Abstract
The pandemic associated to Severe Acute Respiratory Syndrome Coronavirus type 2 (SARS-CoV-2) has resulted in a huge number of deaths and infected people. Although several vaccine programmes are currently underway and have reached phase 3, and a few small size drugs repurposed to aid treatment of severe cases of COVID-19 infections, effective therapeutic options for this disease do not currently exist. NSP16 is a S-adenosyl-L-Methionine (SAM) dependent 2′O-Methyltransferase that converts mRNA cap-0 into cap-1 structure to prevent virus detection by cell innate immunity mechanisms. NSP16 methylates the ribose 2′O-position of the first nucleotide of the mRNA only in the presence of an interacting partner, the protein NSP10. This feature suggests that inhibition of the NSP16 may represent a therapeutic window to treat COVID-19. To test this idea, we performed comparative structural analyses of the NSP16 present in human coronaviruses and developed a sinefungin (SFG) similarity-based virtual screening campaign to assess the druggability of the SARS-CoV-2 NSP16 enzyme. Through these studies, we identified the SFG analogue 44601604 as a promising more potent inhibitor of NSP16 to limit viral replication in infected cells, favouring viral clearance.
1. Introduction
There is urgency to develop effective therapeutics against the SARS-CoV-2, the causing agent of the ongoing COVID-19 pandemic. There are seven types of human Coronavirus (CoVs): 229E, NL63, OC43, HKU1, Middle East Respiratory Syndrome (MERS-CoV) and Severe Acute Respiratory Syndrome type 1 (SARS-CoV) and SARS-CoV-2. SARS-CoV-2 has a high amino acid sequence homology to human betacoronavirus SARS-CoV and MERS and a much lower conservation with other coronaviruses. SARS-CoV-2 is proved to be highly pathogenic and cause severe lung injury and multi-organ failure. Currently, there is no vaccine approved to treat COVID-19. However, a few small size drugs originally developed to treat other diseases, such as remdesivir, which was initially developed for hepatitis C but proved to be ineffective, have been repurposed as an aid to treat severe cases of COVID-19. Although an earlier clinical trial in China (February/March 2020) showed that remdesivir was not effective in reducing deaths or the time for improvement from COVID-19 and caused various adverse effects, resulting in the early termination of the trial, remdesivir has been approved in the European Union for the treatment of COVID-19 in adults and adolescents with pneumonia requiring supplemental oxygen [1].
SARS-CoV-2 contains a single positive stranded-RNA genome of approximately 29,800 bases which encodes four structural proteins—Spike (S) glycoprotein, small Envelope (E) glycoprotein, Membrane (M) glycoprotein, and Nucleocapsid (N)—and sixteen non-structural proteins (NSP1–NSP16) that play important roles in virus transcription and replication [2,3,4,5] (Figure 1). The distinctive feature of eukaryotic mRNAs is characterised by the presence of a methylated 5′-cap structure that is required for mRNA stability. Viruses that infect eukaryotic organisms generally modify the 5′-cap of viral RNAs to mimic the hosts mRNA structure. In this manner, the virus protects itself from degradation by 5′-3′ exoribonucleases, enables efficient translation and escapes recognition by the host immune system. SARS-CoV-2 encodes one SAM-dependent methyltransferase (a 2′O-MTase also known as NSP16) that methylates the RNA cap at ribose 2′O positions. Importantly, this enzyme is only active in the presence of its activating partner, the non-structural protein NSP10. The formation of the NSP16/NSP10 complex, which follows a 1:1 stoichiometry ratio, has only been identified in coronaviruses. Crystal structures of the NSP16/NSP10 heterodimer in complex with the co-substrate and methyl donor SAM (Figure 2) (Protein Data Bank (PDB) IDs 6W4H, 6W61, 6W75 and 7C2J) [6]; with the reaction subproduct S-Adenosyl-L-Homocysteine (SAH) (Figure 2) (PDB ID 6WJT); with the inhibitor SFG (Figure 2) (PDB IDs 6WKQ and 6YZ1); in complex with RNA substrate (GTA) and SAM (PDB IDs 6WKS and 6WVN); and with GTA and SAH (PDB ID 6WQ3) have been reported over the years (Table S1 and Figure 1), providing important insight into the enzyme’s catalysis mechanism and its mode of interaction with small size ligands [7,8]. Interestingly, NSP16 can methylate GTA (mGpppA-RNA) but not GTG (mGpppG-RNA) substrates. One conserved groove region of the enzyme is essential for mRNA binding and defined by KDKE residues. Amino acid residue substitutions of the KDKE residues of NSP16 attenuate virus infection in vitro and in vivo.
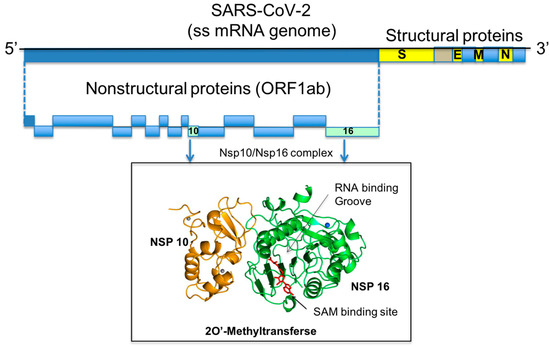
Figure 1.
Genome organisation of SARS-CoV-2. The regions encoding for the NSP10 and NSP16 proteins are highlighted and their 3D structures show in cartoon representation.
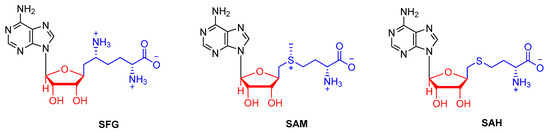
Figure 2.
Chemical structures of SFG, SAM and SAH.
In the present study, we initiated a virtual screening campaign on the SARS-CoV-2 methyltransferase NSP16, a 2′O-MTase with critical roles in viral replication and anti-viral immune response. In brief, we identified the key residues of NSP16 that are required for mRNA and cofactor binding and, therefore, for the regulation of 2′O-MTase catalytic activity (Table S1). Our virtual screening of SARS-CoV-2 NSP16 using a crystal structure of the NSP16/NSP10 complex (PDB ID 6YZ1) enabled us to rank the predicted binding affinities around the cofactor-binding site and identify 4 top hit compounds corresponding to 18 drugs that exhibit high chemical similarity with the NSP16 inhibitor SFG. Our studies confirm the high potential of targeting SARS-CoV-2 NSP16 catalytic activity with small size inhibitors to treat COVID-19 infections.
2. Results
2.1. Comparative Analysis and Structural Characterisation of 2′O-MTase from Coronavirus
Similar to other coronavirus, SARS-CoV-2 replicates in the cytoplasm of infected cells, enabling it to produce its own capping machinery. Its 2′O MTase enzyme, commonly referred to NSP16, in complex with the non-structural protein NSP10 catalyses the formation of viral mRNA cap structures that mimic those present in the host mRNAs [9]. The cap structure consists of a methylated guanosine at position 7 linked to the first transcribed nucleotide by a 5′-5′ triphosphate bridge (7mGpppN). The cap structure plays several key roles in mRNA functionality such as pre-mRNA processing, nuclear export and cap-dependent protein synthesis [10]. The cap structure is also essential for mRNA stability and limits its degradation by cellular 5′-3′ exoribonucleases. Structural similarity analysis of analogues of the natural NSP16 co-substrate SAM suggest that SAM-analogue compounds that act as competitive binders of SAM may be potent inhibitors of 2′O-MTase catalytic activity.
The human betacoronavirus SARS-CoV-2 shares a high amino acid sequence similarity to human betacoronaviruses SARS-CoV and MERS (Figure 3). The high SARS-CoV, MERS CoV and SARS-CoV-2 sequence similarity is reflected in their pathogenicity to the human, causing influenza-like symptoms that range from mild discomfort to severe lung injury and multi-organ failure, eventually leading to death.
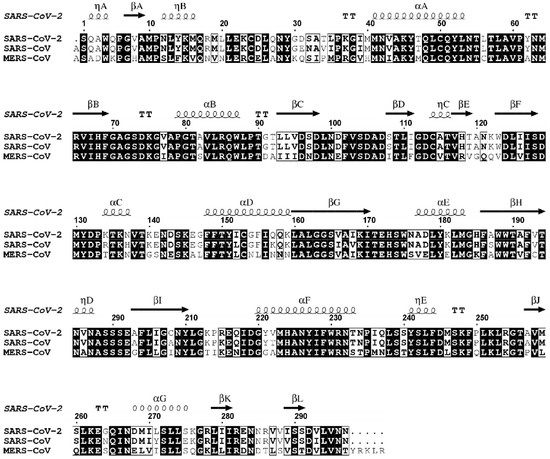
Figure 3.
Sequence alignment of 2′O-MTases (e.g., NSP16) viral proteins from SARS-CoV-2, SARS-CoV and MERS-CoV. Residues that are fully conserved are shown in solid black boxes; those with identity of 70% or higher are shown in white boxes. The secondary structure of SARS-CoV-2 NSP16 derived from the crystal structure (PDB ID 6WKQ) is shown above the alignments. Figure generated with the program ESPript 3.0 [11].
2.1.1. NSP16 is a Suitable Drug Target
Several crystal structures of NSP16 SARS-CoV-2 have been solved recently and the coordinates deposited in the PDB. These include NSP16 in complex with SAM (PDB IDs 6W4H, 6W61, 6W75 and 7C2J), with SAH (PDB ID 6WJT), with SFG (PDB IDs 6WKQ and 6YZ1), with SAM and RNA (PDB IDs 6WKS and 6WVN) and with SAH and RNA (PDB ID 6WQ3). SAH is the by-product of SAM-dependent methyl transfer reaction, whereas SFG is a SAM analogue that functions as a SAM competitive inhibitor (Table S1). The structures revealed that NSP16 adopts the catechol O-MTase fold containing alternating β strands (β1–β7) and α helices (αZ and αA–αE) that form a seven-stranded β sheet with three α helices on each side (Figure 1). Our analysis of these crystal structures confirmed that SAM, SAH and SFG bind to a well-defined pocket in NSP16, which is not surprising given the very slight variations of the chemical structure of these small molecules (Figure 4). SAM contains an additional sulphur atom in comparison to SAH, and, for SFG compared with SAM and SAH, a nitrogen substitutes the sulphur atoms. RNA binds to a region that is in close proximity to the SAM binding pocket with an orientation that allows its ribose ring to become in close proximity to the amino group of the SAM cofactor, which is required for the NSP16-dependant 2′O methylation of the RNA substrate (Figure 4). Moreover, the RNA structure does not seem to be altered upon SAM, SAH and SFG binding. Reciprocally, the mode of binding of these three molecules to NPS16 does not differ in the absence or presence of RNA.
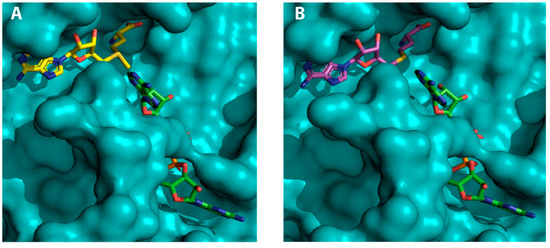
Figure 4.
SAM-dependent methyltransferases (MTases) ligands and RNA in complex with SARS-CoV-2 NSP16: (A) surface representation of the NSP16 (teal), SAM (yellow) and RNA (green) ternary complex; and (B) surface representation of NSP16 (teal), SAH (magenta) and RNA (green) ternary complex.
The deposited atomic coordinates of SARS-CoV-2 NSP16 crystal structures prompted us to investigate the structural similarities of this enzyme with a human cap-specific mRNA (nucleoside-2′O-)-MTase (PDB ID 4N48). Our analysis revealed two highly conserved elements. One is the SAM binding pocket, which defines the relative orientation of the methyl group donor. The other conserved element consists of the residues that define the enzyme active site (Figure 5). Interestingly, although the NPS16 cap-binding site can be mapped onto a similar region in SARS and MERS coronavirus, human and viral 2′O-ribose mRNA MTases interact with the guanosine cap very differently (Figure 6). The substantial difference in the overall topology of human and viral 2′O-ribose mRNA MTases strongly suggests that the latter can be specifically targeted with small molecules that occupy the SAM binding site. In this regard, it would be important to assess whether small-molecule antagonists of SAM binding to NSP16 of SARS-CoV-2 may be repurposed to limit viral replication and/or unmask viral RNA to the host immune system.
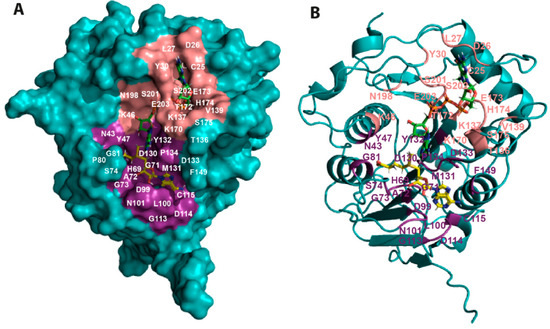
Figure 5.
Mapping the residues implicated in SAM and RNA binding onto the SARS-CoV-2 NSP16 structure. (A) Surface representation of SARS-CoV-2 NSP16 (teal) (PDB ID 6WKS) showing the relative positions of the SAM (yellow) and RNA (green) binding regions. The RNA molecule binds to a groove proximal to the SAM binding pocket. The NSP16 residues located within 4 Å of SAM and RNA are shown in deep salmon and purple colour, respectively. (B) Ribbon representation of the ternary complex in the same orientation as (A).
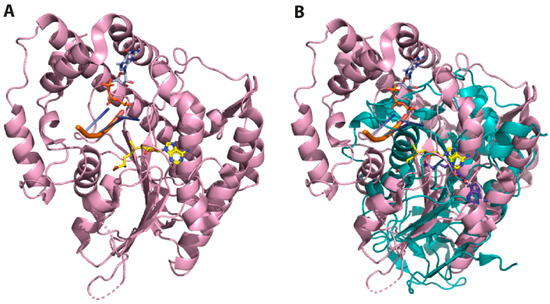
Figure 6.
Superposition of the 3D structures of SARS-CoV-2 and human 2′O-MTase. (A) Ribbon representation of human 2′O-MTase (magenta) in complex with SAM (yellow) and RNA (orange). (B) The superposition of SARS-CoV-2 (teal) and human 2′O-MTase (magenta) reveals substantial differences in their topology. The structures correspond to the PDB IDs 6WKQ and 4N48, respectively.
2.1.2. NSP16 Exhibits a Druggable Pocket
The first crystal structures of the non-structural protein NSP16 from SARS-CoV-2 revealed a well-defined co-substrate binding pocket. More recently, the crystal structure of NSP16 SARS-CoV-2 in complex with the small size compound SFG was solved and the coordinates deposited in the PDB (IDs 6WKQ and 6YZ1) (Figure 7). As 2′O-methylation of viral RNA cap structures limited the infected host’s antiviral response, small size compounds that interfere with NSP16 2′O-MTase catalytic activity are likely to undergo viral clearance in the infected cells.
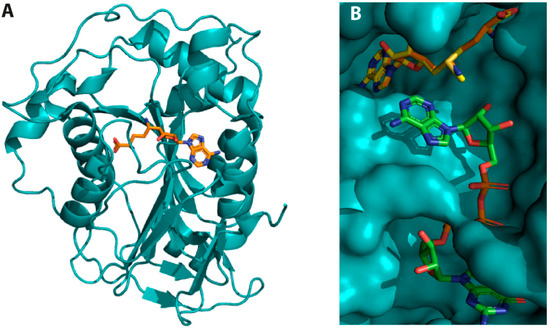
Figure 7.
Crystal structure of SARS-CoV-2 NSP16 complexes. (A) NSP16 in complex with the inhibitor SFG. The inhibitor (shown in orange) binds to the same region as SAM (PDB ID 6WKQ). (B) Structure superposition of SARS-CoV-2 NSP16 in complex with SAM (yellow) and RNA (green) (PDB ID 6WVN) with that of NSP16 in complex with SFG, a SAM analogue in which an amino group substitutes the sulphur atom of the former. The structure superposition shows that SAM and SFG bind to the same NSP16 pocket and that RNA binding implicates a region proximal to the SAM binding pocket.
2.2. Virtual Screening
Herein, we aimed to identify novel NSP16 inhibitors based on the SFG chemotype. For this purpose, we developed a High-Throughput Virtual Screening (HTVS) using the recently solved structure of the NSP10-NSP16 complex bound to the MTase inhibitor SFG (PDB ID 6YZ1) in an attempt to identify novel drug-like molecules with improved NSP16 binding and optimised pharmacokinetics profile.
A chemical library of compounds sharing >0.85 Tanimoto similarity index with SFG and complying Lipinski’s rules was retrieved from PubChem [12]. This database of 1321 molecules was used as the input source for our HTVS upon ligand preparation. The LigPrep module integrated in the Maestro software was used to optimise chemical structure conformations considering the most suitable ionisation state under physiological pH conditions and possible tautomers. This process allowed us to obtain low-energy molecules minimised using the OPLS3 force field [13]. It is worth noting that the Tanimoto similarity is calculated between a reference active molecule structure (commonly referred to as fingerprint) and multiple query fingerprints. Although, in principle, using a (>0.85) cut-off for the Tanimoto similarity measurement may lead to the exclusion of possible candidates with good 3D similarity to SFG, the use of the Schrödinger molecular modelling suite reduced this risk. This is because the Schrödinger modelling suite incorporates a minimised Structural Interaction Fingerprint (SIFt) approach that was originally developed by Deng and collaborators [14]. The SIFt integrated in Maestro (Schrödinger, LLC, New York, NY, 2019) includes nine interaction types (any contact, backbone contact, sidechain contact, polar contact, hydrophobic contact, H-bond donor and acceptor, aromatic and charged interactions).
This library of compounds was then subjected to molecular docking studies using the HTVS Glide-dock module integrated in the Schrödinger package [15,16]. High precision docking calculations using XP Glide (Schrödinger, LLC, New York, NY, 2019) were accomplished for the compounds with the highest docking scores in the HTVS. Selected candidates are shown in Figure 8, and their XP Glide interaction energies are displayed in Table 1. SFG was included in our studies to allow energetic comparison (the binding pose in which SFG conformation matches the crystallographic structure was selected, Figure S1). Docks of the most promising candidates are shown in Figure 9. Ligand interaction diagrams and the dock of 44367977 superimposed to SFG are shown in Figures S2 and S3. These docks show that, while maintaining the crucial interactions exhibited by the adenosine moiety of SFG, structural diversity of the alkyl chain is not only accepted but it even improves the predicted energy of complex formation.
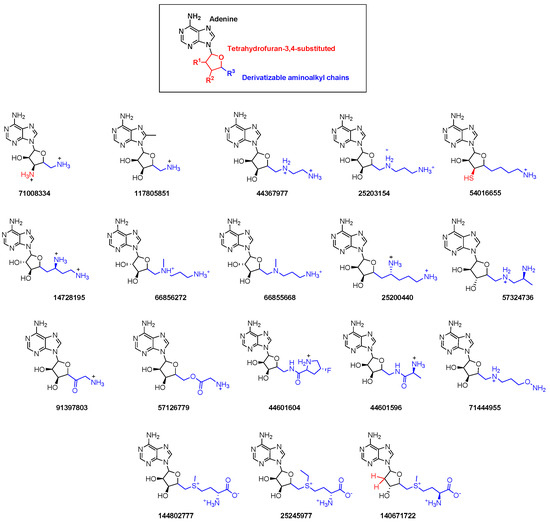
Figure 8.
Chemical structure of newly identified NSP16 inhibitors from the HTVS.

Table 1.
Physicochemical descriptors of selected compounds from the HTVS computed with QikProp and Glide docking score.
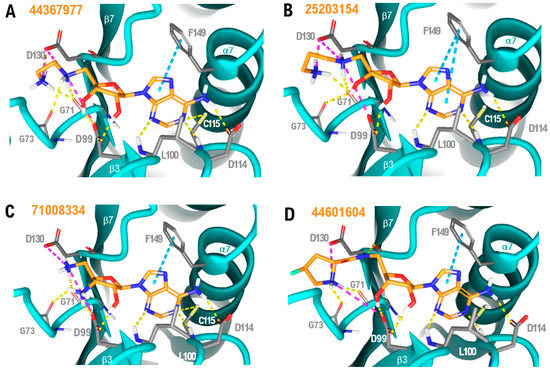
Figure 9.
NSP16-ligand complexes generated using our molecular docking strategy: (A) 44367977/NSP16; (B) 25203154/NSP16; (C) 71008334/NSP16; and (D) 44601604/NSP16. Yellow dashed lines represent hydrogen bonds, magenta dashed lines represent salt bridges and blue dashed lines show aromatic stacking; non-polar hydrogens are not shown. Direct interacting residues are displayed as grey tubes; the ligand chemical backbone is shown in orange with oxygen atoms highlighted in red and nitrogen atoms in blue.
The novel molecules demonstrate that the aminoacidic part of SFG or SAM can be substituted by a simple aminoalkyl chain to improve the number of stabilising interactions with NSP16. Conformationally restricted groups such as pyrrolidine, borne by 44601604, are also accepted in the pocket with an improved docking score when compared to SFG. As shown in Figure 9A–D, an additional salt bridge between the amine at position 2 of the alkyl chain (44367977 and 25203154), the amine of the pentose moiety (71008334) and the pyrrolidine nitrogen (44601604) and the NSP16 residue D99, as well as an extra hydrogen bond with the backbone carbonyl oxygen of NSP16 residue G73, can all contribute to stabilise the NSP16–drug-like compound complexes, resulting in improved docking energies when compared to SFG. It is worth noting that the terminal carboxylic acid group of SFG, SAM or SAH is not present in the alkyl chain of most of the top-ranking molecules. However, losing the hydrogen bond between the carboxylate and the NSP16 residue N43 (Figure S1) is energetically compensated by the aforementioned stabilising interactions. Concerning the pentose moiety, modification of the tetrahydrofuran group including an amino (71008334), a thiol (54016655) or the absence of substituent (140671722) at positions 3 or 4 are all predicted to be tolerated. One of the two hydroxyls is retained in all the best candidate hits. No substantial conformational changes were identified in the adenine group of the top-ranking molecules. Therefore, our results indicate the versatility of the amino acidic-like chain of SFG, whereas the nucleoside moiety seems to highly stabilise the complex. It is worth remarking that molecules with higher docking scores are dicationic (Table 1). This is likely the result of both cations engaging in stabilising polar interactions with the two negatively charged residues facing the binding site (D99 and D130, Figure 9), thus contributing to the global binding energy of the complex.
In the search of effective SARS-CoV-2 NSP16 inhibitors, it is crucial to avoid the presence of potential promiscuous moieties or Pan-Assay Interference compounds (PAINS) [17,18]. Therefore, the selected molecules were subjected to a PAINS identification study using the swissADME webserver [19]. Compounds 144802777, 25245977 and 140671722 bear potential liabilities (a sulfonium ion) and therefore were filtered out during compound selection. Moreover, in silico prediction of properties related to Absorption, Distribution, Metabolism, Excretion and Toxicity (ADMET) were calculated using the QikProp screening tool of Maestro (Schrödinger, LLC, New York, NY, 2019). Our drug-likeness calculations indicate that the molecules identified as potent NSP16 inhibitors abide to the Lipinski and Jorgensen pharmacokinetic rules [20,21]. As depicted in Table 1, the prediction of aqueous solubility, blood–brain barrier permeability, or binding to human serum albumin suggest that these candidates have a suitable drug profile. Predicted skin permeability values of some of the candidates fall outside the range predicted by QikProp for 95% of known oral drugs. However, they present better values than SFG. It is worth noting that the analogue 44601604 (Figure 9D) is the most promising candidate since it exhibits the best theoretical energy value of complex-formation and presents an optimal pharmacokinetics profile.
It is worth mentioning that, while SFG has already been widely studied, no biological activity for most of the identified compounds has been reported (Table S2). PubChem BioAssays for 44601596 and 44601604 showed either inactive or inconclusive results. This may allow the identification of more selective and efficacious hit compounds. Even though the best candidates obtained from our virtual screening shared similar main cores with SFG, a wider structural diversity was also explored, which led to lower score compounds that diverge at the adenine group position. Future work will aim to explore further structural diversity, including the systematic bioisosteric replacement of the molecular scaffold.
3. Discussion
Current efforts around the globe to treat COVID-19, the disease caused by the coronavirus SARS-CoV-2, are focused in the production of vaccines and small size drugs. On the one hand, vaccine development efforts are concentrated in the four main structural proteins: Spike (S) glycoprotein, small Envelope (E) glycoprotein, Membrane (M) glycoprotein and Nucleocapsid (N) protein. Although some vaccine development programmes have successfully completed phase 2 clinical trials, whether a SARS-CoV-2 vaccine will be able to elicit long-term immunity in the human population, regardless of specific ethnicity, gender and/or age group, remains to be seen. On the other hand, independent clinical trials around the world have shown that some drugs originally approved for their use in the human to treat a different disease condition, can be repurposed to aid the treatment of severe cases of COVID-19 infection that required hospitalisation and the use of mechanical respirators. Hence, the use of small size drugs arises as an important therapeutic strategy to treat COVID-19. Building on this knowledge, we developed a chemoinformatics campaign focused on the non-structural protein NSP16, an enzyme that presents 2′O-MTase activity and plays a key role in 2′O-methylation of viral RNA cap structures to limit the antiviral response of the host’s infected cells. Through this in silico study, SFG derivatives with a more potent and druggable profile than SFG were identified as potential NSP16 SARS-CoV-2 inhibitors. Top ranking derivatives bind to a well-defined pocket of the enzyme exhibiting the importance of specific residues of this cleft to establish a productive interaction with the small size ligands. The chemical space in the vicinity of the aminoacidic moiety of SFG was widely explored. It was noted that the most potent compounds preserve the adenosine core, thus confirming its relevance for the establishment of productive interactions with NSP16. The exploitation of these features will pave the way to design further broad-spectrum inhibitors that could not only target SARS-CoV-2 NSP16 but also the orthologue proteins found in other coronaviruses. For instance, the addition of a keto group at the appropriate position of the pyrrolidine ring of compound 44601604 should enable a further stabilising interaction with the NSP16 residue N43. Future work will aim at fine-tuning our chemical scaffold to design molecules that while maintaining desirable pharmacological features can also be extended to occupy the binding crevice.
4. Materials and Methods
4.1. Virtual Screening
The 2′O-MTase structure file from Protein Data Bank (PDB ID 6YZ1) was used in our modelling studies [22]. Initial protein optimisation was accomplished using the Protein Preparation Wizard module integrated in the Maestro suite software (Schrödinger, LLC, New York, NY, USA, 2019).
4.1.1. Ligand Preparation
A chemical library was downloaded from PubChem using the SFG chemotype as the lead structure. This PubChem dataset consists of 1321 compounds that comply the following criteria: Tanimoto similarity index >0.85 to SFG and no violation of Lipinsky’s rules for drug likeness [12]. The selected cut-off value of the Tanimoto coefficient enables the simultaneous optimisation of the SFG chemotype and the exploration of novel functional substitutions.
Low-energy three-dimensional conformations of the molecules were obtained using the LigPrep module of the Maestro suite (Schrödinger, LLC, New York, NY, USA, 2019). Additionally, the Epik software was employed to predict pKa values in the pH range between 7.0 and 7.5 and return all chemically sensible structures using Hammett and Taft methodology [23]. All compounds were minimised using the OPLS3 force field as implemented in Maestro [13].
4.1.2. High-Throughput Virtual Screening (HTVS)
Molecular docking was performed using the HTVS Glide-dock module integrated in the Schrödinger package [15,16]. The aforementioned chemical library of 1321 unique structures (considering diverse low energy conformers for each molecule) was explored in the binding site of our target 2′O-MTase. The docking grid was built using Glide and was centred on the position of the crystallised inhibitor, SFG, setting the dimensions of 20 Å. Ligand flexibility was used to explore an arbitrary number of torsional degrees of freedom, in addition to the six spatial degrees of freedom spanned by the translational and rotational parameters. Ligand poses generated in such a way were run through a series of hierarchical filters to evaluate ligand interactions with the receptor. Docking score, glide gscore, glide emodel, ionisation penalty and Topological Polar Surface Area (TPSA) were used to select the docking poses [15,16]. Molecules with docking scores lower than −10 Kcal/mol were selected for further docking studies. High precision docking calculations using the XP Glide were performed with the best candidates (18 molecules, Figure 8) and the lead compound SFG, which was included to allow comparison. Energy minimisation was performed using the OPLS3 force field 4.1.3 ADMET in silico calculations.
The SwissADME [19] server was used to evaluate the presence of promiscuous moieties in the selected candidates. Molecules 144802777, 25245977 and 140671722 were filtered off due to presence of a sulfonium ion, identified as PAINS, in their structure. The ADME/Tox profiles of the remaining 15 molecules identified by the HTVS were further explored. For this purpose, a set of 34 physicochemical descriptors was computed using QikProp version 3.5 integrated in Maestro (Schrödinger, LLC, New York, NY, USA). The QikProp descriptors are depicted in Table 1.
5. Conclusions
Using a similarity-based virtual screening approach, we propose the use of certain SFG analogues as SARS-CoV-2 NSP16 inhibitors. Best ranking molecules not only present improved docking energies but also better druggable pharmacokinetic profiles compared to the well-known pan 2′O-MTase inhibitor sinefungin (SFG). We anticipate that some of these compounds can be developed into therapeutic drugs against SARS-CoV-2 by limiting viral replication in the infected cells. Inhibition of the viral 2′O-MTase activity might stimulate the detection of viral RNAs by the host RIG-I or Mda5 antiviral pathway and restore the host antiviral response mediated by interferon-stimulated proteins that recognise mis-capped RNA and inhibit its translation. It is our hope that the present study on the identification and chemoinformatics characterisation of SARS-CoV-2 NPS16 MTase drug-like binders will stimulate others to join the global efforts to advance the development of molecules to tackle the pandemic COVID-19 disease.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4344/10/9/1023/s1. Table S1, IDs of the 3D-structure of the 2′O-methyltransferases; Figure S1, Molecular docking of SFG in NSP16; Figure S2, The 44367977/NSP16 complex with SFG superimposed; Figure S3, Ligand interaction diagrams of SFG, 44367977, 25203154, 71008334 and 44601604; Table S2, PubChem indexed biological assays results for the identified molecules and SFG.
Author Contributions
Conceptualisation, V.M.B.-G. and A.B.; methodology, P.M. and N.L.C.; software, P.M. and S.G.Z.; resources, S.G.Z.; writing—review and editing, P.M., N.L.C., S.G.Z., A.B. and V.M.B.-G.; and supervision, V.M.B.-G. and A.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by CSIC (CSIC-COV19-013) and MICINN (PID2019-105337RB-C21).
Acknowledgments
P.M. is grateful to the Comunidad de Madrid program “Atracción de Talento” number 2018-T2BMD-10819. N.L.C. is supported by a Professor Nigel Groome PhD Studentship, Oxford Brookes University. We thank the anonymous referees for their insightful and helpful comments and suggestions. We apologise to those colleagues whose important contributions to this field could not be cited due to space limitations.
Conflicts of Interest
The authors declare no conflict of interest.
References
- European Medicines Agency. First COVID-19 treatment recommended for EU authorisation. In European Medicines Agency EMA/264817; European Medicines Agency: Amsterdam, The Netherlands, 2020; pp. 1–2. [Google Scholar]
- Gorbalenya, A.E.; Baker, S.C.; Baric, R.S.; de Groot, R.J.; Drosten, C.; Gulyaeva, A.A.; Haagmans, B.L.; Lauber, C.; Leontovich, A.M.; Neuman, B.W.; et al. The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar]
- Perlman, S.; Netland, J. Coronaviruses post-SARS: Update on replication and pathogenesis. Nat. Rev. Microbiol. 2009, 7, 439–450. [Google Scholar] [CrossRef]
- Wu, A.; Peng, Y.; Huang, B.; Ding, X.; Wang, X.; Niu, P.; Meng, J.; Zhu, Z.; Zhang, Z.; Wang, J.; et al. Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell Host Microbe 2020, 27, 325–328. [Google Scholar] [CrossRef]
- Snijder, E.J.; Decroly, E.; Ziebuhr, J. The Nonstructural Proteins Directing Coronavirus RNA Synthesis and Processing. Adv. Virus Res. 2016, 96, 59–126. [Google Scholar] [PubMed]
- Chen, Y.; Su, C.; Ke, M.; Jin, X.; Xu, L.; Zhang, Z.; Wu, A.; Sun, Y.; Yang, Z.; Tien, P.; et al. Biochemical and structural insights into the mechanisms of sars coronavirus RNA ribose 2′-O-methylation by nsp16/nsp10 protein complex. PLoS Pathog. 2011, 7, e1002294. [Google Scholar] [CrossRef] [PubMed]
- Decroly, E.; Debarnot, C.; Ferron, F.; Bouvet, M.; Coutard, B.; Imbert, I.; Gluais, L.; Papageorgiou, N.; Sharff, A.; Bricogne, G.; et al. Crystal structure and functional analysis of the SARS-coronavirus RNA cap 2′-o-methyltransferase nsp10/nsp16 complex. PLoS Pathog. 2011, 7, e1002059. [Google Scholar] [CrossRef]
- Rosas-Lemus, M.; Minasov, G.; Shuvalova, L.; Inniss, N.; Brunzelle, J.; Satchell, K.J.F. Structure of SARS-CoV-2 2′-O-methyltransferase heterodimer with RNA Cap analog and sulfates bound reveals new strategies for structure-based inhibitor design. Bio Rxiv 2020, 1–26. [Google Scholar]
- Hyde, J.L.; Diamond, M.S. Innate immune restriction and antagonism of viral RNA lacking 2′-O methylation. Virology 2015, 479–480, 66–74. [Google Scholar] [CrossRef]
- Ramanathan, A.; Robb, G.B.; Chan, S.H. mRNA capping: Biological functions and applications. Nucleic Acids Res. 2016, 44, 7511–7526. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, 320–324. [Google Scholar] [CrossRef]
- Kim, S.; Thiessen, P.A.; Bolton, E.E.; Chen, J.; Fu, G.; Gindulyte, A.; Han, L.; He, J.; He, S.; Shoemaker, B.A.; et al. PubChem substance and compound databases. Nucleic Acids Res. 2016, 44, D1202–D1213. [Google Scholar] [CrossRef] [PubMed]
- Harder, E.; Damm, W.; Maple, J.; Wu, C.; Reboul, M.; Xiang, J.Y.; Wang, L.; Lupyan, D.; Dahlgren, M.K.; Knight, J.L.; et al. OPLS3: A Force Field Providing Broad Coverage of Drug-like Small Molecules and Proteins. J. Chem. Theory Comput. 2016, 12, 281–296. [Google Scholar] [CrossRef]
- Deng, Z.; Chuaqui, C.; Singh, J. Structural Interaction Fingerprint (SIFt): A Novel Method for Analyzing Three-Dimensional Protein-Ligand Binding Interactions. J. Med. Chem. 2004, 47, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Warren, G.L.; Andrews, C.W.; Capelli, A.M.; Clarke, B.; LaLonde, J.; Lambert, M.H.; Lindvall, M.; Nevins, N.; Semus, S.F.; Senger, S.; et al. A critical assessment of docking programs and scoring functions. J. Med. Chem. 2006, 49, 5912–5931. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef]
- Capuzzi, S.J.; Muratov, E.N.; Tropsha, A. Phantom PAINS: Problems with the Utility of Alerts for Pan-Assay Interference Compounds. J. Chem. Inf. Model. 2017, 57, 417–427. [Google Scholar] [CrossRef]
- Baell, J.B.; Holloway, G.A. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 2010, 53, 2719–2740. [Google Scholar] [CrossRef]
- Daina, A.O.M.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Lipinski, C.A. Drug-like properties and the causes of poor solubility and poor permeability. J. Pharmacol. Toxicol. Methods 2001, 44, 235–249. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Duffy, E.M. Prediction of drug solubility from structure. Adv. Drug Deliv. Rev. 2002, 54, 355–366. [Google Scholar] [CrossRef]
- Krafcikova, P.; Silhan, J.; Nencka, R.; Boura, E. Structural analysis of the SARS-CoV-2 methyltransferase complex involved in coronaviral RNA cap creation. Bio Rxiv 2020, 11, 1–7. [Google Scholar]
- Greenwood, J.R.; Calkins, D.; Sullivan, A.P.; Shelley, J.C. Towards the comprehensive, rapid, and accurate prediction of the favorable tautomeric states of drug-like molecules in aqueous solution. J. Comput. Aided. Mol. Des. 2010, 24, 591–604. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).