Facile One-Pot Immobilization of a Novel Thermostable Carboxylesterase from Geobacillus uzenensis for Continuous Pesticide Degradation in a Packed-Bed Column Reactor
Abstract
1. Introduction
2. Results and Discussion
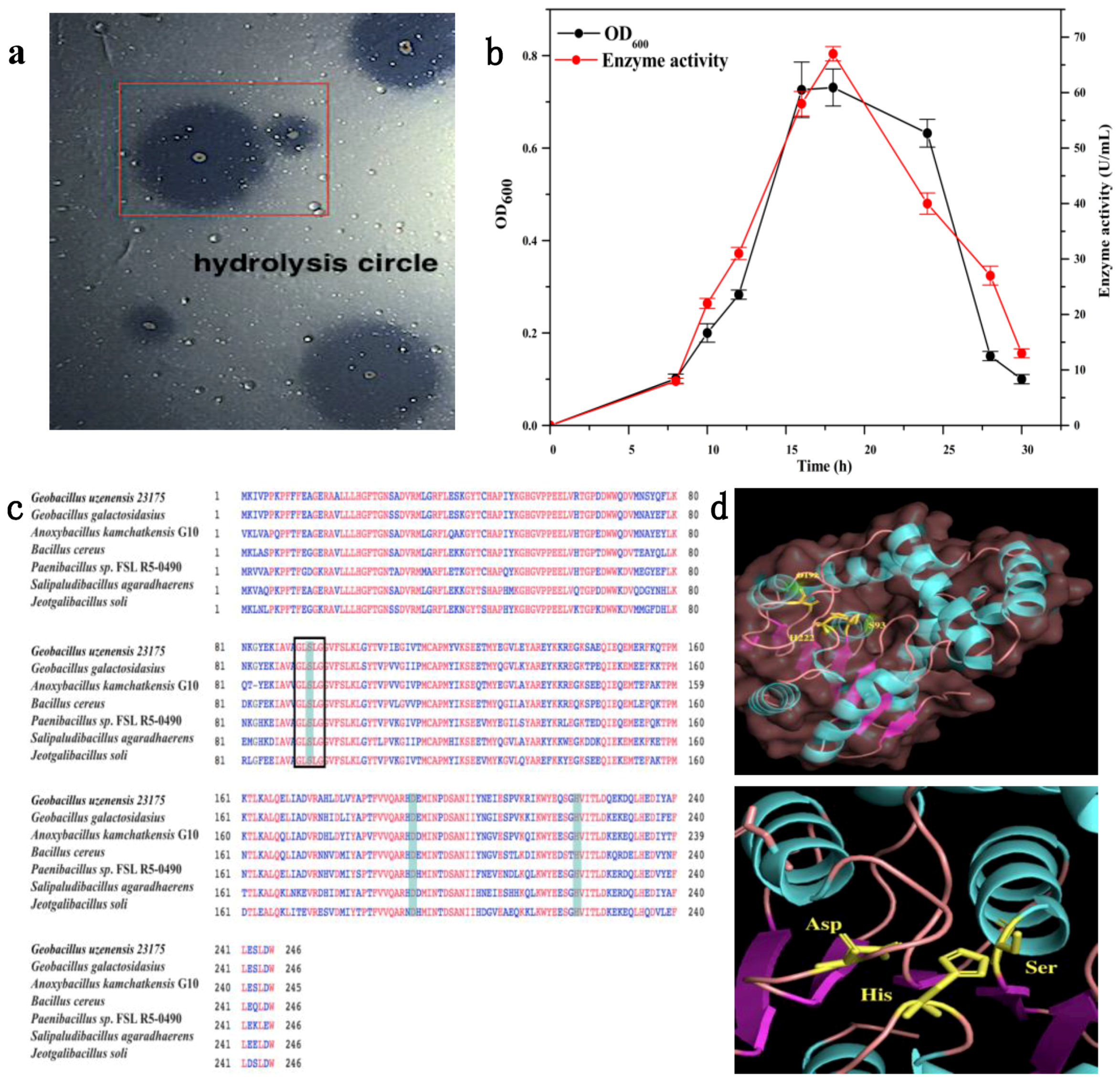
2.1. Fermentation of Wild Strain
2.2. Gene Analysis and Molecular Structure Modeling of Est741
2.3. Cloning, Expression, and Purification of the Original Enzyme Est741 and the Mutant Enzyme EstM160K
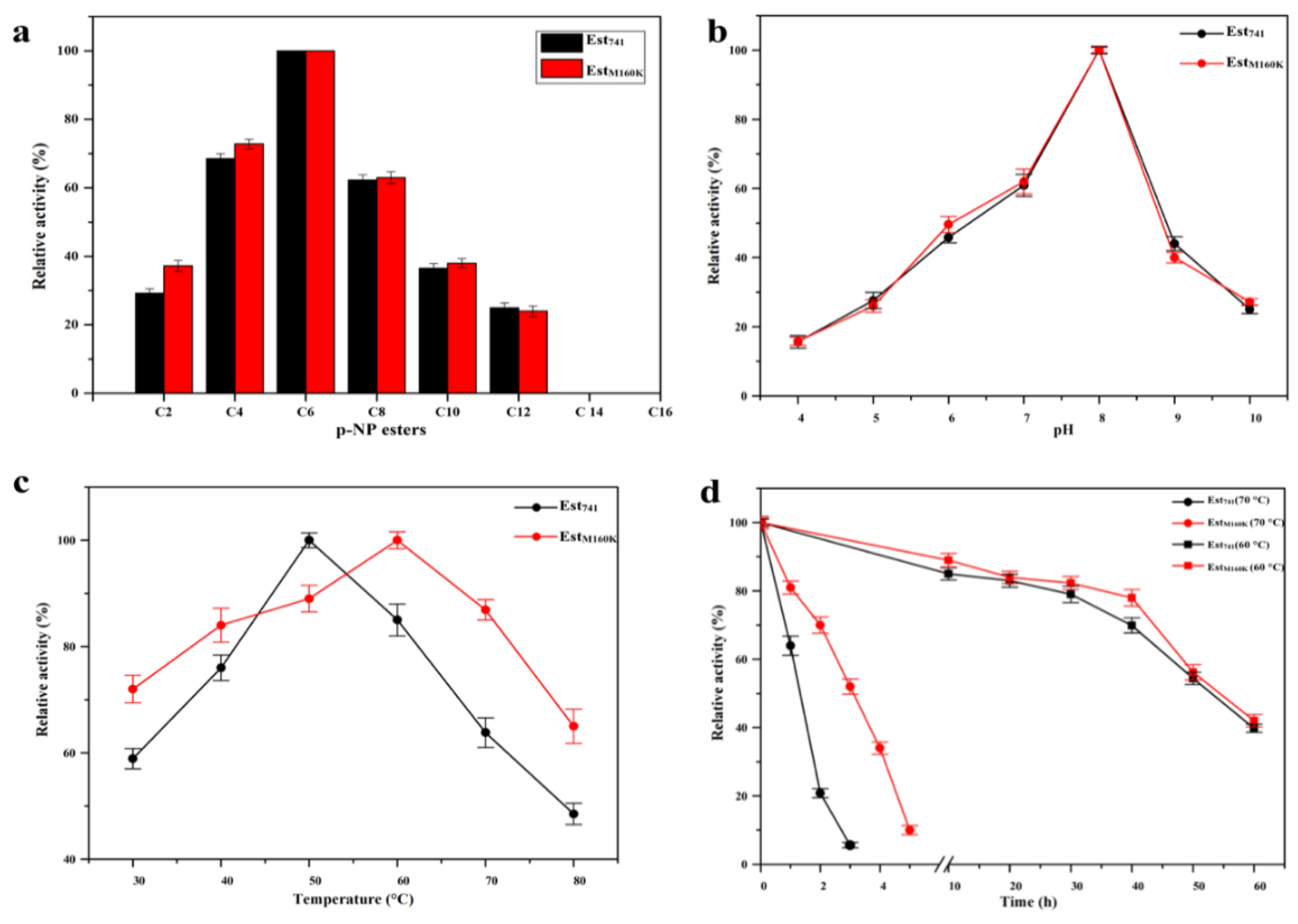
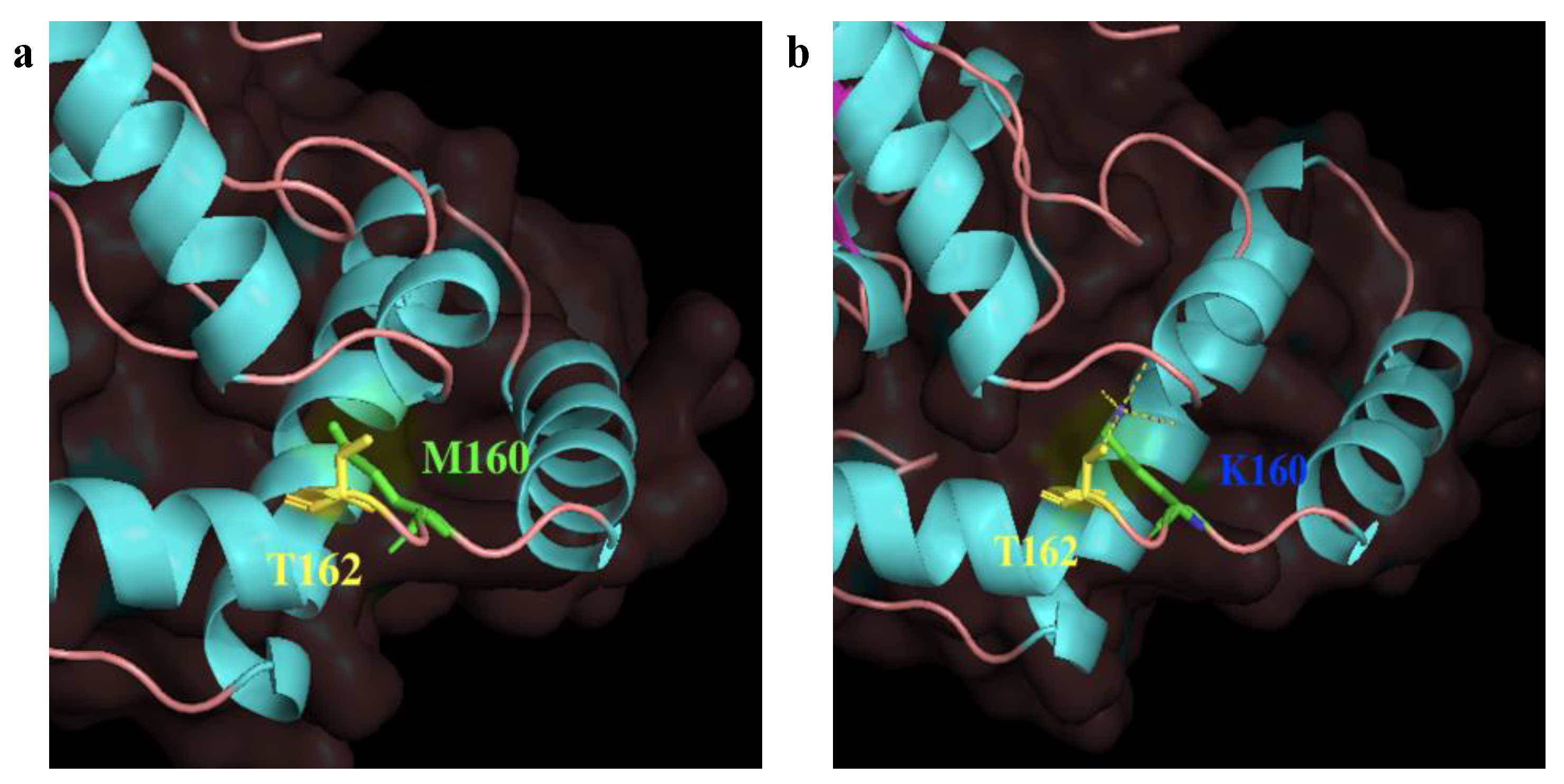
2.4. Enzymatic Properties of Est741/EstM160K and Analysis of the Change of Properties of the Mutant Enzyme EstM160K
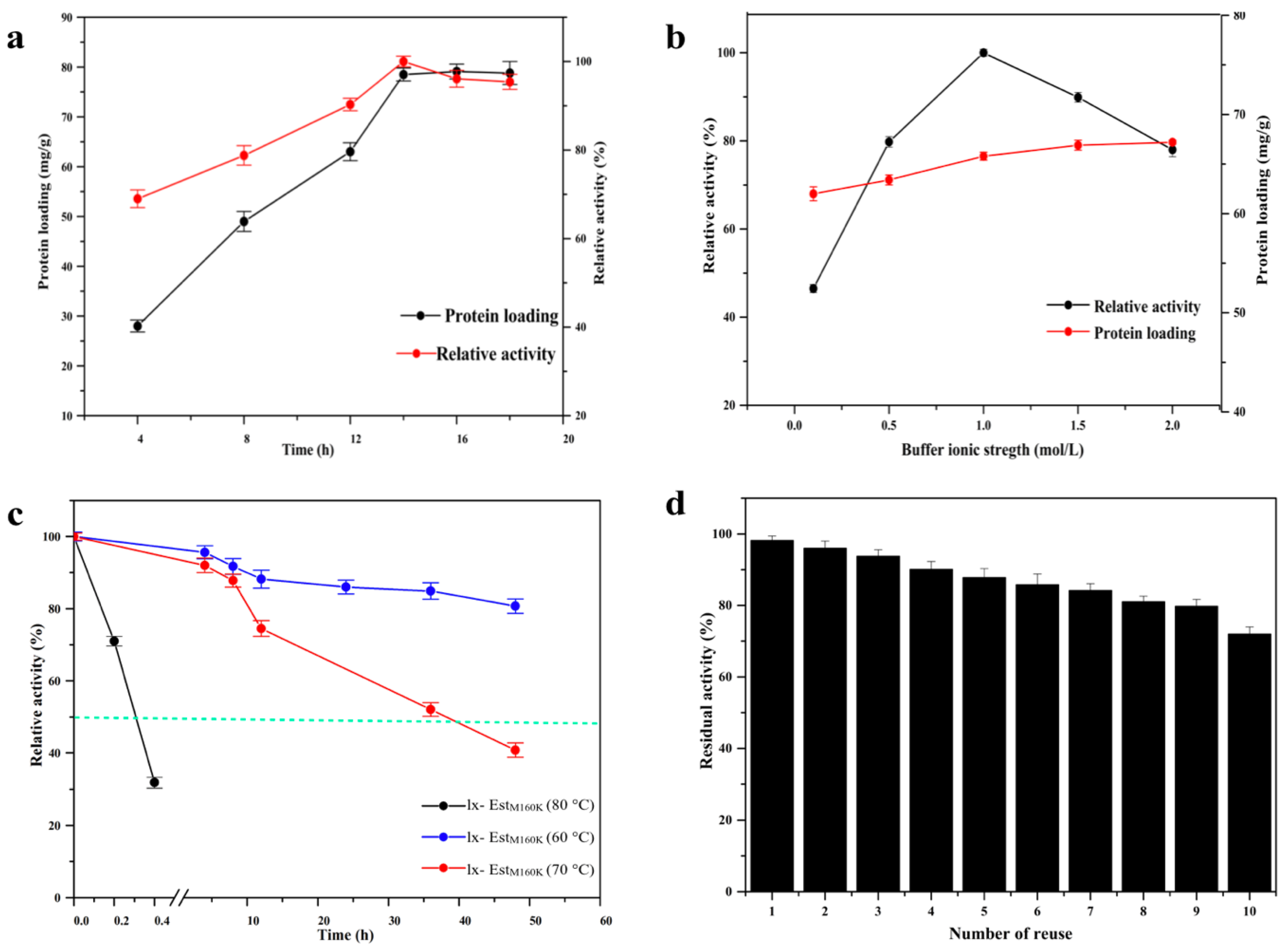
2.5. Study of the Properties of Immobilized lx-EstM160K
2.6. Degradation of Organophosphorus Pesticide Malathion
2.7. Degradation of Pyrethroid Pesticide Wastewater in Packed-Bed Column Reactor
3. Materials and Methods
3.1. Materials
3.2. Plasmids and Strains
3.3. Fermentation of Wild Strain
3.4. Sequence Analysis of Est741
3.5. Expression and Purification of Recombinant Carboxylesterase in E. coli and P. pastoris
3.6. Analysis of the Enzymatic Properties of Est741
3.7. Improving the Enzymatic Properties of Est741 by Site-Directed Mutagenesis
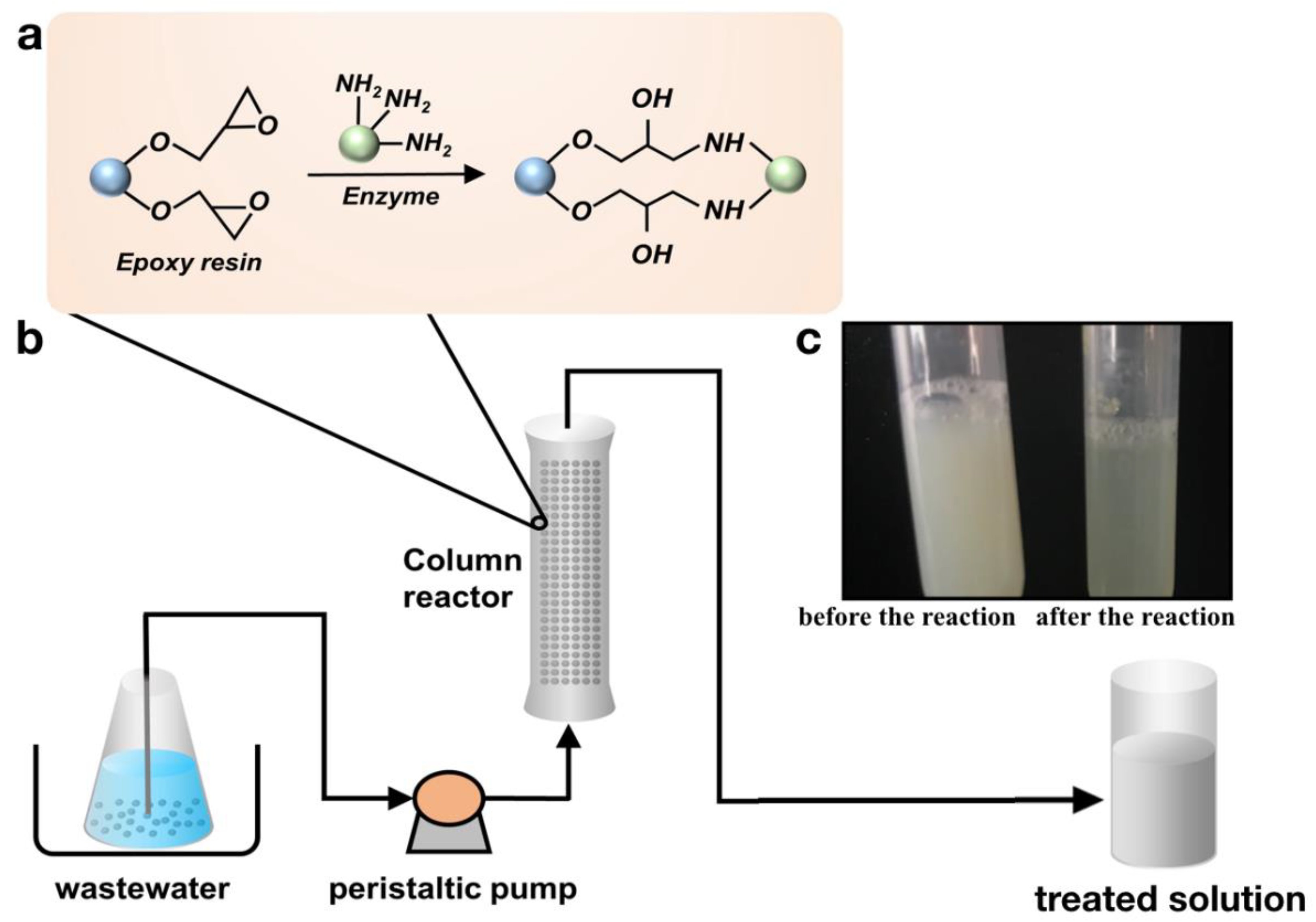
3.8. Immobilization of the Recombinant Esterase on Epoxy Resin and Determination of the Properties of the Enzyme
3.9. Self-Made Radial Packed-Bed Column Reactor and Degradation of Pesticides
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AOX | alcohol oxidase |
| CTAB | cetyltrimethylammonium bromide |
| EDTA | ethylene diamine tetraacetic acid |
| PBR | packed bed reactor |
| SDS | sodium dodecyl sulfate |
| SDS-PAGE | sodium dodecyl sulfate polyacrylamide gel electrophoresis |
| SEM | scanning electron microscopy |
| UV-Vis | ultraviolet and visible spectrophotometry |
References
- Cai, X.; Wang, W.; Lin, L.; He, D.; Huang, G.; Shen, Y.; Wei, W.; Wei, D. Autotransporter domain-dependent enzymatic analysis of a novel extremely thermostable carboxylesterase with high biodegradability towards pyrethroid pesticides. Sci. Rep. 2017, 7, 3461. [Google Scholar] [CrossRef]
- Le, L.T.H.L.; Yoo, W.; Jeon, S.; Kim, K.K.; Kim, T.D. Characterization and Immobilization of a Novel SGNH Family Esterase (LaSGNH1) from Lactobacillus acidophilus NCFM. Int. J. Mol. Sci. 2020, 21, 91. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, P.; Huang, Y.; Zhan, H.; Chen, S. Insight into microbial applications for the biodegradation of pyrethroid insecticides. Front. Microbiol. 2019, 10, 1778. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Li, M.; Yan, Y. Heterologous expression and characterization of a new lipase from Pseudomonas fluorescens Pf0–1 and used for biodiesel production. Sci. Rep. 2017, 7, 15711. [Google Scholar] [CrossRef]
- Kambiranda, D.M.; Asraful-Islam, S.M.; Cho, K.M.; Math, R.K.; Lee, Y.H.; Kim, H.; Yun, H.D. Expression of esterase gene in yeast for organophosphates biodegradation. Pestic. Biochem. Physiol. 2009, 94, 15–20. [Google Scholar] [CrossRef]
- Zuo, Z.; Gong, T.; Che, Y.; Liu, R.; Xu, P.; Jiang, H.; Qiao, C.; Song, C.; Yang, C. Engineering Pseudomonas putida KT2440 for simultaneous degradation of organophosphates and pyrethroids and its application in bioremediation of soil. Biodegradation 2015, 26, 223–233. [Google Scholar] [CrossRef]
- Gudiukaite, R.; Sadauskas, M.; Gegeckas, A.; Malunavicius, V.; Citavicius, D. Construction of a novel lipolytic fusion biocatalyst GDEst-lip for industrial application. J. Ind. Microbiol. Biotechnol. 2017, 44, 799–815. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, M.; Beecher, D.; Mangan, D.; Rowan, A.S.; Monte, A.; Sroka, S.; Modregger, J.; Hundle, B.; Moody, T.S. A novel lipase enzyme panel exhibiting superior activity and selectivity over lipase B from Candida antarctica for the kinetic resolution of secondary alcohols. Tetrahedron Asymmetry 2012, 23, 583–586. [Google Scholar] [CrossRef]
- Kandhari, N.; Sinha, S. Complex network analysis of thermostable mutants of Bacillus subtilis Lipase A. Appl. Netw. Sci. 2017, 2, 18. [Google Scholar] [CrossRef][Green Version]
- Mohammadi, M.; Sepehrizadeh, Z.; Ebrahim-Habibi, A.; Shahverdi, A.R.; Faramarzi, M.A.; Setayesh, N. Enhancing activity and thermostability of lipase A from Serratia marcescens by site-directed mutagenesis. Enzyme. Microb. Technol. 2016, 93–94, 18–28. [Google Scholar] [CrossRef]
- Heidari, R.; Devonshire, A.L.; Campbell, B.E.; Dorrian, S.J.; Oakeshott, J.G.; Russell, R.J. Hydrolysis of pyrethroids by carboxylesterases from Lucilia cuprina and Drosophila melanogaster with active sites modified by in vitro mutagenesis. Insect Biochem. Mol. Biol. 2005, 35, 597–609. [Google Scholar] [CrossRef] [PubMed]
- Angkawidjaja, C.; Koga, Y.; Takano, K. Structure and stability of a thermostable carboxylesterase from the thermoacidophilic archaeon Sulfolobus tokodai. FEBS J. 2012, 279, 3071–3084. [Google Scholar] [CrossRef]
- Chen, B.; Hu, J.; Miller, E.M.; Xie, W.; Cai, M.; Gross, R.A. Candida antarctica lipase B chemically immobilized on epoxy-activated micro- and nanobeads: Catalysts for polyester synthesis. Biomacromolecules 2008, 9, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Mihailović, M.; Stojanović, M.; Banjanac, K.; Carević, M.; Prlainović, N.; Milosavić, N.; Bezbradica, D. Immobilization of lipase on epoxy-activated Purolite ® A109 and its post-immobilization stabilization. Process Biochem. 2014, 49, 637–646. [Google Scholar] [CrossRef]
- Singh, B.; Kaur, J.; Singh, K. Biodegradation of malathion by Brevibacillus sp. strain KB2 and Bacillus cereus strain PU. World J. Microb. Biotechnol. 2012, 28, 1133–1141. [Google Scholar] [CrossRef]
- Tazdaït, D.; Abdi, N.; Lounici, H.; Grib, H.; Mameri, N.; Pauss, A. Biodegradation of malathion with indigenous acclimated activated sludge in batch mode and in continuous-flow packed-bed reactor. Bioremediation J. 2013, 17, 294–304. [Google Scholar] [CrossRef]
- Salum, T.F.C.; Villeneuve, P.; Barea, B.; Yamamoto, C.I.; Côcco, L.C.; Mitchell, D.A.; Krieger, N. Synthesis of biodiesel in column fixed-bed bioreactor using the fermented solid produced by Burkholderia cepacia LTEB11. Process Biochem. 2010, 45, 1348–1354. [Google Scholar] [CrossRef]
- Nguyen, L.N.; Hai, F.I.; Dosseto, A. Continuous adsorption and biotransformation of micropollutants by granular activated carbon-bound laccase in a packed-bed enzyme reactor. Bioresour. Technol. 2016, 210, 108–116. [Google Scholar] [CrossRef]
- Karpushova, A.; Brümmer, F.; Barth, S.; Lange, S.; Schmid, R.D. Cloning, recombinant expression and biochemical characterisation of novel esterases from Bacillus sp. associated with the marine sponge Aplysina aerophoba. Appl. Microbiol. Biotechnol. 2005, 67, 59–69. [Google Scholar] [CrossRef]
- Chiş, L.; Hriscu, M.; Bica, A.; Toşa, M.; Nagy, G.; Róna, G.; G Vértessy, B.; Dan, I.F. Molecular cloning and characterization of a thermostable esterase/lipase produced by a novel Anoxybacillus flavithermus strain. J. Gen. Appl. Microbiol. 2013, 59, 119. [Google Scholar] [CrossRef]
- Unrean, P. Pathway analysis of Pichia pastoris to elucidate methanol metabolism and its regulation for production of recombinant proteins. Biotechnol. Progr. 2014, 30, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Madan, B.; Mishra, P. Directed evolution of Bacillus licheniformis lipase for improvement of thermostability. Biochem. Eng. J. 2014, 91, 276–282. [Google Scholar] [CrossRef]
- Kumar, R.; Singh, R.; Kaur, J. Characterization and molecular modelling of an engineered organic solvent tolerant, thermostable lipase with enhanced enzyme activity. J. Mol. Catal. B Enzym. 2013, 97, 243–251. [Google Scholar] [CrossRef]
- Li, C.; Tan, T.; Zhang, H.; Feng, W. Analysis of the conformational stability and activity of Candida antarctica lipase B in organic solvents insight from molecular dynamics and quantum mechanics/simulations. J. Biol. Chem. 2010, 285, 28434–28441. [Google Scholar] [CrossRef] [PubMed]
- Souza, S.P.D.; Almeida, R.A.D.; Garcia, G.G.; Leão, R.A.; Bassut, J.; Souza, R.O.D.; Itabaiana, I., Jr. Immobilization of lipase B from Candida antarctica on epoxy-functionalized silica: Characterization and improving biocatalytic parameters. J. Chem. Technol. Biotechnol. 2018, 93, 105–111. [Google Scholar] [CrossRef]
- Li, X.; Li, D.; Wang, W.; Durrani, R.; Bo, Y.; Wang, Y. Immobilization of SMG1-F278N lipase onto a novel epoxy resin: Characterization and its application in synthesis of partial glycerides. J. Mol. Catal. B Enzym. 2016, 133, 154–160. [Google Scholar] [CrossRef]
- Ren, Y.; He, B.; Yan, F.; Wang, H.; Cheng, Y.; Lin, L.; Feng, Y.; Li, J. Continuous biodiesel production in a fixed bed reactor packed with anion-exchange resin as heterogeneous catalyst. Bioresour. Technol. 2011, 113, 19–22. [Google Scholar] [CrossRef]
- Mateo, C.; Grazu, V.; Palomo, J.M.; Lopezgallego, F.; Fernandezlafuente, R.; Guisan, J.M. Immobilization of enzymes on heterofunctional epoxy supports. Nat. Protoc. 2007, 2, 1022–1033. [Google Scholar] [CrossRef]
- Palomo, J.M.; Muñoz, G.; Gloria, F.L. Modulation of Mucor miehei lipase properties via directed immobilization on different hetero-functional epoxy resins: Hydrolytic resolution of (R, S)-2-butyroyl-2-phenylacetic acid. J. Mol. Catal. B Enzym. 2003, 21, 201–210. [Google Scholar] [CrossRef]
- Rodrigues, J.; Canet, A.; Rivera, I. Biodiesel production from crude Jatropha oil catalyzed by non-commercial immobilized heterologous Rhizopus oryzae and Carica papaya lipases. Bioresour. Technol. 2016, 213, 88–95. [Google Scholar] [CrossRef]
- Goda, S.K.; Elsayed, I.E.; Khodair, T.A.; El-Sayed, W.; Mohamed, M.E. Screening for and isolation and identification of malathion-degrading bacteria: Cloning and sequencing a gene that potentially encodes the malathion-degrading enzyme, carboxylestrase in soil bacteria. Biodegradation 2010, 21, 903–913. [Google Scholar] [CrossRef]
- Xie, Z.; Xu, B.; Ding, J.; Liu, L.; Zhang, X.; Li, J.; Huang, Z. Heterologous expression and characterization of a malathion-hydrolyzing carboxylesterase from a thermophilic bacterium, Alicyclobacillus tengchongensis. Biotechnol. Lett. 2013, 35, 1283–1289. [Google Scholar] [CrossRef]
- Schofield, D.A.; Dinovo, A.A. Generation of a mutagenized organophosphorus hydrolase for the biodegradation of the organophosphate pesticides malathion and demeton-S. J. Appl. Microbiol. 2010, 109, 548–557. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.P.; Jiang, T.; Liu, X.; Zheng, Y.G. Efficient production of S-(+)-2-chlorophenylglycine by immobilized penicillin G acylase in a recirculating packed bed reactor. Biochem. Eng. J. 2013, 74, 88–94. [Google Scholar] [CrossRef]
- Feng, Y.; Zhang, A.; Li, J.; He, B. A continuous process for biodiesel production in a fixed bed reactor packed with cation-exchange resin as heterogeneous catalyst. Bioresour. Technol. 2012, 113, 19–22. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Basu, A.; Kashyap, V.; Roberts, D. Experimental and numerical analysis of biological regeneration of perchlorate laden ion-exchange resins in batch reactors. Environ. Eng. Sci. 2010, 27, 75–83. [Google Scholar] [CrossRef]
- Priji, P.; Unni, K.N.; Sajith, S.; Binod, P.; Benjamin, S. Production, optimization, and partial purification of lipase from Pseudomonas sp. strain BUP6, a novel rumen bacterium characterized from Malabari goat. Biotechnol. Appl. Biochem. 2015, 62, 71–78. [Google Scholar] [CrossRef]
- Babaki, M.; Yousefi, M.; Habibi, Z.; Brask, J.; Mohammadi, M. Preparation of highly reusable biocatalysts by immobilization of lipases on epoxy-functionalized silica for production of biodiesel from canola oil. Biochem. Eng. J. 2015, 101, 23–31. [Google Scholar] [CrossRef]
- Jeganathan, J.; Nakhla, G.; Bassi, A. Hydrolytic pretreatment of oily wastewater by immobilized lipase. J. Hazard. Mater. 2007, 145, 127–135. [Google Scholar] [CrossRef]
- Harshit, D.; Charmy, K.; Nrupesh, P. Organophosphorus pesticides determination by novel HPLC and spectrophotometric method. Food Chem. 2017, 230, 448–453. [Google Scholar] [CrossRef]
- Peng, G.; He, Q.; Lu, Y.; Mmereki, D.; Zhong, Z. Determination of organophosphorus pesticides and their major degradation product residues in food samples by HPLC-UV. Environ. Sci. Pollut. Res. Int. 2016, 23, 19409–19416. [Google Scholar] [CrossRef] [PubMed]
- Bouri, M.; Salghi, R.; Bazzi, L.; Zarrouk, A.; Rios, A.; Zougagh, M. Pesticide residue levels in green beans cultivated in Souss Masa valley (Morocco) after multiple applications of bifenthrin and λ-cyhalothrin. Bull. Environ. Contam. Toxicol. 2012, 89, 638–643. [Google Scholar] [CrossRef] [PubMed]
| Methanol Concentration (W/V, %) | Dry Cell Weight (g/L) | Relative Activity (%) |
|---|---|---|
| 0.5 | 86.30 ± 0.28 | 86.2 ± 0.3 |
| 1.0 | 90.09 ± 0.43 | 100.0 |
| 1.5 | 82.86 ± 0.41 | 91.3 ± 0.6 |
| Organic Solvents | Residual Activity (%) | Ion | Residual Activity (%) | Surfactants and EDTA | Residual Activity (%) |
|---|---|---|---|---|---|
| Control | 100 ± 0 | Control | 100 ± 0 | Control | 100 ± 0 |
| Methanol | 91.5 ± 2.5 | Mg2+ | 89.5 ± 3.1 | Tween20 | 93.8 ± 2.2 |
| Ethanol | 75.6 ± 2.3 | Cu2+ | 85.2 ± 2.6 | Tween60 | 90.8 ± 2.0 |
| Acetonitrile | 63.8 ± 2.3 | Ca2+ | 103.4 ± 1.8 | Tween80 | 87.8 ± 1.9 |
| Isopropanol | 71.1 ± 1.9 | Mn2+ | 97.0 ± 2.2 | SDS | 92.4 ± 2.5 |
| Acetone | 87.9 ± 1.5 | Zn2+ | 76.1 ± 3.7 | TritonX-100 | 43.5 ± 3.5 |
| n-hexane | 73.2 ± 2.9 | Co2+ | 92.7 ± 2.9 | EDTA | 103.2 ± 3.1 |
| n-propanol | 72.5 ± 3.2 | Na+ | 94.5 ± 3.3 | ||
| Isooctane | 76.4 ± 2.6 | Fe3+ | 102.8 ± 2.4 | ||
| Benzene | 61.8 ± 2.2 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, X.; Tang, X.; Dong, F.; Lin, L.; Wei, W.; Wei, D. Facile One-Pot Immobilization of a Novel Thermostable Carboxylesterase from Geobacillus uzenensis for Continuous Pesticide Degradation in a Packed-Bed Column Reactor. Catalysts 2020, 10, 518. https://doi.org/10.3390/catal10050518
Yang X, Tang X, Dong F, Lin L, Wei W, Wei D. Facile One-Pot Immobilization of a Novel Thermostable Carboxylesterase from Geobacillus uzenensis for Continuous Pesticide Degradation in a Packed-Bed Column Reactor. Catalysts. 2020; 10(5):518. https://doi.org/10.3390/catal10050518
Chicago/Turabian StyleYang, Xiaohui, Xudong Tang, Fengying Dong, Lin Lin, Wei Wei, and Dongzhi Wei. 2020. "Facile One-Pot Immobilization of a Novel Thermostable Carboxylesterase from Geobacillus uzenensis for Continuous Pesticide Degradation in a Packed-Bed Column Reactor" Catalysts 10, no. 5: 518. https://doi.org/10.3390/catal10050518
APA StyleYang, X., Tang, X., Dong, F., Lin, L., Wei, W., & Wei, D. (2020). Facile One-Pot Immobilization of a Novel Thermostable Carboxylesterase from Geobacillus uzenensis for Continuous Pesticide Degradation in a Packed-Bed Column Reactor. Catalysts, 10(5), 518. https://doi.org/10.3390/catal10050518