Expression, Characterisation and Homology Modelling of a Novel Hormone-Sensitive Lipase (HSL)-Like Esterase from Glaciozyma antarctica
Abstract
1. Introduction
2. Results and Discussion
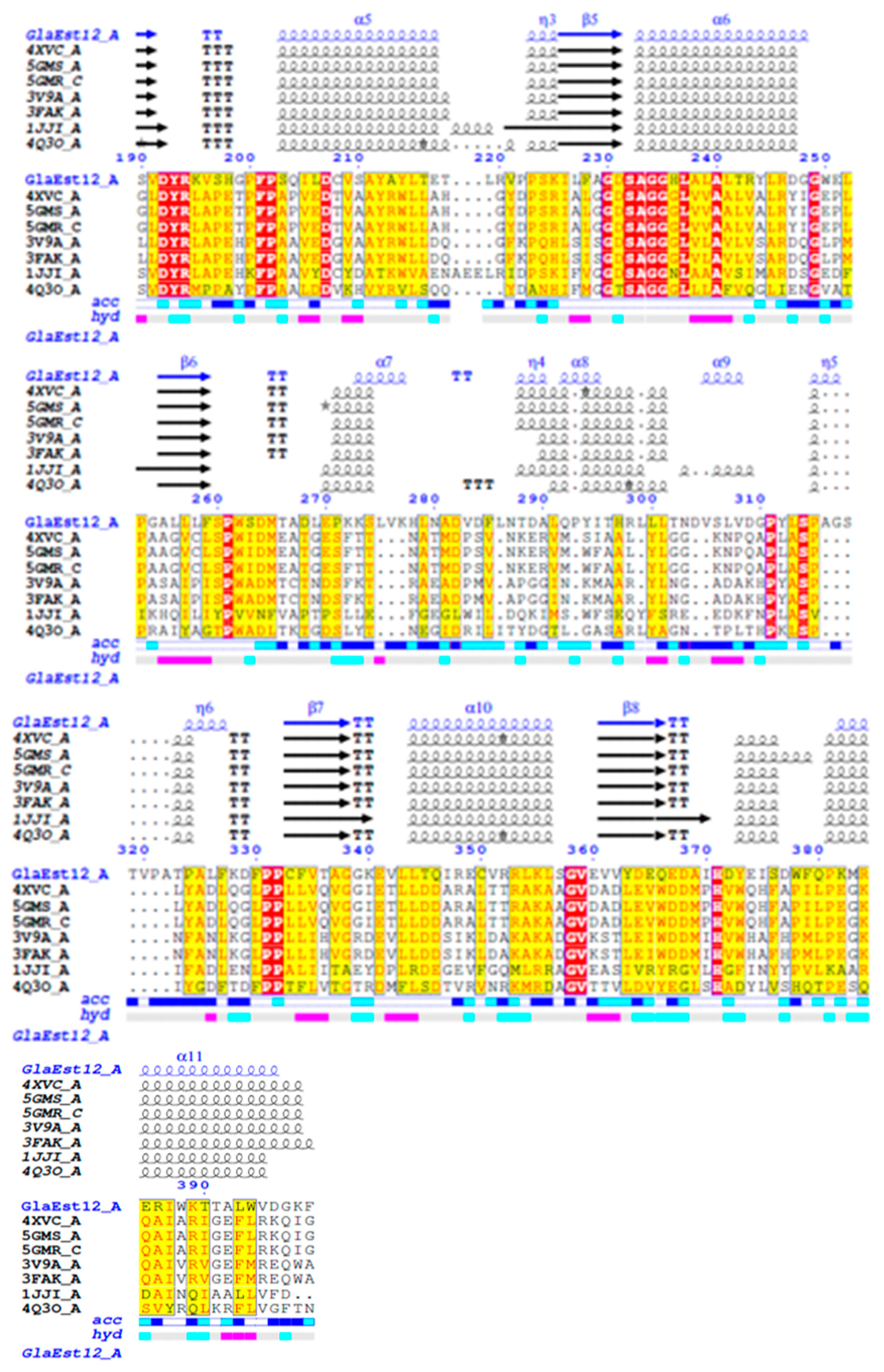
2.1. Sequence Analysis of GlaEst12
2.2. Expression and Purification of Recombinant GlaEst12
2.3. Characterisation of Purified GlaEst12
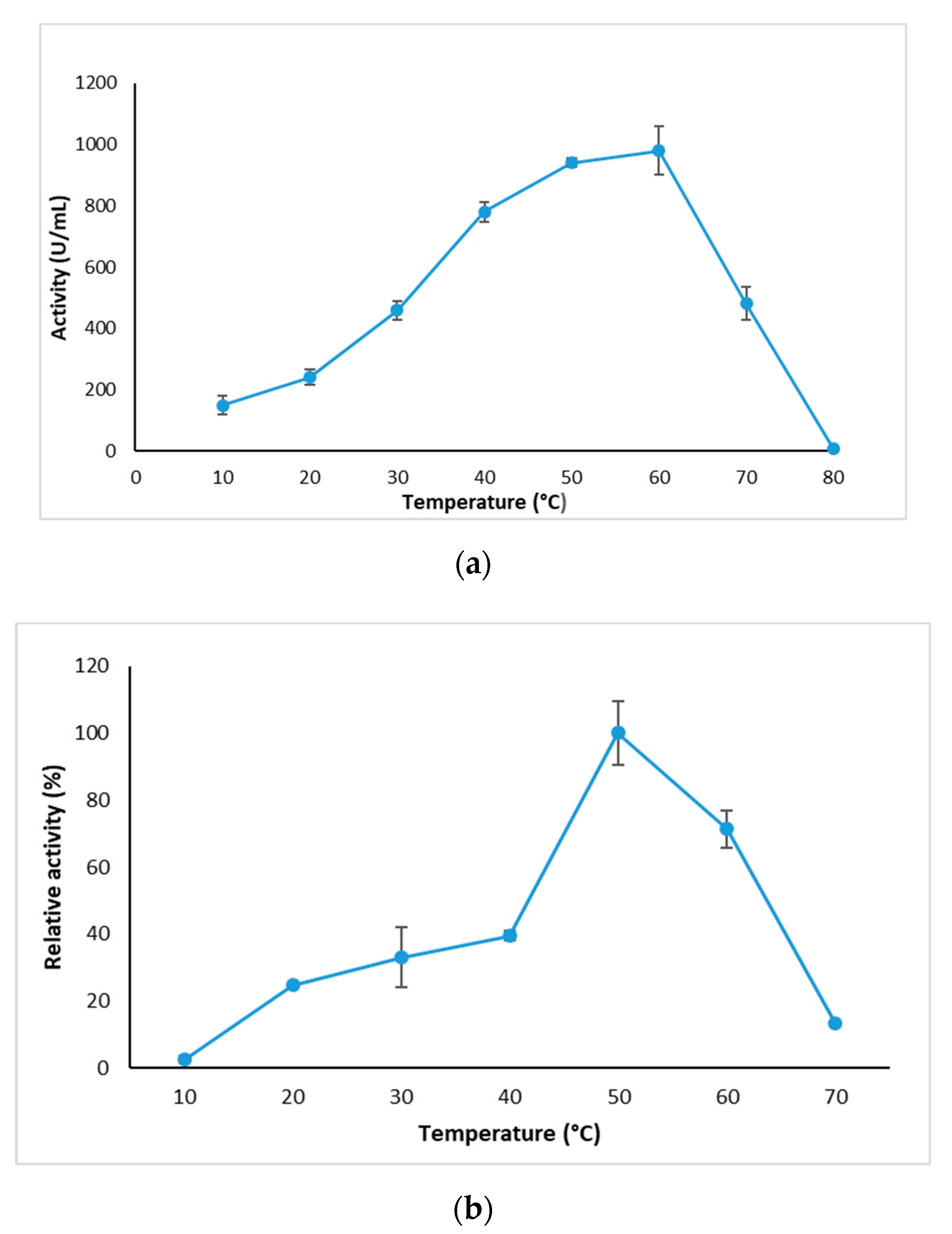
2.3.1. Effect of Temperature on GlaEst12 Esterase Activity and Stability
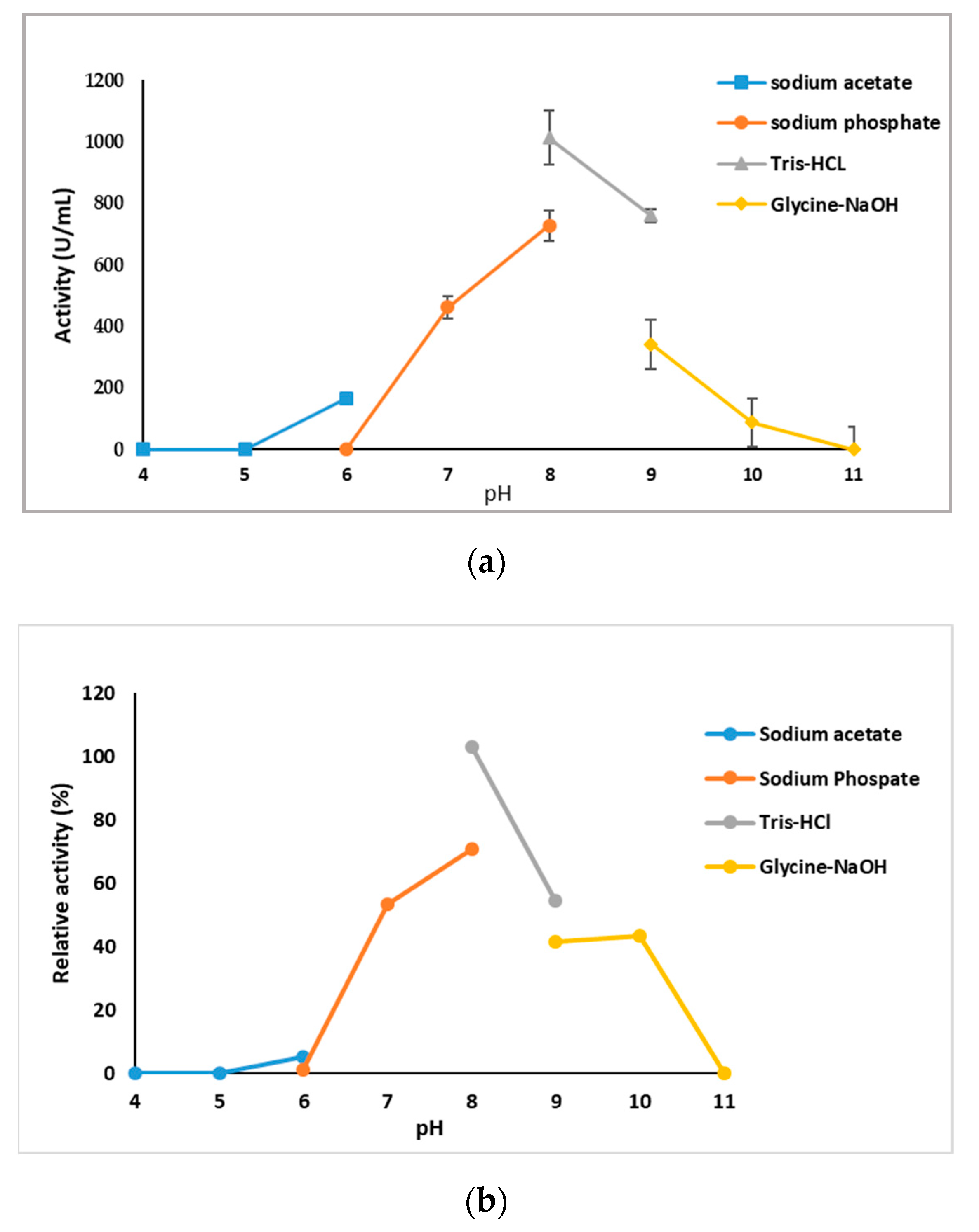
2.3.2. Effects of pH on GlaEst12 Activity and Stability
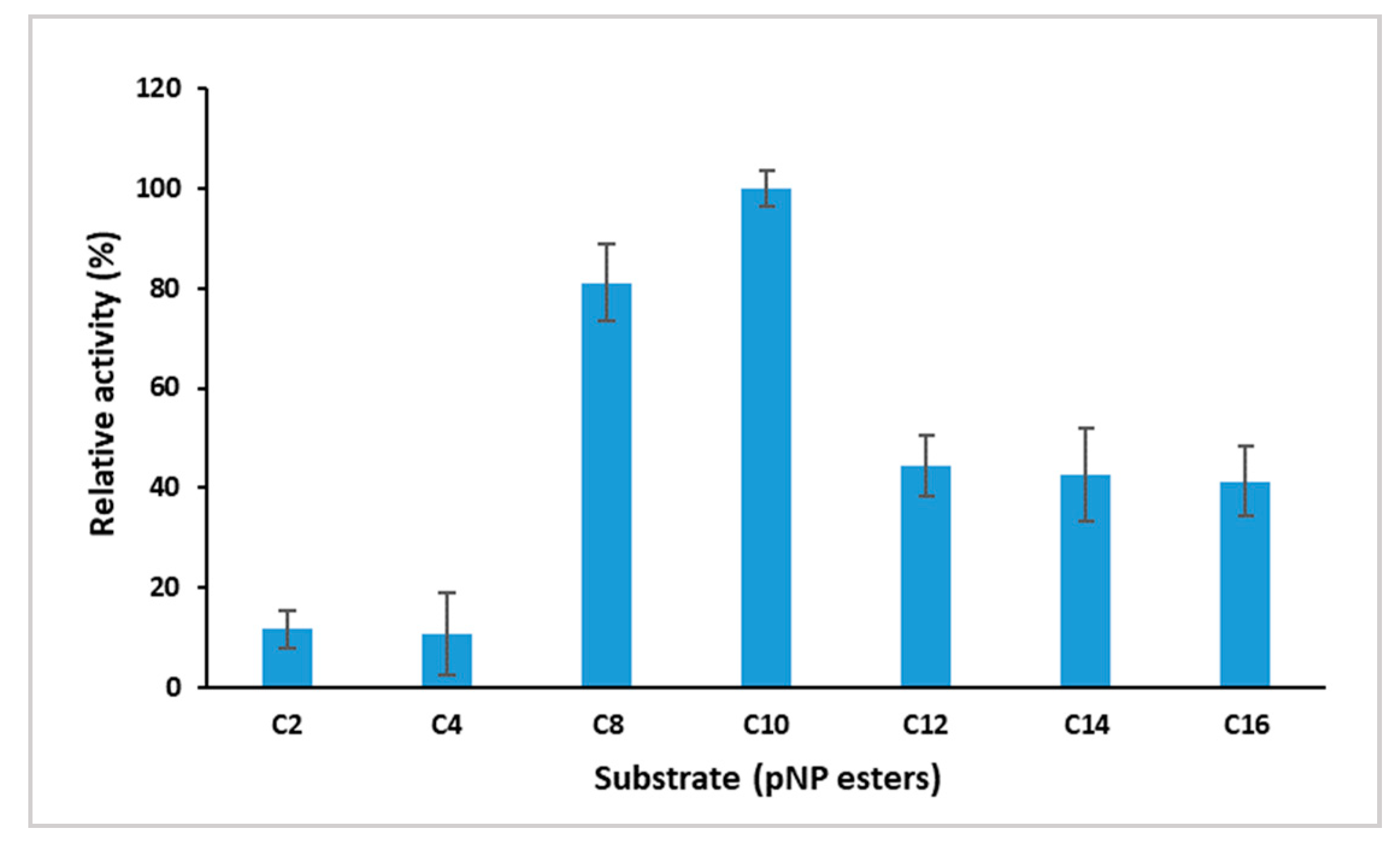
2.3.3. Substrate Specificity of GlaEst12 Esterase
2.3.4. Effect of Metal Ions on Esterase Activity
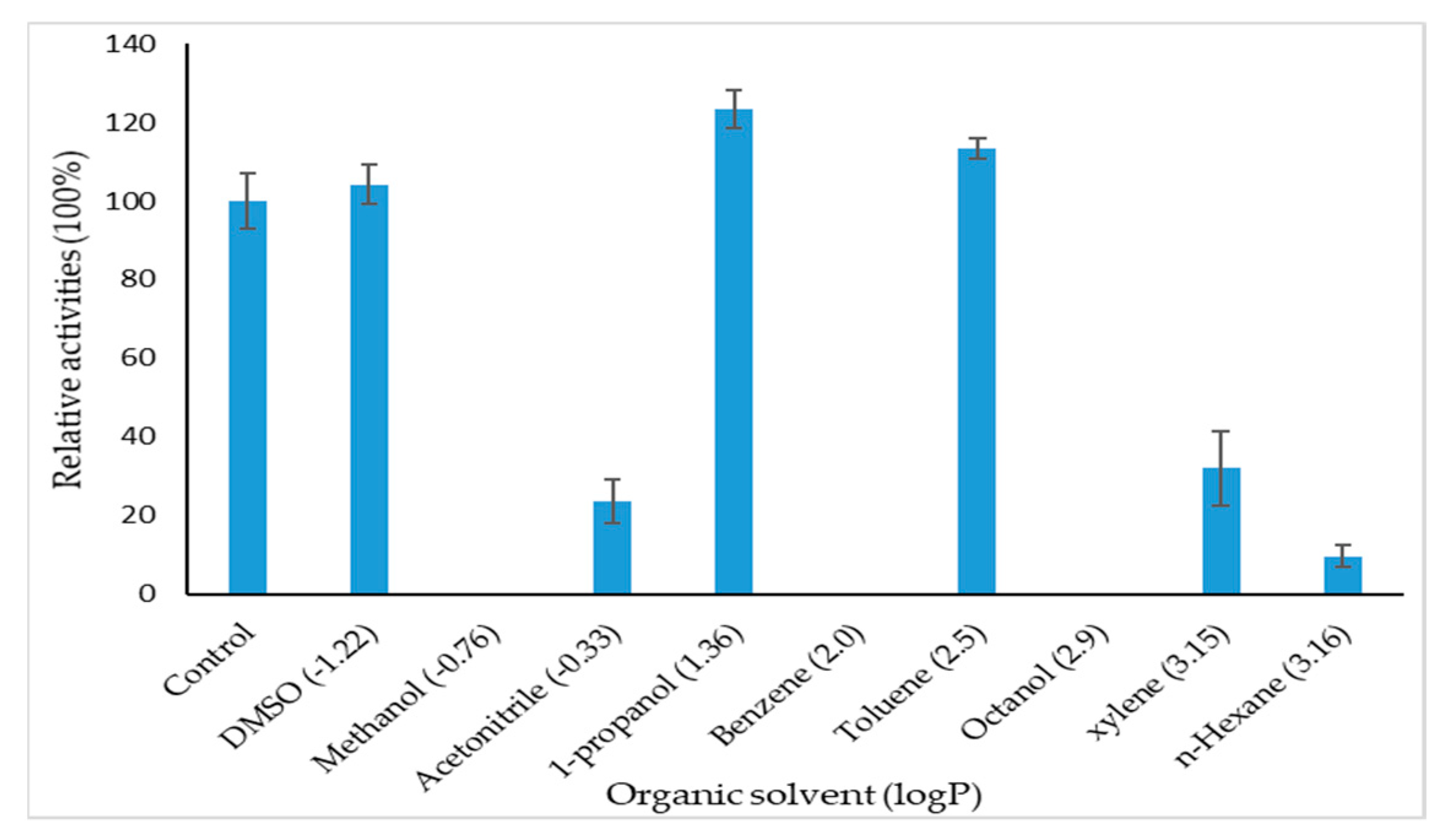
2.3.5. Effect of Organic Solvents on GlaEst12
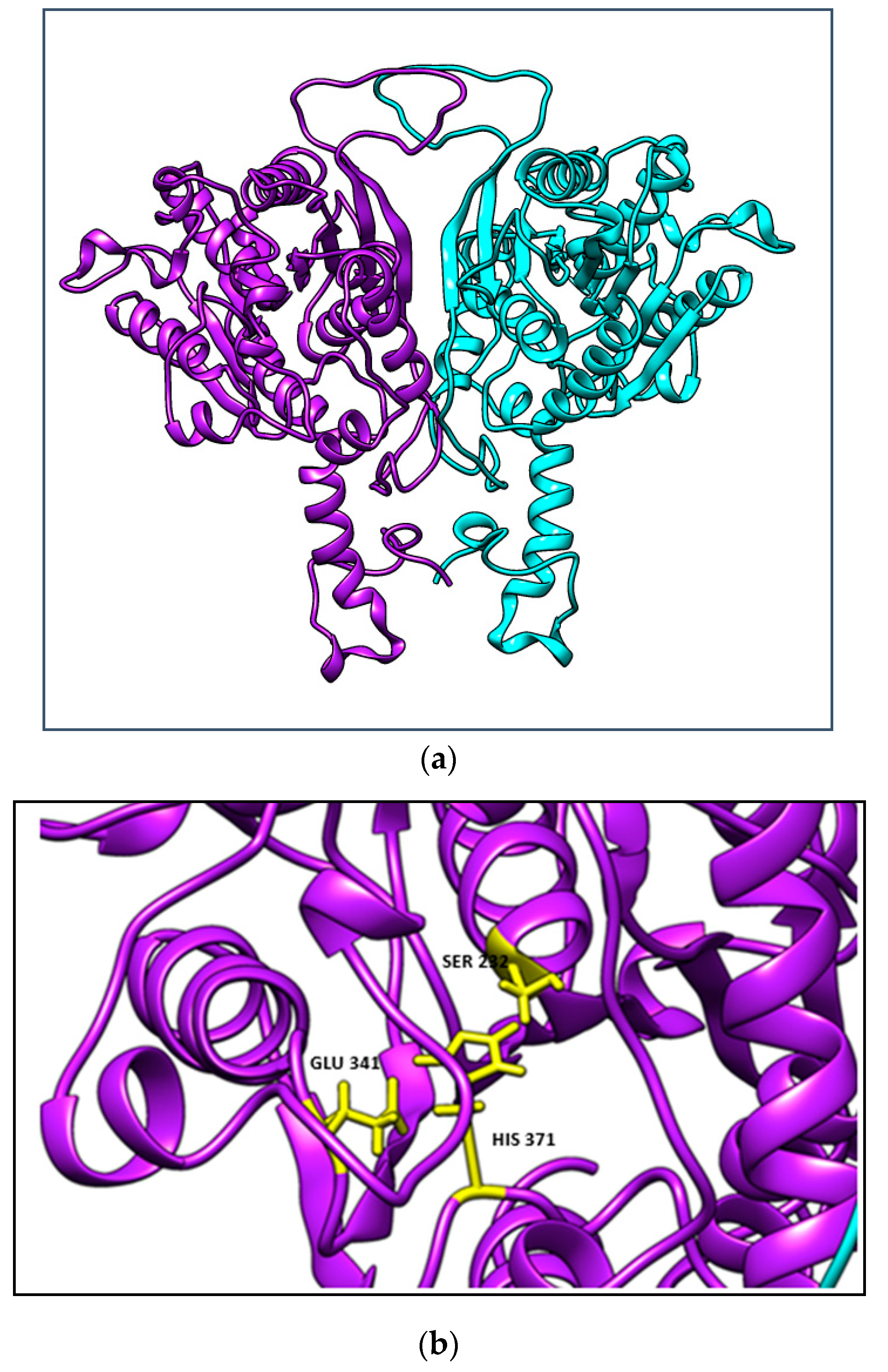
2.4. Homology Modelling and Validation of GlaEst12
3. Materials and Methods
3.1. Sequence Analysis of GlaEst12
3.2. Gene Synthesis, Bacteria Strains, and Plasmids
3.3. Cloning of GlaEst12 in E. coli
3.4. Expression, Solubilisation, and Refolding of GlaEst12 Inclusion Bodies
3.5. Purification of Recombinant of GlaEst12-Like Esterase
3.6. Enzyme Assay
3.7. Characterisation of Purified GlaEst12
3.7.1. Effect of Temperature on Activity and Stability
3.7.2. Effect of pH and pH Stability
3.7.3. Effect of Substrate Specificity
3.7.4. Effect of Metals Ions
3.7.5. Effect of Organic Solvents
3.8. Homology Modelling and Structure Validation
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jensen, M.B.V.; Horsfall, L.E.; Wardrope, C.; Togneri, P.D.; Marles-Wright, J.; Rosser, S.J. Characterisation of a New Family of Carboxyl Esterases with an OsmC Domain. PLoS ONE 2016, 11, e0166128. [Google Scholar] [CrossRef]
- Fojan, P. What Distinguishes an Esterase from a Lipase: A Novel Structural Approach. Biochimie 2000, 82, 1033–1041. [Google Scholar] [CrossRef]
- De Simone, G.; Mandrich, L.; Menchise, V.; Giordano, V.; Febbraio, F.; Rossi, M.; Pedone, C.; Manco, G. A Substrate-Induced Switch in the Reaction Mechanism of a Thermophilic Esterase: KINETIC EVIDENCES AND STRUCTURAL BASIS. J. Biol. Chem. 2004, 279, 6815–6823. [Google Scholar] [CrossRef]
- Lampidonis, A.D.; Rogdakis, E.; Voutsinas, G.E.; Stravopodis, D.J. The Resurgence of Hormone-Sensitive Lipase (HSL) in Mammalian Lipolysis. Gene 2011, 477, 1–11. [Google Scholar] [CrossRef]
- Langin, D.; Laurell, H.; Holst, L.S.; Belfrage, P.; Holm, C. Gene Organization and Primary Structure of Human Hormone-Sensitive Lipase: Possible Significance of a Sequence Homology with a Lipase of Moraxella TA144, an Antarctic Bacterium. Proc. Natl. Acad. Sci. USA 1993, 90, 4897–4901. [Google Scholar] [CrossRef]
- Kim, T.D. Bacterial Hormone-Sensitive Lipases (BHSLs): Emerging Enzymes for Biotechnological Applications. J. Microbiol. Biotechnol. 2017, 27, 1907–1915. [Google Scholar] [CrossRef]
- Pöhlmann, C.; Wang, Y.; Humenik, M.; Heidenreich, B.; Gareis, M.; Sprinzl, M. Rapid, Specific and Sensitive Electrochemical Detection of Foodborne Bacteria. Biosens. Bioelectron. 2009, 24, 2766–2771. [Google Scholar] [CrossRef]
- Febbraio, F.; Merone, L.; Cetrangolo, G.P.; Rossi, M.; Nucci, R.; Manco, G. Thermostable Esterase 2 from Alicyclobacillus acidocaldarius as Biosensor for the Detection of Organophosphate Pesticides. Anal. Chem. 2011, 83, 1530–1536. [Google Scholar] [CrossRef]
- Jaeger, K.E.; Eggert, T. Lipases for Biotechnology. Curr. Opin. Biotechnol. 2002, 13, 390–397. [Google Scholar] [CrossRef]
- Bassegoda, A.; Fillat, A.; Pastor, F.I.J.; Diaz, P. Special Rhodococcus sp. CR-53 Esterase Est4 Contains a GGG(A)X-Oxyanion Hole Conferring Activity for the Kinetic Resolution of Tertiary Alcohols. Appl. Microbiol. Biotechnol. 2013, 97, 8559–8568. [Google Scholar] [CrossRef]
- Yan, Q.; Yang, S.; Duan, X.; Xu, H.; Liu, Y.; Jiang, Z. Characterization of a Novel Hormone-Sensitive Lipase Family Esterase from Rhizomucor miehei with Tertiary Alcohol Hydrolysis Activity. J. Mol. Catal. B Enzym. 2014, 109, 76–84. [Google Scholar] [CrossRef]
- Petrovskaya, L.E.; Novototskaya-Vlasova, K.A.; Gapizov, S.S.; Spirina, E.V.; Durdenko, E.V.; Rivkina, E.M. New Member of the Hormone-Sensitive Lipase Family from the Permafrost Microbial Community. Bioengineered 2017, 8, 420–423. [Google Scholar] [CrossRef]
- Li, P.Y.; Ji, P.; Li, C.Y.; Zhang, Y.; Wang, G.L.; Zhang, X.Y.; Xie, B.B.; Qin, Q.L.; Chen, X.L.; Zhou, B.C.; et al. Structural Basis for Dimerization and Catalysis of a Novel Esterase from the GTSAG Motif Subfamily of the Bacterial Hormone-Sensitive Lipase Family. J. Biol. Chem. 2014, 289, 19031–19041. [Google Scholar] [CrossRef]
- Turchetti, B.; Thomas Hall, S.R.; Connell, L.B.; Branda, E.; Buzzini, P.; Theelen, B.; Müller, W.H.; Boekhout, T. Psychrophilic Yeasts from Antarctica and European Glaciers: Description of Glaciozyma Gen. Nov., Glaciozyma martinii sp. Nov. and Glaciozyma watsonii sp. Nov. Extremophiles 2011, 15, 573–586. [Google Scholar] [CrossRef]
- Boo, S.Y.; Wong, C.M.V.L.; Rodrigues, K.F.; Najimudin, N.; Murad, A.M.A.; Mahadi, N.M. Thermal Stress Responses in Antarctic Yeast, Glaciozyma antarctica PI12, Characterized by Real-Time Quantitative PCR. Polar Biol. 2013, 36, 381–389. [Google Scholar] [CrossRef]
- Alias, N.; Mazian, A.; Salleh, A.B.; Basri, M.; Rahman, R.N.Z.R. Molecular Cloning and Optimization for High Level Expression of Cold-Adapted Serine Protease from Antarctic Yeast Glaciozyma antarctica PI12. Enzym. Res. 2014, 2014, 1–20. [Google Scholar] [CrossRef]
- Hashim, N.H.F.; Sulaiman, S.; Abu Bakar, F.D.; Illias, R.M.; Kawahara, H.; Najimudin, N.; Mahadi, N.M.; Murad, A.M.A. Molecular Cloning, Expression and Characterisation of Afp4, an Antifreeze Protein from Glaciozyma antarctica. Polar Biol. 2014, 37, 1495–1505. [Google Scholar] [CrossRef]
- Ramli, A.N.M.; Azhar, M.A.; Shamsir, M.S.; Rabu, A.; Murad, A.M.A.; Mahadi, N.M.; Illias, R.M. Sequence and Structural Investigation of a Novel Psychrophilic α-Amylase from Glaciozyma antarctica PI12 for Cold-Adaptation Analysis. J. Mol. Model. 2013, 19, 3369–3383. [Google Scholar] [CrossRef]
- Ramli, A.; Mahadi, N.; Rabu, A.; Murad, A.; Bakar, F.; Illias, R. Molecular Cloning, Expression and Biochemical Characterisation of a Cold-Adapted Novel Recombinant Chitinase from Glaciozyma antarctica PI12. Microb. Cell Fact. 2011, 10, 94. [Google Scholar] [CrossRef]
- De Santi, C.; Tutino, M.L.; Mandrich, L.; Giuliani, M.; Parrilli, E.; Del Vecchio, P.; de Pascale, D. The Hormone-Sensitive Lipase from Psychrobacter sp. TA144: New Insight in the Structural/Functional Characterization. Biochimie 2010, 92, 949–957. [Google Scholar] [CrossRef]
- Li, Z.; Su, L.; Wang, L.; Liu, Z.; Gu, Z.; Chen, J.; Wu, J. Novel Insight into the Secretory Expression of Recombinant Enzymes in Escherichia coli. Process Biochem. 2014, 49, 599–603. [Google Scholar] [CrossRef]
- Deb, C. A Novel Lipase Belonging to the Hormone-Sensitive Lipase Family Induced under Starvation to Utilize Stored Triacylglycerol in Mycobacterium tuberculosis. J. Biol. Chem. 2006, 281, 3866–3875. [Google Scholar] [CrossRef] [PubMed]
- Bornhorst, J.A.; Falke, J.J. Purification of Proteins Using Polyhistidine Affinity Tags. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2000; Volume 326, pp. 245–254. [Google Scholar]
- Liu, Y.; Xu, H.; Yan, Q.; Yang, S.; Duan, X.; Jiang, Z. Biochemical Characterization of a First Fungal Esterase from Rhizomucor miehei Showing High Efficiency of Ester Synthesis. PLoS ONE 2013, 8, e77856. [Google Scholar] [CrossRef][Green Version]
- Jeon, J.H.; Lee, H.S.; Kim, J.T.; Kim, S.J.; Choi, S.H.; Kang, S.G.; Lee, J.H. Identification of a New Subfamily of Salt-Tolerant Esterases from a Metagenomic Library of Tidal Flat Sediment. Appl. Microbiol. Biotechnol. 2012, 93, 623–631. [Google Scholar] [CrossRef]
- Jin, P.; Pei, X.; Du, P.; Yin, X.; Xiong, X.; Wu, H.; Zhou, X.; Wang, Q. Overexpression and Characterization of a New Organic Solvent-Tolerant Esterase Derived from Soil Metagenomic DNA. Bioresour. Technol. 2012, 116, 234–240. [Google Scholar] [CrossRef]
- Salwoom, L.; Rahman, R.A.; Zaliha, R.N.; Salleh, A.B.; Convey, P.; Ali, M.; Shukuri, M. New Recombinant Cold-Adapted and Organic Solvent Tolerant Lipase from Psychrophilic Pseudomonas sp. LSK25, Isolated from Signy Island Antarctica. Int. J. Mol. Sci. 2019, 20, 1264. [Google Scholar] [CrossRef]
- Latip, W.; Rahman, R.N.; Leow, A.T.; Shariff, F.M.; Ali, M.S. Expression and Characterization of Thermotolerant Lipase with Broad PH Profiles Isolated from an Antarctic Pseudomonas sp. Strain AMS3. PeerJ 2016, 4, e2420. [Google Scholar] [CrossRef]
- Liu, Z.Q.; Zheng, X.B.; Zhang, S.P.; Zheng, Y.G. Cloning, Expression and Characterization of a Lipase Gene from the Candida antarctica ZJB09193 and Its Application in Biosynthesis of Vitamin A Esters. Microbiol. Res. 2012, 167, 452–460. [Google Scholar] [CrossRef]
- Joseph, B.; Ramteke, P.W.; Thomas, G. Cold Active Microbial Lipases: Some Hot Issues and Recent Developments. Biotechnol. Adv. 2008, 26, 457–470. [Google Scholar] [CrossRef]
- Lopez-Lopez, O.; Cerdan, M.; Siso, M. New Extremophilic Lipases and Esterases from Metagenomics. Curr. Protein Pept. Sci. 2014, 15, 445–455. [Google Scholar] [CrossRef]
- Li, P.Y.; Chen, X.L.; Ji, P.; Li, C.Y.; Wang, P.; Zhang, Y.; Xie, B.B.; Qin, Q.L.; Su, H.N.; Zhou, B.C.; et al. Interdomain Hydrophobic Interactions Modulate the Thermostability of Microbial Esterases from the Hormone-Sensitive Lipase Family. J. Biol. Chem. 2015, 290, 11188–11198. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Huo, Y.Y.; Ji, R.; Kuang, S.; Ji, C.; Xu, X.W.; Li, J. Structural Insights of a Hormone Sensitive Lipase Homologue Est22. Sci. Rep. 2016, 6, 28550. [Google Scholar] [CrossRef] [PubMed]
- Andreini, C.; Bertini, I.; Cavallaro, G.; Holliday, G.L.; Thornton, J.M. Metal Ions in Biological Catalysis: From Enzyme Databases to General Principles. J. Biol. Inorg. Chem. 2008, 13, 1205–1218. [Google Scholar] [CrossRef] [PubMed]
- Gricajeva, A.; Bikutė, I.; Kalėdienė, L. Atypical Organic-Solvent Tolerant Bacterial Hormone Sensitive Lipase-like Homologue EstAG1 from Staphylococcus Saprophyticus AG1: Synthesis and Characterization. Int. J. Biol. Macromol. 2019, 130, 253–265. [Google Scholar] [CrossRef] [PubMed]
- Virk, A.P. A New Esterase, Belonging to Hormone-Sensitive Lipase Family, Cloned from Rheinheimera sp. Isolated from Industrial Effluent. J. Microbiol. Biotechnol. 2011, 21, 667–674. [Google Scholar] [CrossRef] [PubMed]
- Bansal, V.; Delgado, Y.; Fasoli, E.; Ferrer, A.; Griebenow, K.; Secundo, F.; Barletta, G.L. Effect of Prolonged Exposure to Organic Solvents on the Active Site Environment of Subtilisin Carlsberg. J. Mol. Catal. B Enzym. 2010, 64, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.E.; Chivian, D.; Baker, D. Protein Structure Prediction and Analysis Using the Robetta Server. Nucleic Acids Res. 2004, 32, W526–W531. [Google Scholar] [CrossRef]
- D’Amico, S.; Collins, T.; Marx, J.C.; Feller, G.; Gerday, C. Psychrophilic Microorganisms: Challenges for Life. EMBO Rep. 2006, 7, 385–389. [Google Scholar] [CrossRef]
- Rehdorf, J.; Behrens, G.A.; Nguyen, G.S.; Kourist, R.; Bornscheuer, U.T. Pseudomonas Putida Esterase Contains a GGG(A)X-Motif Confering Activity for the Kinetic Resolution of Tertiary Alcohols. Appl. Microbiol. Biotechnol. 2012, 93, 1119–1126. [Google Scholar] [CrossRef]
- Ali, M.S.M.; Ganasen, M.; Rahman, R.N.Z.R.A.; Chor, A.L.T.; Salleh, A.B.; Basri, M. Cold-Adapted RTX Lipase from Antarctic Pseudomonas sp. Strain AMS8: Isolation, Molecular Modeling and Heterologous Expression. Protein J. 2013, 32, 317–325. [Google Scholar] [CrossRef]
- Firdaus-Raih, M.; Hashim, N.H.F.; Bharudin, I.; Abu Bakar, M.F.; Huang, K.K.; Alias, H.; Lee, B.K.B.; Mat Isa, M.N.; Mat-Sharani, S.; Sulaiman, S.; et al. The Glaciozyma antarctica Genome Reveals an Array of Systems That Provide Sustained Responses towards Temperature Variations in a Persistently Cold Habitat. PLoS ONE 2018, 13, e0189947. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Sumby, K.M.; Matthews, A.H.; Grbin, P.R.; Jiranek, V. Cloning and Characterization of an Intracellular Esterase from the Wine-Associated Lactic Acid Bacterium Oenococcus oeni. Appl. Environ. Microbiol. 2009, 75, 6729–6735. [Google Scholar] [CrossRef] [PubMed]
- Colovos, C.; Yeates, T.O. Verification of Protein Structures: Patterns of Nonbonded Atomic Interactions. Protein Sci. 1993, 2, 1511–1519. [Google Scholar] [CrossRef] [PubMed]
- Luthy, R.; Bowie, J.U.; Eisenberg, D. Assesment of Protein Models with 3 Dimensional Profile. Nature 1992, 356, 83–85. [Google Scholar] [CrossRef]




| Validation Tools | Score (%) |
|---|---|
| (A) Verify 3D | 87.8 |
| (B) Errat | 91.3 |
| (C) Ramachandran Plot | |
| Most favoured region | 84.8 |
| Additional allowed region | 14.8 |
| Generously allowed region | 0.3 |
| Disallowed region | 0.1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohamad Tahir, H.; Raja Abd Rahman, R.N.Z.; Chor Leow, A.T.; Mohamad Ali, M.S. Expression, Characterisation and Homology Modelling of a Novel Hormone-Sensitive Lipase (HSL)-Like Esterase from Glaciozyma antarctica. Catalysts 2020, 10, 58. https://doi.org/10.3390/catal10010058
Mohamad Tahir H, Raja Abd Rahman RNZ, Chor Leow AT, Mohamad Ali MS. Expression, Characterisation and Homology Modelling of a Novel Hormone-Sensitive Lipase (HSL)-Like Esterase from Glaciozyma antarctica. Catalysts. 2020; 10(1):58. https://doi.org/10.3390/catal10010058
Chicago/Turabian StyleMohamad Tahir, Hiryahafira, Raja Noor Zaliha Raja Abd Rahman, Adam Thean Chor Leow, and Mohd Shukuri Mohamad Ali. 2020. "Expression, Characterisation and Homology Modelling of a Novel Hormone-Sensitive Lipase (HSL)-Like Esterase from Glaciozyma antarctica" Catalysts 10, no. 1: 58. https://doi.org/10.3390/catal10010058
APA StyleMohamad Tahir, H., Raja Abd Rahman, R. N. Z., Chor Leow, A. T., & Mohamad Ali, M. S. (2020). Expression, Characterisation and Homology Modelling of a Novel Hormone-Sensitive Lipase (HSL)-Like Esterase from Glaciozyma antarctica. Catalysts, 10(1), 58. https://doi.org/10.3390/catal10010058





