomyces mobaraensis | E. coli | 0.2 | [12] |
| Streptomyces chromofuscus | E. coli | 3.94 | [35] |
| Streptomyces sp. YU100 | E. coli | ND | [11] |
| Streptomyces antibioticus | E. coli | ND | [17] |
| Streptomyces PMF | E. coli | 30.2 | This study |
| Strains or Plasmids | Characteristics | Sources |
|---|---|---|
| Strains | - | - |
| Streptomyces PMF | Source of PLDPMF gene | ATCC |
| Streptomyces racemochromogenes | Source of PLDSR gene | ATCC |
| E. coli DH5α | Used as cloning vector | Invitrogen |
| E. coli BL21(DE3) | Used as expression host | Invitrogen |
| BL21(DE3)/pET28a-PLDPMF | Express plasmid: pET28a-PLDPMF | This study |
| BL21(DE3)/pET28a-PLDSR | Express plasmid: pET28a-PLDSR | This study |
| BL21(DE3)/pET22b-PLDPMF | Express plasmid: pET22b-PLDPMF | This study |
| BL21(DE3)/Nat-PLD * | Express plasmid: Nat-PLD * | This study |
| BL21(DE3)/OmpA-PLD * | Express plasmid: OmpA-PLD * | This study |
| BL21(DE3)/OmpA-Nat-PLD * | Express plasmid: OmpA-Nat-PLD * | This study |
| BL21(DE3)/OmpC-Nat-PLD * | Express plasmid: OmpC-Nat-PLD * | This study |
| BL21(DE3)/OmpF-Nat-PLD * | Express plasmid: OmpF-Nat-PLD * | This study |
| BL21(DE3)/OmpT-Nat-PLD * | Express plasmid: OmpT-Nat-PLD * | This study |
| BL21(DE3)/LamB-Nat-PLD * | Express plasmid: LamB-Nat-PLD * | This study |
| BL21(DE3)/PhoA-Nat-PLD * | Express plasmid: PhoA-Nat-PLD * | This study |
| BL21(DE3)/MalE-Nat-PLD * | Express plasmid: MalE-Nat-PLD * | This study |
| BL21(DE3)/PelB-Nat-PLD * | Express plasmid: PelB-Nat-PLD * | This study |
| Plasmids | - | - |
| pET28a | pBR322 ori, PT7, KanR | Our lab |
| pET22b | pBR322 ori, PT7, OmpA signal peptide, AmpR | Our lab |
| pET28a-PLDPMF | pET28a derivative; PT7-lacO-PLDPMF | This study |
| pET28a-PLDSR | pET28a derivative; PT7-lacO-PLDSR | This study |
| pET22b-PLDPMF | pET22b derivative; PT7-lacO-OmpA-PLDPMF | This study |
| Nat-PLD * | pET22b derivative; PT7-lacO-Nat-PLD * | This study |
| OmpA-PLD * | pET22b derivative; PT7-lacO-OmpA-PLD * | This study |
| OmpA-Nat-PLD * | pET22b derivative; PT7-lacO-OmpA-Nat-PLD * | This study |
| OmpC-Nat-PLD * | pET22b derivative; PT7-lacO-OmpC-Nat-PLD * | This study |
| OmpF-Nat-PLD * | pET22b derivative; PT7-lacO-OmpF-Nat-PLD * | This study |
| OmpT-Nat-PLD * | pET22b derivative; PT7-lacO-OmpT-Nat-PLD * | This study |
| LamB-Nat-PLD * | pET22b derivative; PT7-lacO-LamB-Nat-PLD * | This study |
| PhoA-Nat-PLD * | pET22b derivative; PT7-lacO-PhoA-Nat-PLD * | This study |
| MalE-Nat-PLD * | pET22b derivative; PT7-lacO-MalE-Nat-PLD * | This study |
| PelB-Nat-PLD * | pET22b derivative; PT7-lacO-PelB-Nat-PLD * | This study |
| Name | Primers | Sequences (5′–3′) |
|---|---|---|
| P1 | NcoI-PLDPMF-F | CATGCCATGGCAGCTGACTCTGCTACCCCG |
| P2 | EcoRI-PLDPMF-R | CCGGAATTCTCAGGCGTTGCAGATCCC |
| P3 | NcoI-PLDSR-F | CATGCCATGGGTGCGGAGGTGTGGTCGTAC |
| P4 | EcoRI-PLDSR-R | CCGGAATTCTCAGGCCTGGCAGAGG |
| P5 | NdeI-Nat-PLD * | GGAATTCCATATGCTACATGGGTCACACCT |
| P6 | XhoI-Nat-PLD * | CTCGAGCGGAGCGTTGCAGATACCAC |
| P7 | NcoI-OmpA-Nat-PLD * | CCATGGGCTACATGGGTCACA |
| P8 | XhoI-OmpA-Nat-PLD * | CTCGAGCGGAGCGTTGCAGATACCAC |
| P9 | NcoI-OmpA-PLD * | CATGCCATGGCAGCTGACTCTGCTACCCCG |
| P10 | XhoI-OmpA-PLD * | CTCGAGCGGAGCGTTGCAGATACCAC |
| P11 | LamB-F | GGAATTCCATATGATTACTCTGCGCAAACTTCCTCTGGCGGTTGCCGTCGCAGCGGGCGTAATGTCTGCTCAGGCAATGGCTCCATGGGCTACATGGGTCACA |
| P12 | MalE-F | GGAATTCCATATGAAAATAAAAACAGGTGCACGCATCCTCGCATTATCCGCATTAACGACGATGATGTTTTCCGCCTCGGCTCTCGCCCCATGGGCTACATGGGTCACA |
| P13 | OmpC-F | GGAATTCCATATGAAAGTTAAAGTACTGTCCCTCCTGGTCCCAGCTCTGCTGGTAGCAGGCGCAGCAAACGCTCCATGGGCTACATGGGTCACA |
| P14 | OmpF-F | GGAATTCCATATGAAGCGCAATATTCTGGCAGTGATCGTCCCTGCTCTGTTAGTAGCAGGTACTGCAAACGCTCCATGGGCTACATGGGTCACA |
| P15 | OmpT-F | GGAATTCCATATGCGGGCGAAACTTCTGGGAATAGTCCTGACAACCCCTATTGCGATCAGCTCTTTTGCTCCATGGGCTACATGGGTCACA |
| P16 | PhoA-F | GGAATTCCATATGAAACAAAGCACTATTGCACTGGCACTCTTACCGTTACTGTTTACCCCTGTGACAAAAGCCCCATGGGCTACATGGGTCACA |
| P17 | PelB-F | GGAATTCCATATGAAATACCTGCTGCCGACCGCTGCTGCTGGTCTGCTGCTCCTCGCTGCCCAGCCGGCGATGGCCATGGGCTACATGGGTCACA |
| P18 | General reverse primer | CTCGAGCG+GAGCGTTGCAGATACCAC |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Xu, S.; Pang, Y.; Wang, X.; Chen, K.; Ouyang, P. Highly Efficient Extracellular Production of Recombinant Streptomyces PMF Phospholipase D in Escherichia coli. Catalysts 2020, 10, 1057. https://doi.org/10.3390/catal10091057
Wang J, Xu S, Pang Y, Wang X, Chen K, Ouyang P. Highly Efficient Extracellular Production of Recombinant Streptomyces PMF Phospholipase D in Escherichia coli. Catalysts. 2020; 10(9):1057. https://doi.org/10.3390/catal10091057
Chicago/Turabian StyleWang, Jing, Sheng Xu, Yang Pang, Xin Wang, Kequan Chen, and Pingkai Ouyang. 2020. "Highly Efficient Extracellular Production of Recombinant Streptomyces PMF Phospholipase D in Escherichia coli" Catalysts 10, no. 9: 1057. https://doi.org/10.3390/catal10091057
APA StyleWang, J., Xu, S., Pang, Y., Wang, X., Chen, K., & Ouyang, P. (2020). Highly Efficient Extracellular Production of Recombinant Streptomyces PMF Phospholipase D in Escherichia coli. Catalysts, 10(9), 1057. https://doi.org/10.3390/catal10091057