Characterization and Catalytic-Site-Analysis of an Aldo-Keto Reductase with Excellent Solvent Tolerance
Abstract
:1. Introduction
2. Results
2.1. Cloning and Sequencing Analysis of AKR3-2-9
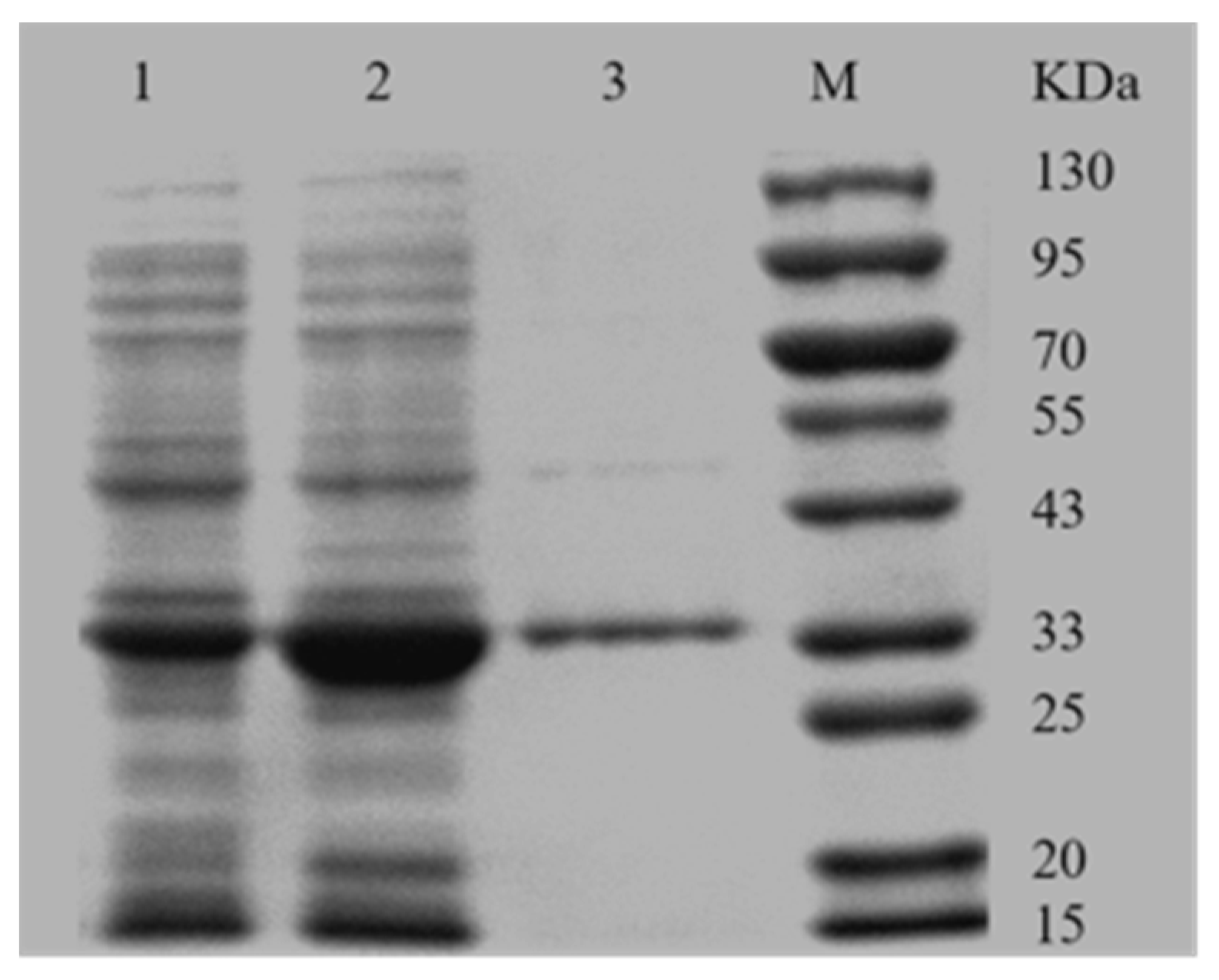
2.2. Expression and the Purification of AKR3-2-9
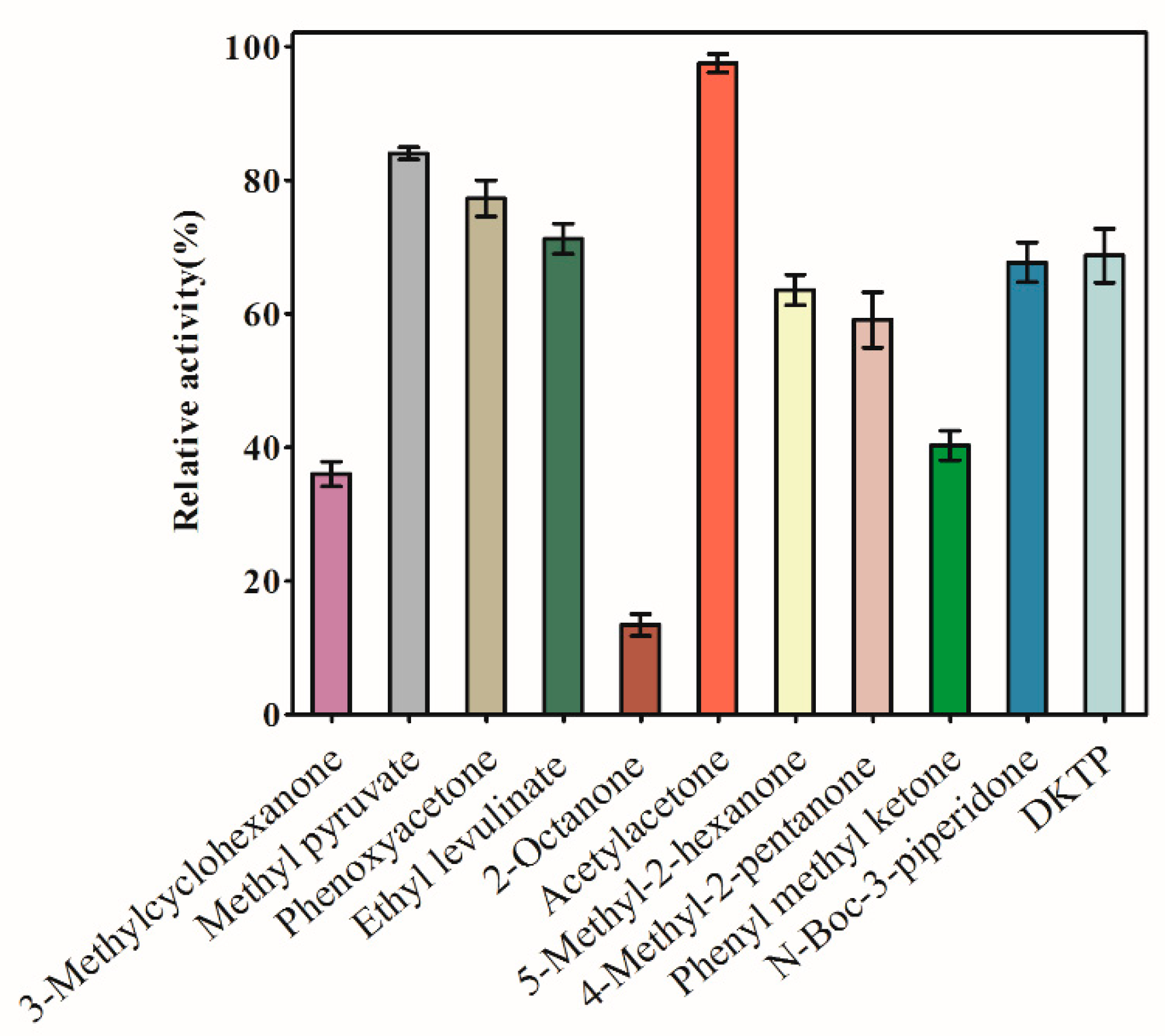
2.3. Substrate Specificity
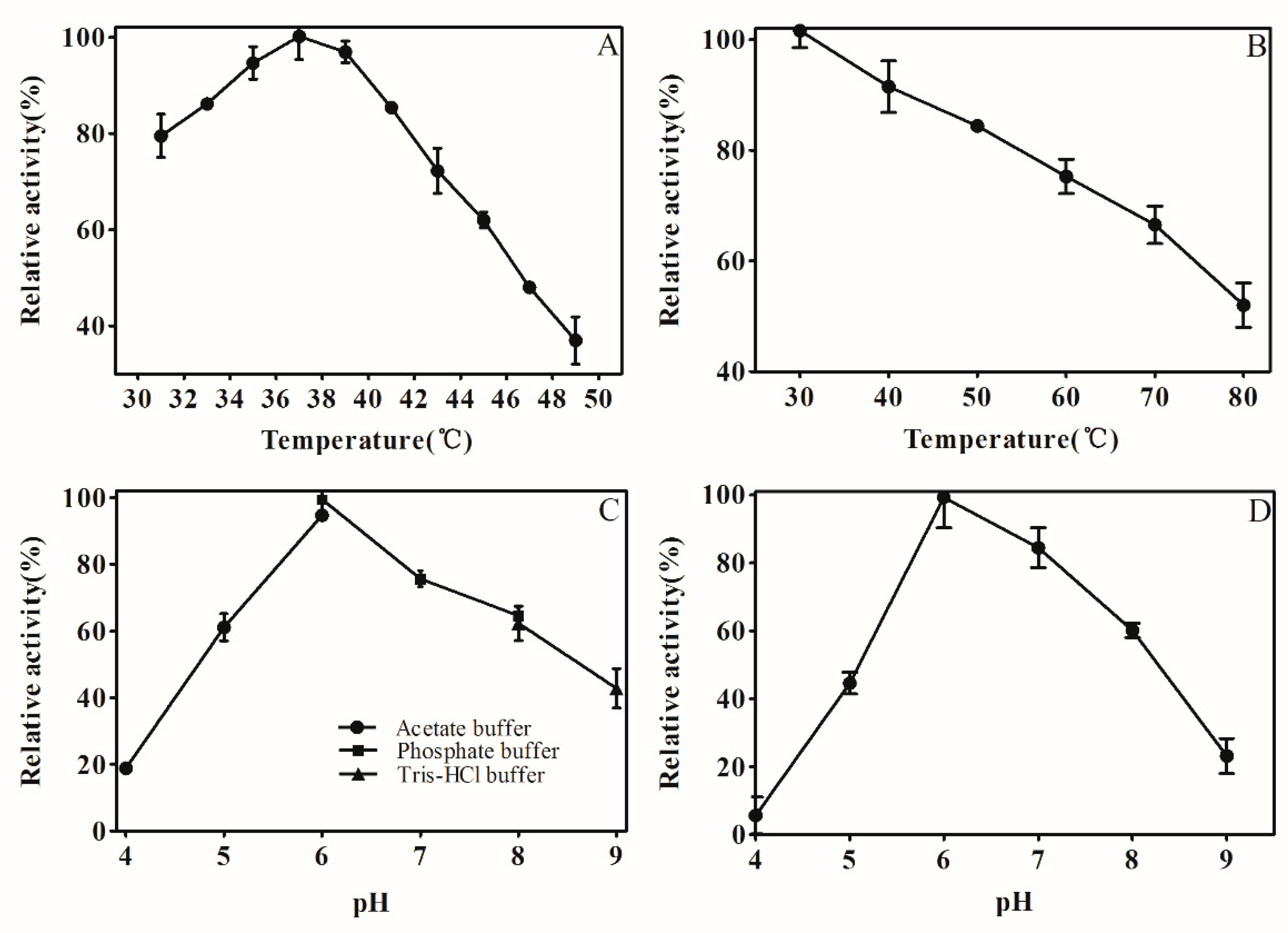
2.4. Effect of Temperature and pH
2.5. Detection of AKR3-2-9 Tolerant Organic Solvent Properties
2.6. Kinetic Analysis
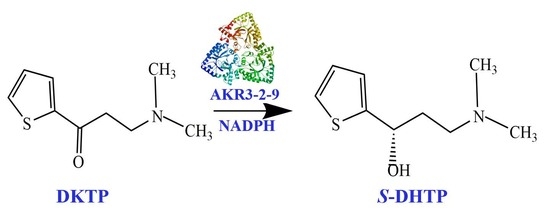
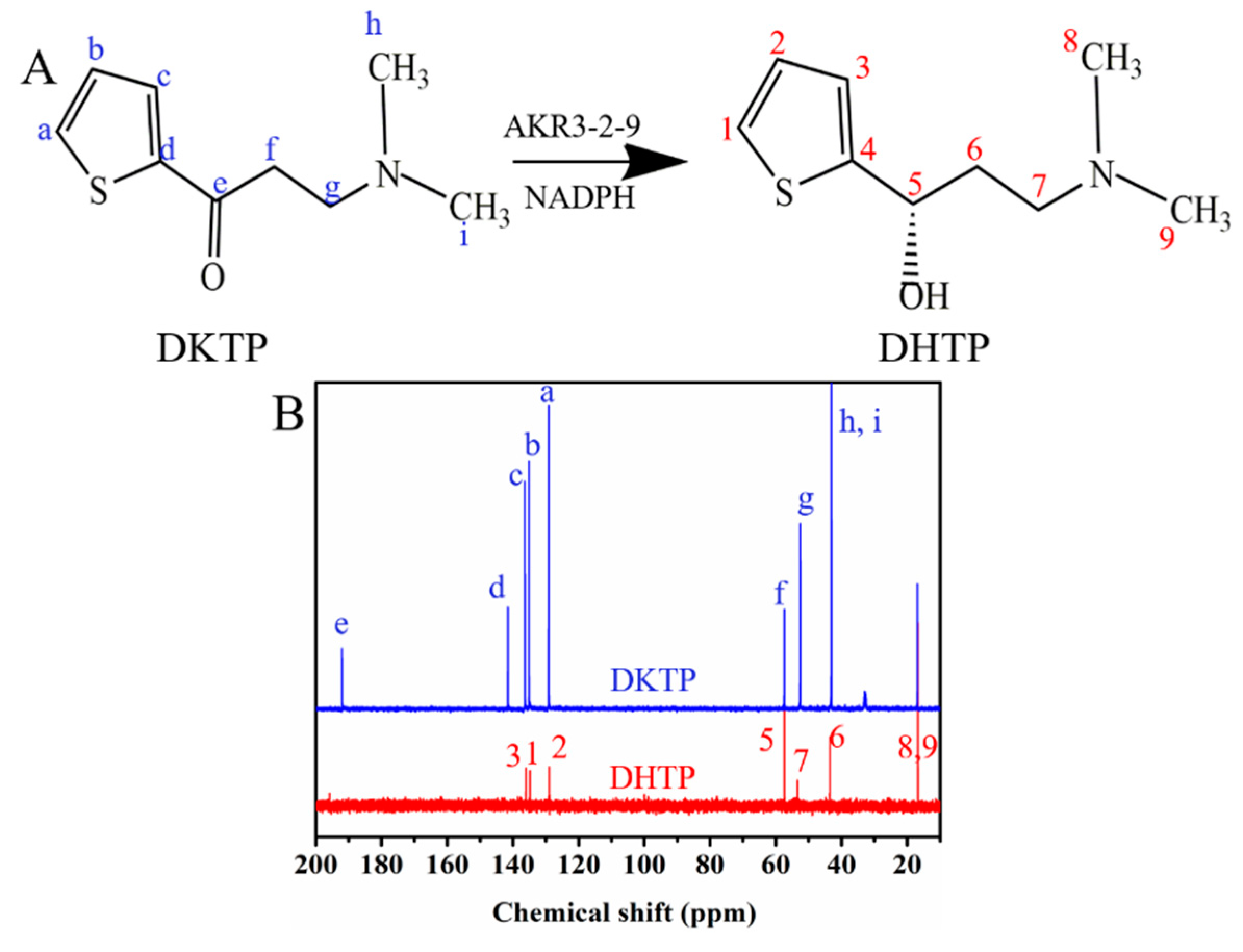
2.7. The Activity towards DKTP
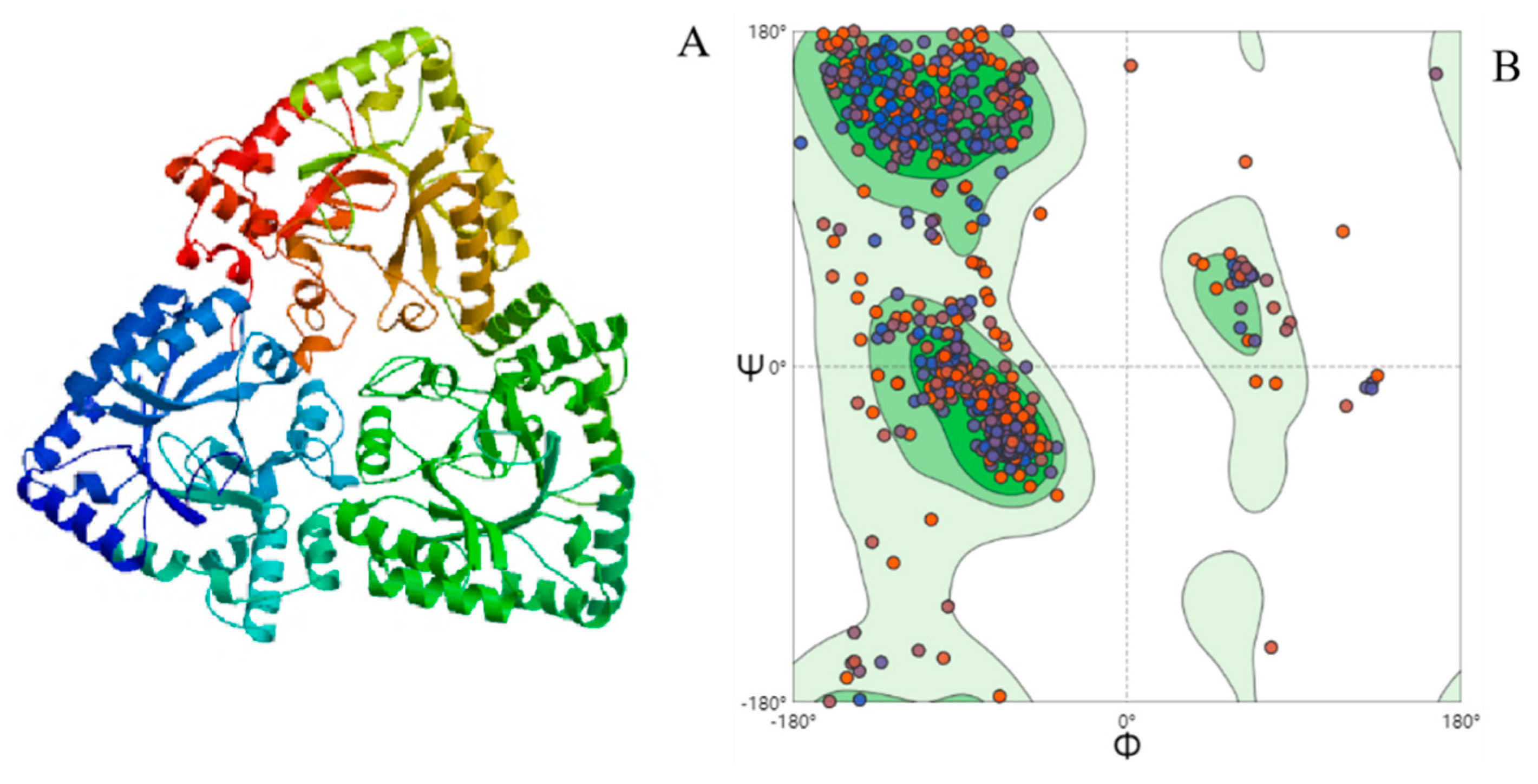
2.8. Molecular Modeling
3. Discussion
4. Materials and Methods
4.1. Strains, Vectors, Chemicals
4.2. Homologous Protein-Searching Analysis and Homology Modeling
4.3. Cloning, Expression and Purification of AKR3-2-9
4.4. Enzyme Activity Assays and Kinetic Parameter Studies
4.5. Characterization of the AKR 3-2-9
4.5.1. Substrate Specificity of AKR3-2-9
4.5.2. Effect of Temperature and pH
4.5.3. Organic Solvent Tolerance of AKR3-2-9
4.6. The Activity towards DKTP
4.7. The GenBank Accession Number of the AKR3-2-9
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Khan, I.A.; Saxena, A.K. Metal-Free, mild, nonepimerizing, chemo-and enantio- or diastereoselective N-Alkylation of amines by alcohols via oxidation/imine-iminium formation/reductive amination: A pragmatic synthesis of octahydropyrazinopyridoindoles and higher ring analogues. Cheminform 2014, 45, 11656–11669. [Google Scholar] [CrossRef]
- Zhou, X.; Zheng, D.; Cui, B.; Han, W.; Chen, Y. Novozyme 435 lipase mediated enantioselective kinetic resolution: A facile method for the synthesis of chiral tetrahydroquinolin-4-ol and tetrahydro-1H-benzo[b]azepin-5-ol derivatives. Tetrahedron 2015, 71, 4738–4744. [Google Scholar] [CrossRef]
- Fish, R.H. 1,4-NADH biomimetic co-factors with horse liver alcohol dehydrogenase (HLADH), utilizing [Cp*Rh(bpy)H](OTf) for co-factor regeneration, do in fact, produce chiral alcohols from reactions with achiral ketones. Catalysts 2019, 9, 562. [Google Scholar] [CrossRef] [Green Version]
- Tekedar, H.C.; Sanli-Mohamed, G. Molecular cloning, over expression and characterization of thermoalkalophilic esterases isolated from Geobacillus sp. Extremophiles 2011, 15, 203–211. [Google Scholar] [CrossRef] [Green Version]
- Woodley, J.M. New opportunities for biocatalysis: Making pharmaceutical processes greener. Trends Biotechnol. 2008, 26, 321–327. [Google Scholar] [CrossRef]
- Kataoka, M.; Kita, K.; Wada, M.; Yasohara, Y.; Hasegawa, J.; Shimizu, S. Novel bioreduction system for the production of chiral alcohols. Appl. Microbiol. Biotechnol. 2003, 62, 437. [Google Scholar] [CrossRef]
- Deechongkit, S.; You, S.L.; Kelly, J.W. Synthesis of all nineteen appropriately protected chiral alpha-hydroxy acid equivalents of the alpha-amino acids for boc solid-phase depsi-peptide synthesis. Org. Lett. 2004, 6, 497–500. [Google Scholar] [CrossRef]
- Li, B.; Li, Y.; Bai, D.; Zhang, X.; Yang, H.; Wang, J.; Liu, G.; Yue, J.; Ling, Y.; Zhou, D. Whole-cell biotransformation systems for reduction of prochiral carbonyl compounds to chiral alcohol in Escherichia coli. Sci. Rep. 2014, 4, 6750. [Google Scholar] [CrossRef]
- Goldberg, K.; Schroer, K.; Lutz, S.A. Biocatalytic ketone reduction-a powerful tool for the production of chiral alcohols-part II: Whole-cell reductions. Appl. Microbiol. Biotechnol. 2007, 76, 249–255. [Google Scholar] [CrossRef]
- Gröger, H.; Chamouleau, F.; Orologas, N.; Rollmann, C.; Drauz, K.; Hummel, W.; Weckbecker, A.; May, O. Enantioselective reduction of ketones with “designer cells” at high substrate concentrations: Highly efficient access to functionalized optically active alcohols. Angew. Chem. Int. Ed. Engl. 2010, 45, 5677–5681. [Google Scholar]
- Junhua, T.; Jian-He, X. Biocatalysis in development of green pharmaceutical processes. Curr. Opin. Chem. Biol. 2009, 13, 43–50. [Google Scholar]
- Zhang, Y.; Ji, F.L.; Wang, J.Y.; Pu, Z.J.; Jiang, B.; Bao, Y.M. Purification and characterization of a novel organic solvent-tolerant and cold-adapted lipase from Psychrobacter sp. ZY124. Extremophiles 2018, 22, 287–300. [Google Scholar] [CrossRef]
- Penning, T.M. The aldo-keto reductases (AKRs): Overview. Chem.-Biol. Interact. 2015, 234, 236–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hyndman, D.; Bauman, D.R.; Heredia, V.V.; Penning, T.M. The aldo-keto reductase superfamily homepage. Chem.-Biol. Interact. 2003, 143, 621–631. [Google Scholar] [CrossRef]
- Lv, D.-Q.; Zhou, S.; Lai, D.-Y.; Chen, Z.-M.; Ma, Y.-H. Characterization of an aldo-keto reductase from Thermotoga maritima with high thermostability and a broad substrate spectrum. Biotechnol. Lett. 2013, 35, 757–762. [Google Scholar]
- Hee, C.Y.; Hye Jeong, C.; Dooil, K.; Ki-Nam, U.; Hyung-Kwoun, K. Asymmetric synthesis of (S)-3-chloro-1-phenyl-1-propanol using Saccharomyces cerevisiae reductase with high enantioselectivity. Appl. Microbiol. Biotechnol. 2010, 87, 185–193. [Google Scholar]
- Asako, H.; Wakita, R.K.; Shimizu, M.; Sakai, J.; Itoh, N. Purification and cDNA cloning of NADPH-dependent aldoketoreductase, involved in asymmetric reduction of methyl 4-bromo-3-oxobutyrate, from Penicillium citrinum IFO4631. Appl. Environ. Microbiol. 2005, 71, 1101–1104. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Yao, N.; Mu, X.; Yan, X. Gene mining-based identification of aldo–keto reductases for highly stereoselective reduction of bulky ketones. Bioresour. Bioprocess. 2018, 5, 33. [Google Scholar]
- Luo, X.; Wang, Y.J.; Zheng, Y.G. Cloning and characterization of a NADH-dependent aldo-keto reductase from a newly isolated Kluyveromyces lactis XP1461. Enzym. Microb. Technol. 2015, 77, 68–77. [Google Scholar] [CrossRef]
- Cao, C.; Fukae, T.; Yamamoto, T.; Kanamaru, S.; Matsuda, T. Purification and characterization of fluorinated ketone reductase from Geotrichum candidum NBRC 5767. Biochem. Eng. J. 2013, 76, 13–16. [Google Scholar] [CrossRef] [Green Version]
- Zheng, G.W.; Xu, J.H. New opportunities for biocatalysis: Driving the synthesis of chiral chemicals. Curr. Opin. Biotechnol. 2011, 22, 784–792. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Tingting, Y.E.; Zhuanzhuan, M.A.; Chen, R.; Xie, T.; Yin, X. Characterization and site-directed mutation of a novel aldo-keto reductase from Lodderomyces elongisporus NRRL YB-4239 with high production rate of ethyl (R)-4-chloro-3-hydroxybutanoate. J. Ind. Microbiol. Biotechnol. 2014, 41, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ellis, E.M. Microbial aldo-keto reductases. FEMS Microbiol. Lett. 2002, 216, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Barski, O.A.; Gabbay, K.H.; Grimshaw, C.E.; Bohren, K.M. Mechanism of human aldehyde reductase: Characterization of the active site pocket. Biochemistry 1995, 34, 11264–11275. [Google Scholar] [CrossRef] [PubMed]
- Regina, K.; Bernd, N. Electrostatic stabilization in a pre-organized polar active site: The catalytic role of Lys-80 in Candida tenuis xylose reductase (AKR2B5) probed by site-directed mutagenesis and functional complementation studies. Biochem. J. 2005, 389, 507–515. [Google Scholar]
- Bohren, K.M.; Grimshaw, C.E.; Lai, C.J.; Harrison, D.H.; Ringe, D.; Petsko, G.A.; Gabbay, K.H. Tyrosine-48 is the proton donor and histidine-110 directs substrate stereochemical selectivity in the reduction reaction of human aldose reductase: Enzyme kinetics and crystal structure of the Y48H mutant enzyme. Biochemistry 1994, 33, 2021–2023. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, C.; Zhang, L.; Yao, Z.; Cui, D.; Wu, L.; Lin, J.; Yuan, Y.R.; Wei, D. Structural and mutational studies on an aldo-keto reductase AKR5C3 from Gluconobacter oxydans. Protein Sci. A Publ. Protein Soc. 2015, 23, 1540–1549. [Google Scholar] [CrossRef] [Green Version]
- Schlegel, B.P.; Jez, J.M.; Penning, T.M. Mutagenesis of 3 alpha-hydroxysteroid dehydrogenase reveals a “push-pull” mechanism for proton transfer in aldo-keto reductases. Biochemistry 1998, 37, 3538–3548. [Google Scholar] [CrossRef]
- Knadler, M.P.; Lobo, E.; Chappell, J.; Bergstrom, R. Duloxetine. Clin. Pharmacokinet. 2011, 50, 281–294. [Google Scholar] [CrossRef]
- Soni, P.; Banerjee, U.C. Biotransformations for the production of the chiral drug (S)-Duloxetine catalyzed by a novel isolate of Candida tropicalis. Appl. Microbiol. Biotechnol. 2005, 67, 771–777. [Google Scholar] [CrossRef]
- Eisenthal, R.; Danson, M.J.; Hough, D.W. Catalytic efficiency and kcat/Km: A useful comparator? Trends Biotechnol. 2007, 25, 247–249. [Google Scholar] [CrossRef] [PubMed]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Cassarino, T.G.; Bertoni, M.; Bordoli, L. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014, 42, 252–258. [Google Scholar] [CrossRef] [PubMed]


| Substrate | Km (mM) | kcat (s−1) | kcat/Km (s−1 mM−1) |
|---|---|---|---|
| Acetylacetone | 6.29 ± 0.36 | (4.195 ± 0.08) × 103 | 0.667 × 103 |
| Methyl pyruvate | 7.431 ± 0.47 | (4.24 ± 0.09) × 103 | 0.571 × 103 |
| Phenoxyacetone | 9.844 ± 0.82 | (3.648 ± 0.12) × 103 | 0.371 × 103 |
| DKTP | 2.81 ± 0.78 | (1.601 ± 0.19) × 103 | 0.569 × 103 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pei, R.; Wu, W.; Zhang, Y.; Tian, L.; Jiang, W.; Zhou, S.-F. Characterization and Catalytic-Site-Analysis of an Aldo-Keto Reductase with Excellent Solvent Tolerance. Catalysts 2020, 10, 1121. https://doi.org/10.3390/catal10101121
Pei R, Wu W, Zhang Y, Tian L, Jiang W, Zhou S-F. Characterization and Catalytic-Site-Analysis of an Aldo-Keto Reductase with Excellent Solvent Tolerance. Catalysts. 2020; 10(10):1121. https://doi.org/10.3390/catal10101121
Chicago/Turabian StylePei, Rui, Weiliang Wu, Yuqian Zhang, Libing Tian, Wei Jiang, and Shu-Feng Zhou. 2020. "Characterization and Catalytic-Site-Analysis of an Aldo-Keto Reductase with Excellent Solvent Tolerance" Catalysts 10, no. 10: 1121. https://doi.org/10.3390/catal10101121