The “Green” FMOs: Diversity, Functionality and Application of Plant Flavoproteins
Abstract
1. Introduction
2. Monooxygenase Enzymes Play Critical Roles in Plant Metabolism
3. Plants possess Class B Flavoproteins
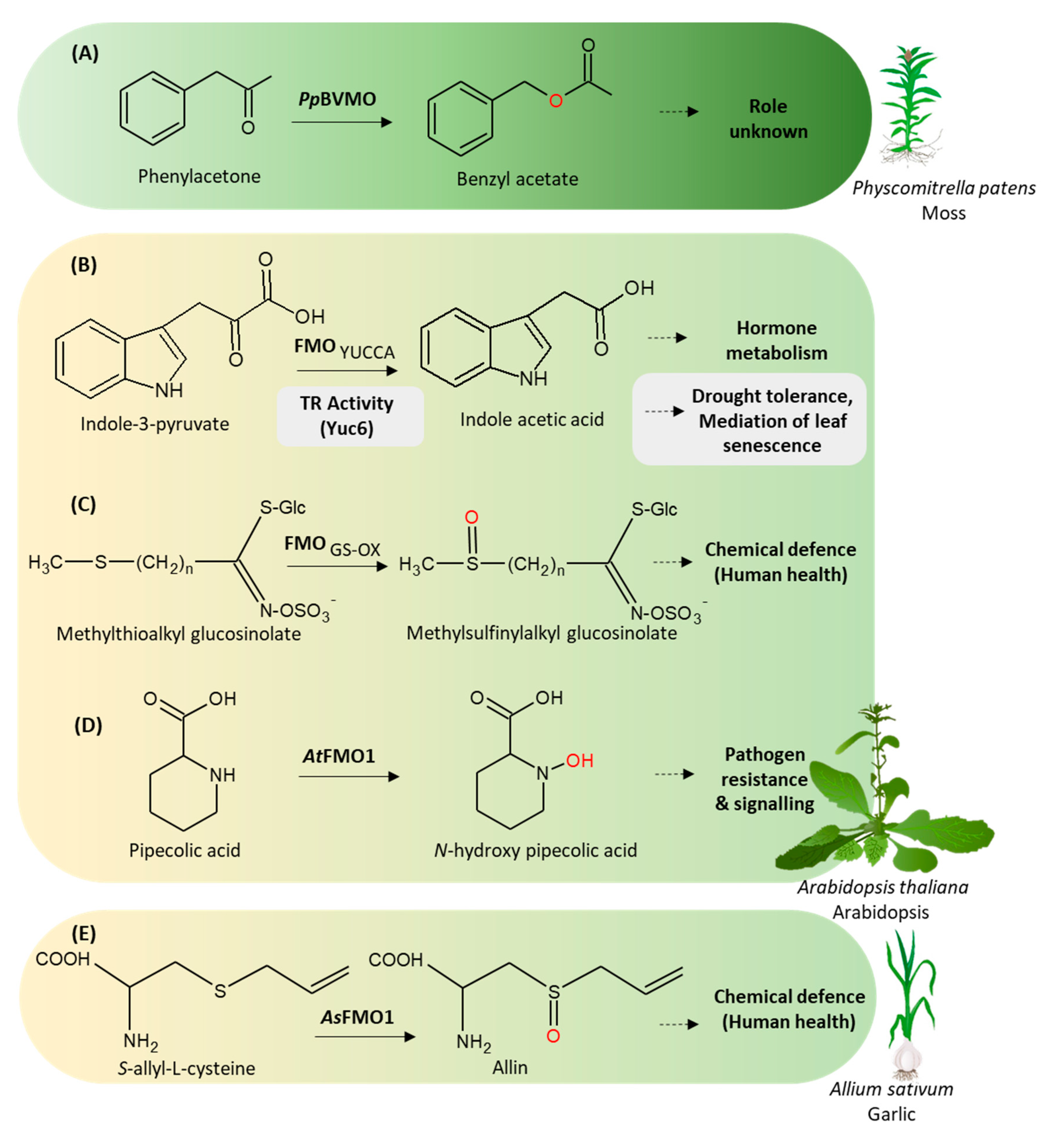
3.1. N-hydroxylating Monooxygenases
3.2. Baeyer-Villiger Monooxygenases
BVMO in Moss (Physcomitrella Patens)
3.3. Class B Flavoproteins (BFMOs)
3.3.1. YUCCAs
3.3.2. S-oxidizing FMOs
3.3.3. N-oxidizing FMOs
4. The Road Ahead for Plant FMO Research
4.1. Identification of Substrates
4.2. A Need for Structural Insights of Plant FMOs
4.3. Optimizing Strategies for Plant FMO Biochemical Studies
4.4. What’s in a Name? FMO Nomenclature
5. The Unexploited Potential of Plant FMOs for Agriculture and Biotechnology
6. Conclusions
Supplementary Materials
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BFMO | Class B Flavin-containing monooxygenase |
| BVMO | Baeyer-Villiger monooxygenase |
| CYP | Cytochrome P450 monooxygenase |
| FAD | Flavin Adenine Dinucleotide |
| IAA | Indole acetic acidIPA: Indole-3-pyruvate IPTG: Isopropyl β-D-1-thiogalactopyranoside |
| NADH/NADPH | Nicotinamide coenzyme |
| N-OH-Pip | N-hydroxy pipecolic acid |
| NHMO | N-hydroxylating monooxygenase |
| PAMO | Phenylacetone monooxygenase |
| Pip | Pipecolic acid |
| ROS | Reactive oxygen species |
| SAR | Systemic acquired resistance |
| TAM | Tryptamine |
| TR | thiol-reductase |
References
- Van Berkel, W.J.H.; Kamerbeek, N.M.; Fraaije, M.W. Flavoprotein monooxygenases, a diverse class of oxidative biocatalysts. J. Biotechnol. 2006, 124, 670–689. [Google Scholar] [CrossRef]
- Gul, T.; Krzek, M.; Permentier, H.P.; Fraaije, M.W.; Bischoff, R. Microbial flavoprotein monooxygenases as mimics of mammalian flavin-containing monooxygenases for the enantioselective preparation of drug metabolites. Drug Metab. Dispos. 2016, 44, 1270–1276. [Google Scholar] [CrossRef]
- Ceccoli, R.D.; Bianchi, D.A.; Rial, D. V Flavoprotein monooxygenases for oxidative biocatalysis: Recombinant expression in microbial hosts and applications. Front. Microbiol. 2014, 5, 25. [Google Scholar] [CrossRef]
- Mthethwa, K.S.; Kassier, K.; Engel, J.; Kara, S.; Smit, M.S.; Opperman, D.J. Fungal BVMOs as alternatives to cyclohexanone monooxygenase. Enzyme Microb. Technol. 2017, 106, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Van Beilen, J.B.; Duetz, W.A.; Schmid, A.; Witholt, B. Practical issues in the application of oxygenases. Trends Biotechnol. 2003, 21, 170–177. [Google Scholar] [CrossRef]
- Mitchell, A.J.; Weng, J.K. Unleashing the synthetic power of plant oxygenases: From mechanism to application. Plant Physiol. 2019, 179, 813–829. [Google Scholar] [CrossRef] [PubMed]
- Krueger, S.K.; Williams, D.E. Mammalian flavin-containing monooxygenases: Structure/function, genetic polymorphisms and role in drug metabolism. Pharmacol. Ther. 2005, 106, 357–387. [Google Scholar] [CrossRef] [PubMed]
- Hyland, R.; Jones, B.C.; Smith, D.A. Identification of the Cytochrome P450 Enzymes Involved in the N-Oxidation of Voriconazole. Drug Metab. Dispos. 2003, 31, 540–547. [Google Scholar] [CrossRef] [PubMed]
- Yanni, S.B.; Annaert, P.P.; Augustijns, P.; Bridges, A.; Gao, Y.; Benjamin, D.K.; Thakker, D.R. Role of Flavin-Containing Monooxygenase in Oxidative Metabolism of Voriconazole by Human Liver Microsomes. Drug Metab. Dispos. 2008, 36, 1119–1125. [Google Scholar] [CrossRef]
- Cashman, J.R. Some distinctions between flavin-containing and cytochrome P450 monooxygenases. Biochem. Biophys. Res. Commun. 2005, 338, 599–604. [Google Scholar] [CrossRef]
- Ziegler, D.M. An overview of the mechanism, substrate specificities, and structure of FMOs. Drug Metab. Rev. 2002, 34, 503–511. [Google Scholar] [CrossRef]
- Riebel, A.; De Gonzalo, G.; Fraaije, M.W. Expanding the biocatalytic toolbox of flavoprotein monooxygenases from Rhodococcus jostii RHA1. J. Mol. Catal. B Enzym. 2013, 88, 20–25. [Google Scholar] [CrossRef]
- Butinar, L.; Mohorčič, M.; Deyris, V.; Duquesne, K.; Iacazio, G.; Claeys-Bruno, M.; Friedrich, J.; Alphand, V. Prevalence and specificity of Baeyer–Villiger monooxygenases in fungi. Phytochemistry 2015, 117, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Phillips, I.R.; Shephard, E.A. Drug metabolism by flavin-containing monooxygenases of human and mouse. Expert Opin. Drug Metab. Toxicol. 2017, 13, 167–181. [Google Scholar] [CrossRef] [PubMed]
- Cashman, J.R.; Park, S.B.; Berkman, C.E.; Cashman, L.E. Role of hepatic flavin-containing monooxygenase 3 in drug and chemical metabolism in adult humans. Chem. Biol. Interact. 1995, 96, 33–46. [Google Scholar] [CrossRef]
- Rossner, R.; Kaeberlein, M.; Leiser, S.F. Flavin-containing monooxygenases in aging and disease: Emerging roles for ancient enzymes. J. Biol. Chem. 2017, 292, 11138–11146. [Google Scholar] [CrossRef] [PubMed]
- Cashman, J.R. Role of flavin-containing monooxygenase in drug development. Expert Opin. Drug Metab. Toxicol. 2008, 4, 1507–1521. [Google Scholar] [CrossRef] [PubMed]
- Canyelles, M.; Tondo, M.; Cedó, L.; Farràs, M.; Escolà-Gil, J.C.; Blanco-Vaca, F. Trimethylamine N-Oxide: A Link among Diet, Gut Microbiota, Gene Regulation of Liver and Intestine Cholesterol Homeostasis and HDL Function. Int. J. Mol. Sci. 2018, 19, 3228. [Google Scholar] [CrossRef]
- Han, G.H.; Shin, H.-J.; Kim, S. Optimization of bio-indigo production by recombinant E. coli harboring fmo gene. Enzym. Microb. Technol. - Enzym. MICROB TECHNOL 2008, 42, 617–623. [Google Scholar] [CrossRef]
- Meyer, A.; Held, M.; Schmid, A.; Kohler, H.-P.E.; Witholt, B. Synthesis of 3-tert-butylcatechol by an engineered monooxygenase. Biotechnol. Bioeng. 2003, 81, 518–524. [Google Scholar] [CrossRef]
- Schlaich, N.L. Flavin-containing monooxygenases in plants: Looking beyond detox. Trends Plant Sci. 2007, 12, 412–418. [Google Scholar] [CrossRef] [PubMed]
- Pichersky, E.; Lewinsohn, E. Convergent Evolution in Plant Specialized Metabolism. Annu. Rev. Plant Biol. 2011, 62, 549–566. [Google Scholar] [CrossRef] [PubMed]
- Weng, J.K. The evolutionary paths towards complexity: A metabolic perspective. New Phytol. 2014, 201, 1141–1149. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.; Werck-Reichhart, D. A P450-centric view of plant evolution. Plant J. 2011, 66, 194–211. [Google Scholar] [CrossRef]
- Mascotti, M.L.; Juri Ayub, M.; Furnham, N.; Thornton, J.M.; Laskowski, R.A. Chopping and Changing: The Evolution of the Flavin-dependent Monooxygenases. J. Mol. Biol. 2016, 428, 3131–3146. [Google Scholar] [CrossRef]
- Ziegler, D.M. Flavin-containing monooxygenases: Catalytic mechanism and substrate specificities. Drug Metab. Rev. 1988, 19, 1–32. [Google Scholar] [CrossRef]
- Furnham, N.; Sillitoe, I.; Holliday, G.L.; Cuff, A.L.; Laskowski, R.A.; Orengo, C.A.; Thornton, J.M. Exploring the evolution of novel enzyme functions within structurally defined protein superfamilies. PLoS Comput. Biol. 2012, 8. [Google Scholar] [CrossRef]
- Reeves, G.A.; Dallman, T.J.; Redfern, O.C.; Akpor, A.; Orengo, C.A. Structural Diversity of Domain Superfamilies in the CATH Database. J. Mol. Biol. 2006, 360, 725–741. [Google Scholar] [CrossRef]
- Fraaije, M.W.; Kamerbeek, N.M.; Van Berkel, W.J.H.; Janssen, D.B. Identification of a Baeyer-Villiger monooxygenase sequence motif. FEBS Lett. 2002, 518, 43–47. [Google Scholar] [CrossRef]
- Hansen, B.G.; Kliebenstein, D.J.; Halkier, B.A. Identification of a flavin-monooxygenase as the S-oxygenating enzyme in aliphatic glucosinolate biosynthesis in Arabidopsis. Plant J. 2007, 50, 902–910. [Google Scholar] [CrossRef]
- Catucci, G.; Zgrablic, I.; Lanciani, F.; Valetti, F.; Minerdi, D.; Ballou, D.P.; Gilardi, G.; Sadeghi, S.J. Characterization of a new Baeyer-Villiger monooxygenase and conversion to a solely N-or S-oxidizing enzyme by a single R292 mutation. Biochim. Biophys. Acta - Proteins Proteomics 2016, 1864, 1177–1187. [Google Scholar] [CrossRef] [PubMed]
- Malito, E.; Alfieri, A.; Fraaije, M.W.; Mattevi, A. Crystal structure of a Baeyer—Villiger monooxygenase. Proc. Natl. Acad. Sci. USA 2004, 101, 13157–13162. [Google Scholar] [CrossRef] [PubMed]
- Stehr, M.; Diekmann, H.; Smau, L.; Seth, O.; Ghisla, S.; Singh, M.; Macheroux, P. A hydrophobic sequence motif common to N-hydroxylating enzymes. Trends Biochem. Sci. 1998, 23, 56–57. [Google Scholar] [CrossRef]
- Beneventi, E.; Niero, M.; Motterle, R.; Fraaije, M.; Bergantino, E. Discovery of Baeyer–Villiger monooxygenases from photosynthetic eukaryotes. J. Mol. Catal. B Enzym. 2013, 98, 145–154. [Google Scholar] [CrossRef]
- Badieyan, S.; Bach, R.D.; Sobrado, P. Mechanism of N -hydroxylation catalyzed by flavin-dependent monooxygenases. J. Org. Chem. 2015, 80, 2139–2147. [Google Scholar] [CrossRef] [PubMed]
- Olucha, J.; Lamb, A.L. Mechanistic and structural studies of the N-hydroxylating flavoprotein monooxygenases. Bioorg. Chem. 2011, 39, 171–177. [Google Scholar] [CrossRef]
- Meneely, K.M.; Lamb, A.L. Biochemical characterization of a flavin adenine dinucleotide-dependent monooxygenase, ornithine hydroxylase from Pseudomonas aeruginosa, suggests a novel reaction mechanism. Biochemistry 2007, 46, 11930–11937. [Google Scholar] [CrossRef]
- Albelda-Berenguer, M.; Monachon, M.; Joseph, E. Siderophores: From natural roles to potential applications, 1st ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2019; Volume 106, ISBN 9780128169759. [Google Scholar]
- De Serrano, L.O.; Camper, A.K.; Richards, A.M. An overview of siderophores for iron acquisition in microorganisms living in the extreme. BioMetals 2016, 29, 551–571. [Google Scholar] [CrossRef]
- Renshaw, J.C.; Robson, G.D.; Trinci, A.P.J.; Wiebe, M.G.; Livens, F.R.; Collison, D.; Taylor, R.J. Fungal siderophores: Structures, functions and applications. Mycol. Res. 2002, 106, 1123–1142. [Google Scholar] [CrossRef]
- Nguyen, K.; DeSieno, M.A.; Bae, B.; Johannes, T.W.; Cobb, R.E.; Zhao, H.; Nair, S.K. Characterization of the flavin monooxygenase involved in biosynthesis of the antimalarial FR-900098. Org. Biomol. Chem. 2019, 17, 1506–1518. [Google Scholar] [CrossRef]
- Mascotti, M.L.; Lapadula, W.J.; Ayub, M.J. The origin and evolution of Baeyer - Villiger Monooxygenases (BVMOs): An ancestral family of flavin monooxygenases. PLoS ONE 2015, 10, 0132689. [Google Scholar] [CrossRef] [PubMed]
- Riebel, A.; Dudek, H.M.; De Gonzalo, G.; Stepniak, P.; Rychlewski, L.; Fraaije, M.W. Expanding the set of rhodococcal Baeyer-Villiger monooxygenases by high-throughput cloning, expression and substrate screening. Appl. Microbiol. Biotechnol. 2012, 95, 1479–1489. [Google Scholar] [CrossRef] [PubMed]
- Baeyer, A.; Villiger, V. The Effect of Caro’s Reagent on Ketones. Ber. Dtsch. Chem. Ges 1899, 32, 3625–3633. [Google Scholar] [CrossRef]
- Renz, M.; Meunier, B. 100 Years of Baeyer–Villiger Oxidations. European J. Org. Chem. 1999, 1999, 737–750. [Google Scholar] [CrossRef]
- van Beek, H.L.; de Gonzalo, G.; Fraaije, M.W. Blending Baeyer–Villiger monooxygenases: Using a robust BVMO as a scaffold for creating chimeric enzymes with novel catalytic properties. Chem. Commun. 2012, 48, 3288–3290. [Google Scholar] [CrossRef]
- Tolmie, C.; Smit, M.S.; Opperman, D.J. Native roles of Baeyer-Villiger monooxygenases in the microbial metabolism of natural compounds. Nat. Prod. Rep. 2019, 36, 326–353. [Google Scholar] [CrossRef]
- Fraaije, M.W.; Wu, J.; Heuts, D.P.H.M.; van Hellemond, E.W.; Spelberg, J.H.L.; Janssen, D.B. Discovery of a thermostable Baeyer–Villiger monooxygenase by genome mining. Appl. Microbiol. Biotechnol. 2005, 66, 393–400. [Google Scholar] [CrossRef]
- Biochem, J. Steroid Monooxygenase of Rhodococcus rhodochrous the Genomic DNA, and Hyperexpression, Purification, Characterization of the Recombinant: Sequencing and of Steroid monooxygenase of Rhodococcus rhodochrous is a Baeyer-Villigerase catalyzing the inserti. J. Biochem. 1999, 631, 624–631. [Google Scholar]
- Huijbers, M.M.E.; Montersino, S.; Westphal, A.H.; Tischler, D.; Van Berkel, W.J.H. Flavin dependent monooxygenases. Arch. Biochem. Biophys. 2014, 544, 2–17. [Google Scholar] [CrossRef]
- Zane, N.R.; Chen, Y.; Wang, M.Z.; Thakker, D.R. Cytochrome P450 and flavin-containing monooxygenase families: Age-dependent differences in expression and functional activity. Pediatr. Res. 2018, 83, 527–535. [Google Scholar] [CrossRef]
- Zhao, Y.; Christensen, S.K.; Fankhauser, C.; Cashman, J.R.; Cohen, J.D.; Weigel, D.; Chory, J. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 2001, 291, 306–309. [Google Scholar] [CrossRef]
- Cheng, Y.; Dai, X.; Zhao, Y. Auxin Synthesized by the YUCCA Flavin Monooxygenases Is Essential for Embryogenesis and Leaf Formation in Arabidopsis. Plant Cell 2007, 19, 2430–2439. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Dai, X.; Zhao, Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 2006, 20, 1790–1799. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Jung, J.-H.; Han, D.-Y.; Seo, P.J.; Park, W.J.; Park, C.-M. Activation of a flavin monooxygenase gene YUCCA7 enhances drought resistance in Arabidopsis. Planta 2012, 235, 923–938. [Google Scholar] [CrossRef] [PubMed]
- Müller-Moulï, P.; Nozue, K.; Pytlak, M.L.; Palmer, C.M.; Covington, M.F.; Wallace, A.D.; Harmer, S.L.; Maloof, J.N. YUCCA auxin biosynthetic genes are required for Arabidopsis shade avoidance. PeerJ 2016, 2016, 1–18. [Google Scholar]
- Tivendale, N.D.; Ross, J.J.; Cohen, J.D. The shifting paradigms of auxin biosynthesis. Trends Plant Sci. 2014, 19, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Stepanova, A.N.; Yun, J.; Robles, L.M.; Novak, O.; He, W.; Guo, H.; Ljung, K.; Alonso, J.M. The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell 2011, 23, 3961–3973. [Google Scholar] [CrossRef]
- Won, C.; Shen, X.; Mashiguchi, K.; Zheng, Z.; Dai, X.; Cheng, Y.; Kasahara, H.; Kamiya, Y.; Chory, J.; Zhao, Y. Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011. [Google Scholar] [CrossRef]
- Mashiguchi, K.; Tanaka, K.; Sakai, T.; Sugawara, S.; Kawaide, H.; Natsume, M.; Hanada, A.; Yaeno, T.; Shirasu, K.; Yao, H.; et al. The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 18512–18517. [Google Scholar] [CrossRef]
- Marsch-martinez, N.; Greco, R.; Van Arkel, G.; Herrera-estrella, L.; Pereira, A. Activation Tagging Using the. Society 2002, 129, 1544–1556. [Google Scholar]
- Woodward, C.; Bemis, S.M.; Hill, E.J.; Sawa, S.; Koshiba, T.; Torii, K.U. Interaction of Auxin and ERECTA in Elaborating Arabidopsis Inflorescence Architecture Revealed by the Activation Tagging of a New Member of the YUCCA Family Putative Flavin Monooxygenases. Plant Physiol. 2005, 139, 192–203. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.I.; Sharkhuu, A.; Jin, J.B.; Li, P.; Jeong, J.C.; Baek, D.; Lee, S.Y.; Blakeslee, J.J.; Murphy, A.S.; Bohnert, H.J.; et al. yucca6, a dominant mutation in Arabidopsis, affects auxin accumulation and auxin-related phenotypes. Plant Physiol. 2007, 145, 722–735. [Google Scholar] [CrossRef] [PubMed]
- Tobeña-Santamaria, R.; Bliek, M.; Ljung, K.; Sandberg, G.; Mol, J.N.M.; Souer, E.; Koes, R. Floozy of petunia is a flavin mono-oxygenase-like protein required for the specification of leaf and flower architecture. Genes Dev. 2002, 16, 753–763. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Kamiya, N.; Morinaka, Y.; Matsuoka, M.; Sazuka, T. Auxin Biosynthesis by the YUCCA Genes in Rice. Plant Physiol. 2007, 143, 1362–1371. [Google Scholar] [CrossRef]
- Fujino, K.; Matsuda, Y.; Ozawa, K.; Nishimura, T.; Koshiba, T.; Fraaije, M.W.; Sekiguchi, H. NARROW LEAF 7 controls leaf shape mediated by auxin in rice. Mol. Genet. Genomics 2008, 279, 499–507. [Google Scholar] [CrossRef]
- Expósito-Rodríguez, M.; Borges, A.A.; Borges-Pérez, A.; Hernández, M.; Pérez, J.A. Cloning and Biochemical Characterization of ToFZY, a Tomato Gene Encoding a Flavin Monooxygenase Involved in a Tryptophan-dependent Auxin Biosynthesis Pathway. J. Plant Growth Regul. 2007, 26, 329–340. [Google Scholar] [CrossRef]
- Gallavotti, A.; Barazesh, S.; Malcomber, S.; Hall, D.; Jackson, D.; Schmidt, R.J.; McSteen, P. sparse inflorescence1 encodes a monocot-specific YUCCA-like gene required for vegetative and reproductive development in maize. Proc. Natl. Acad. Sci. USA 2008, 105, 15196–15201. [Google Scholar] [CrossRef]
- Park, H.C.; Cha, J.Y.; Yun, D.J. Roles of YUCCAs in auxin biosynthesis and drought stress responses in plants. Plant Signal. Behav. 2013, 8. [Google Scholar]
- Liu, H.; Xie, W.-F.; Zhang, L.; Valpuesta, V.; Ye, Z.-W.; Gao, Q.-H.; Duan, K. Auxin biosynthesis by the YUCCA6 flavin monooxygenase gene in woodland strawberry. J. Integr. Plant Biol. 2014, 56, 350–363. [Google Scholar] [CrossRef]
- Yan, S.; Che, G.; Ding, L.; Chen, Z.; Liu, X.; Wang, H.; Zhao, W.; Ning, K.; Zhao, J.; Tesfamichael, K.; et al. Different cucumber CsYUC genes regulate response to abiotic stresses and flower development. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef]
- Cao, X.; Yang, H.; Shang, C.; Ma, S.; Liu, L.; Cheng, J. The Roles of Auxin Biosynthesis YUCCA Gene Family in Plants. Int. J. Mol. Sci. 2019, 20, 6343. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Mashiguchi, K.; Chen, Q.; Kasahara, H.; Kamiya, Y.; Ojha, S.; DuBois, J.; Ballou, D.; Zhao, Y. The biochemical mechanism of auxin biosynthesis by an arabidopsis YUCCA flavin-containing monooxygenase. J. Biol. Chem. 2013, 288, 1448–1457. [Google Scholar] [CrossRef] [PubMed]
- Hentrich, M.; Böttcher, C.; Düchting, P.; Cheng, Y.; Zhao, Y.; Berkowitz, O.; Masle, J.; Medina, J.; Pollmann, S. The jasmonic acid signaling pathway is linked to auxin homeostasis through the modulation of YUCCA8 and YUCCA9 gene expression. Plant J. 2013, 74, 626–637. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Dai, X.; De-Paoli, H.; Cheng, Y.; Takebayashi, Y.; Kasahara, H.; Kamiya, Y.; Zhao, Y. Auxin overproduction in shoots cannot rescue auxin deficiencies in arabidopsis roots. Plant Cell Physiol. 2014, 55, 1072–1079. [Google Scholar] [CrossRef]
- Hentrich, M.; Sanchez-Parra, B.; Perez Alonso, M.-M.; Carrasco Loba, V.; Carrillo, L.; Vicente-Carbajosa, J.; Medina, J.; Pollmann, S. YUCCA8 and YUCCA9 overexpression reveals a link between auxin signaling and lignification through the induction of ethylene biosynthesis. Plant Signal. Behav. 2013, 8, e26363. [Google Scholar] [CrossRef]
- Cha, J.-Y.; Kim, W.-Y.; Kang, S.B.; Kim, J.I.; Baek, D.; Jung, I.J.; Kim, M.R.; Li, N.; Kim, H.-J.; Nakajima, M.; et al. A novel thiol-reductase activity of Arabidopsis YUC6 confers drought tolerance independently of auxin biosynthesis. Nat. Commun. 2015, 6, 8041. [Google Scholar] [CrossRef]
- Cha, J.-Y.; Kim, M.R.; Jung, I.J.; Kang, S.B.; Park, H.J.; Kim, M.G.; Yun, D.-J.; Kim, W.-Y. The Thiol Reductase Activity of YUCCA6 Mediates Delayed Leaf Senescence by Regulating Genes Involved in Auxin Redistribution. Front. Plant Sci. 2016, 7, 626. [Google Scholar]
- Krönauer, C.; Kilian, J.; Strauß, T.; Stahl, M.; Lahaye, T. Cell Death Triggered by the YUCCA-like Bs3 Protein Coincides with Accumulation of Salicylic Acid and Pipecolic Acid But Not of Indole-3-Acetic Acid 1. Plant Physiol. 2019, 180, 1647–1659. [Google Scholar]
- Mayfield, J.A.; Frederick, R.E.; Streit, B.R.; Wencewicz, T.A.; Ballou, D.P.; DuBois, J.L. Comprehensive spectroscopic, steady state, and transient kinetic studies of a representative siderophore-associated flavin monooxygenase. J. Biol. Chem. 2010, 285, 30375–30388. [Google Scholar] [CrossRef]
- Chocklett, S.W.; Sobrado, P. Aspergillus fumigatus SidA is a highly specific ornithine hydroxylase with bound flavin cofactor. Biochemistry 2010, 49, 6777–6783. [Google Scholar] [CrossRef]
- Kong, W.; Li, J.; Yu, Q.; Cang, W.; Xu, R.; Wang, Y.; Ji, W. Two Novel Flavin-Containing Monooxygenases Involved in Biosynthesis of Aliphatic Glucosinolates. Front. Plant Sci. 2016, 7, 1292. [Google Scholar] [CrossRef]
- Halkier, B.A.; Gershenzon, J. Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 2006, 57, 303–333. [Google Scholar] [CrossRef] [PubMed]
- Van Poppel, G.; Verhoeven, D.T.; Verhagen, H.; Goldbohm, R.A. Brassica vegetables and cancer prevention. Epidemiology and mechanisms. Adv. Exp. Med. Biol. 1999, 472, 159–168. [Google Scholar] [PubMed]
- Li, J.; Hansen, B.G.; Ober, J.A.; Kliebenstein, D.J.; Halkier, B.A. Subclade of flavin-monooxygenases involved in aliphatic glucosinolate biosynthesis. Plant Physiol. 2008, 148, 1721–1733. [Google Scholar] [CrossRef] [PubMed]
- Byrne, S.L.; Erthmann, P.Ø.; Agerbirk, N.; Bak, S.; Hauser, T.P.; Nagy, I.; Paina, C.; Asp, T. The genome sequence of Barbarea vulgaris facilitates the study of ecological biochemistry. Sci. Rep. 2017, 7, 40728. [Google Scholar] [CrossRef] [PubMed]
- Pedras, M.S.C.; Alavi, M.; To, Q.H. Expanding the nasturlexin family: Nasturlexins C and D and their sulfoxides are phytoalexins of the crucifers Barbarea vulgaris and B. verna. Phytochemistry 2015, 118, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Yoshimoto, N.; Onuma, M.; Mizuno, S.; Sugino, Y.; Nakabayashi, R.; Imai, S.; Tsuneyoshi, T.; Sumi, S.I.; Saito, K. Identification of a flavin-containing S-oxygenating monooxygenase involved in alliin biosynthesis in garlic. Plant J. 2015, 83, 941–951. [Google Scholar] [CrossRef]
- Yoshimoto, N.; Saito, K. S-Alk(en)ylcysteine sulfoxides in the genus Allium: Proposed biosynthesis, chemical conversion, and bioactivities. J. Exp. Bot. 2019, 70, 4123–4137. [Google Scholar] [CrossRef]
- Upadhyay, R.K. Garlic: A potential source of pharmaceuticals and pesticides: A review. Int. J. Green Pharm. 2016, 10, S1–S28. [Google Scholar]
- Ravilious, G.E.; Jez, J.M. Structural biology of plant sulfur metabolism: From assimilation to biosynthesis. Nat. Prod. Rep. 2012, 29, 1138–1152. [Google Scholar] [CrossRef]
- Mishina, T.E.; Zeier, J. The Arabidopsis Flavin-Dependent Monooxygenase FMO1 Is an Essential Component of Biologically Induced Systemic Acquired Resistance. Plant Physiol. 2006, 141, 1666. [Google Scholar] [CrossRef] [PubMed]
- Návarová, H.; Bernsdorff, F.; Döring, A.C.; Zeier, J. Pipecolic acid, an endogenous mediator of defense amplification and priming, is a critical regulator of inducible plant immunity. Plant Cell 2013, 24, 5123–5141. [Google Scholar] [CrossRef] [PubMed]
- Bernsdorff, F.; Döring, A.-C.; Gruner, K.; Schuck, S.; Bräutigam, A.; Zeier, J. Pipecolic Acid Orchestrates Plant Systemic Acquired Resistance and Defense Priming via Salicylic Acid-Dependent and -Independent Pathways. Plant Cell 2016, 28, 102–129. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, M.; Zeier, T.; Bernsdorff, F.; Reichel-Deland, V.; Kim, D.; Hohmann, M.; Scholten, N.; Schuck, S.; Bräutigam, A.; Hölzel, T.; et al. Flavin Monooxygenase-Generated N-Hydroxypipecolic Acid Is a Critical Element of Plant Systemic Immunity. Cell 2018, 173, 456–469. [Google Scholar] [CrossRef]
- Chen, Y.C.; Holmes, E.C.; Rajniak, J.; Kim, J.G.; Tang, S.; Fischer, C.R.; Mudgett, M.B.; Sattely, E.S. N-hydroxy-pipecolic acid is a mobile metabolite that induces systemic disease resistance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, E4920–E4929. [Google Scholar] [CrossRef]
- Wang, X.; Bi, W.-S.; Gao, J.; Xiu-Mei, Y.; Wang, H.-Y.; Da-Qun, L. Systemic acquired resistance, NPR1, and pathogenesis-related genes in wheat and barley. J. Integr. Agric. 2018, 17, 60345–60347. [Google Scholar] [CrossRef]
- Holmes, E.C.; Chen, Y.-C.; Sattely, E.S.; Mudgett, M.B. An engineered pathway for N-hydroxy-pipecolic acid synthesis enhances systemic acquired resistance in tomato. Sci. Signal. 2019, 12. [Google Scholar] [CrossRef]
- Tian, H.; Zhang, Y. The emergence of a mobile signal for systemic acquired resistance. Plant Cell 2019, 31, 1414–1415. [Google Scholar] [CrossRef]
- Bjarnholt, N.; Li, B.; D’Alvise, J.; Janfelt, C. Mass spectrometry imaging of plant metabolites—principles and possibilities. Nat. Prod. Rep. 2014, 31, 818–837. [Google Scholar] [CrossRef]
- Kompauer, M.; Heiles, S.; Spengler, B. Atmospheric pressure MALDI mass spectrometry imaging of tissues and cells at 1.4-μm lateral resolution. Nat. Methods 2017, 14, 90–96. [Google Scholar] [CrossRef]
- Boughton, B.A.; Thinagaran, D.; Sarabia, D.; Bacic, A.; Roessner, U. Mass spectrometry imaging for plant biology: A review. Phytochem. Rev. 2016, 15, 445–488. [Google Scholar] [CrossRef] [PubMed]
- Leebens-Mack, J.H.; Barker, M.S.; Carpenter, E.J.; Deyholos, M.K.; Gitzendanner, M.A.; Graham, S.W.; Grosse, I.; Li, Z.; Melkonian, M.; Mirarab, S.; et al. One thousand plant transcriptomes and the phylogenomics of green plants. Nature 2019, 574, 679–685. [Google Scholar]
- Eswaramoorthy, S.; Bonanno, J.B.; Burley, S.K.; Swaminathan, S. Mechanism of action of a flavin-containing monooxygenase. Proc. Natl. Acad. Sci. USA 2006, 103, 9832–9837. [Google Scholar] [CrossRef] [PubMed]
- Alfieri, A.; Malito, E.; Orru, R.; Fraaije, M.W.; Mattevi, A. Revealing the moonlighting role of NADP in the structure of a flavin-containing monooxygenase. Proc. Natl. Acad. Sci. USA 2008, 105, 6572–6577. [Google Scholar] [CrossRef]
- Kubitza, C.; Faust, A.; Gutt, M.; Gäth, L.; Ober, D.; Scheidig, A.J. Crystal structure of pyrrolizidine alkaloid N-oxygenase from the grasshopper Zonocerus variegatus. Acta Crystallogr. Sect. D Struct. Biol. 2018, 74, 422–432. [Google Scholar] [CrossRef]
- Nicoll, C.R.; Bailleul, G.; Fiorentini, F.; Mascotti, M.L.; Fraaije, M.W.; Mattevi, A. Ancestral-sequence reconstruction unveils the structural basis of function in mammalian FMOs. Nat. Struct. Mol. Biol. 2020, 27, 14–24. [Google Scholar] [CrossRef]
- Zhu, Y.; Li, H.; Su, Q.; Wen, J.; Wang, Y.; Song, W.; Xie, Y.; He, W.; Yang, Z.; Jiang, K.; et al. A phenotype-directed chemical screen identifies ponalrestat as an inhibitor of the plant flavin monooxygenase YUCCA in auxin biosynthesis. J. Biol. Chem. 2019. [Google Scholar] [CrossRef]
- Nelson, D.R. Cytochrome P450 Nomenclature. In Cytochrome P450 Protocols. Methods in Molecular Biology; Phillips, I.R., Shephard, E.A., Eds.; Humana Press: Totowa, NJ, USA, 2006; ISBN 978-1-59259-998-1. [Google Scholar]
- Korver, R.A.; Koevoets, I.T.; Testerink, C. Out of Shape During Stress: A Key Role for Auxin. Trends Plant Sci. 2018, 23, 783–793. [Google Scholar] [CrossRef]
- Velasquez, A.C.; Castroverde, C.D.M.; He, S.Y. Plant-Pathogen Warfare under Changing Climate Conditions. Curr. Biol. 2018, 28, R619–R634. [Google Scholar] [CrossRef]
- Park, J.S.; Kim, H.J.; Cho, H.S.; Jung, H.W.; Cha, J.-Y.; Yun, D.-J.; Oh, S.-W.; Chung, Y.-S. Overexpression of AtYUCCA6 in soybean crop results in reduced ROS production and increased drought tolerance. Plant Biotechnol. Rep. 2019, 13, 161–168. [Google Scholar] [CrossRef]
- Park, S.-C.; Kim, H.S.; Lee, H.-U.; Kim, Y.-H.; Kwak, S.-S. Overexpression of Arabidopsis YUCCA6 enhances environment stress tolerance and inhibits storage root formation in sweetpotato. Plant Biotechnol. Rep. 2019, 13, 345–352. [Google Scholar] [CrossRef]
- Ke, Q.; Wang, Z.; Ji, C.Y.; Jeong, J.C.; Lee, H.-S.; Li, H.; Xu, B.; Deng, X.; Kwak, S.-S. Transgenic poplar expressing Arabidopsis YUCCA6 exhibits auxin-overproduction phenotypes and increased tolerance to abiotic stress. Plant Physiol. Biochem. 2015, 94, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.I.; Baek, D.; Park, H.C.; Chun, H.J.; Oh, D.-H.; Lee, M.K.; Cha, J.-Y.; Kim, W.-Y.; Kim, M.C.; Chung, W.S.; et al. Overexpression of Arabidopsis YUCCA6 in potato results in high-auxin developmental phenotypes and enhanced resistance to water deficit. Mol. Plant 2013, 6, 337–349. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Weng, Y.; Dickey, A.; Wang, K.Y. Plants as Factories for Human Pharmaceuticals: Applications and Challenges. Int. J. Mol. Sci. 2015, 16, 28549–28565. [Google Scholar] [CrossRef] [PubMed]
- Mikkelsen, M.D.; Olsen, C.E.; Halkier, B.A. Production of the cancer-preventive glucoraphanin in tobacco. Mol. Plant 2010, 3, 751–759. [Google Scholar] [CrossRef] [PubMed]
- Moreno, D.A.; Carvajal, M.; López-Berenguer, C.; García-Viguera, C. Chemical and biological characterisation of nutraceutical compounds of broccoli. J. Pharm. Biomed. Anal. 2006, 41, 1508–1522. [Google Scholar] [CrossRef] [PubMed]
- Juge, N.; Mithen, R.F.; Traka, M. Molecular basis for chemoprevention by sulforaphane: A comprehensive review. Cell. Mol. Life Sci. 2007, 64, 1105. [Google Scholar] [CrossRef]
- De Gonzalo, G.; Mihovilovic, M.D.; Fraaije, M.W. Recent Developments in the Application of Baeyer-Villiger Monooxygenases as Biocatalysts. ChemBioChem 2010, 11, 2208–2231. [Google Scholar] [CrossRef]
- Urlacher, V.B.; Eiben, S. Cytochrome P450 monooxygenases: Perspectives for synthetic application. Trends Biotechnol. 2006, 24, 324–330. [Google Scholar] [CrossRef]
- Panke, S.; Wubbolts, M.G.; Schmid, A.; Witholt, B. Production of enantiopure styrene oxide by recombinant Escherichia coli synthesizing a two-component styrene monooxygenase. Biotechnol. Bioeng. 2000, 69, 91–100. [Google Scholar] [CrossRef]
- Hummel, W.; Gröger, H. Strategies for regeneration of nicotinamide coenzymes emphasizing self-sufficient closed-loop recycling systems. J. Biotechnol. 2014, 191, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Liu, C.; Yang, D. Methods for the regeneration of nicotinamide coenzymes. Green Chem. 2013, 15, 1773–1789. [Google Scholar] [CrossRef]
- Willetts, A.J.; Knowles, C.J.; Levitt, M.S.; Roberts, S.M.; Sandey, H.; Shipston, N.F. Biotransformation of endo-bicyclo[2.2.1]heptan-2-ols and endo-bicyclo[3.2.0]hept-2-en-6-ol into the corresponding lactones. J. Chem. Soc. Perkin Trans. 1 1991, 1608–1610. [Google Scholar] [CrossRef]
- Maid, H.; Böhm, P.; Huber, S.M.; Bauer, W.; Hummel, W.; Jux, N.; Gröger, H. Iron Catalysis for In Situ Regeneration of Oxidized Cofactors by Activation and Reduction of Molecular Oxygen: A Synthetic Metalloporphyrin as a Biomimetic NAD(P)H Oxidase. Angew. Chemie Int. Ed. 2011, 50, 2397–2400. [Google Scholar] [CrossRef] [PubMed]
- Brondani, P.B.; Dudek, H.M.; Martinoli, C.; Mattevi, A.; Fraaije, M.W. Finding the Switch: Turning a Baeyer—Villiger Monooxygenase into a NADPH Oxidase. J. Am. Chem. Soc. 2014, 136, 16966–16969. [Google Scholar] [CrossRef] [PubMed]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thodberg, S.; Jakobsen Neilson, E.H. The “Green” FMOs: Diversity, Functionality and Application of Plant Flavoproteins. Catalysts 2020, 10, 329. https://doi.org/10.3390/catal10030329
Thodberg S, Jakobsen Neilson EH. The “Green” FMOs: Diversity, Functionality and Application of Plant Flavoproteins. Catalysts. 2020; 10(3):329. https://doi.org/10.3390/catal10030329
Chicago/Turabian StyleThodberg, Sara, and Elizabeth H. Jakobsen Neilson. 2020. "The “Green” FMOs: Diversity, Functionality and Application of Plant Flavoproteins" Catalysts 10, no. 3: 329. https://doi.org/10.3390/catal10030329
APA StyleThodberg, S., & Jakobsen Neilson, E. H. (2020). The “Green” FMOs: Diversity, Functionality and Application of Plant Flavoproteins. Catalysts, 10(3), 329. https://doi.org/10.3390/catal10030329



