An Intelligent Computer-Aided Scheme for Classifying Multiple Skin Lesions
Abstract
1. Introduction
2. Literature Review
3. Materials
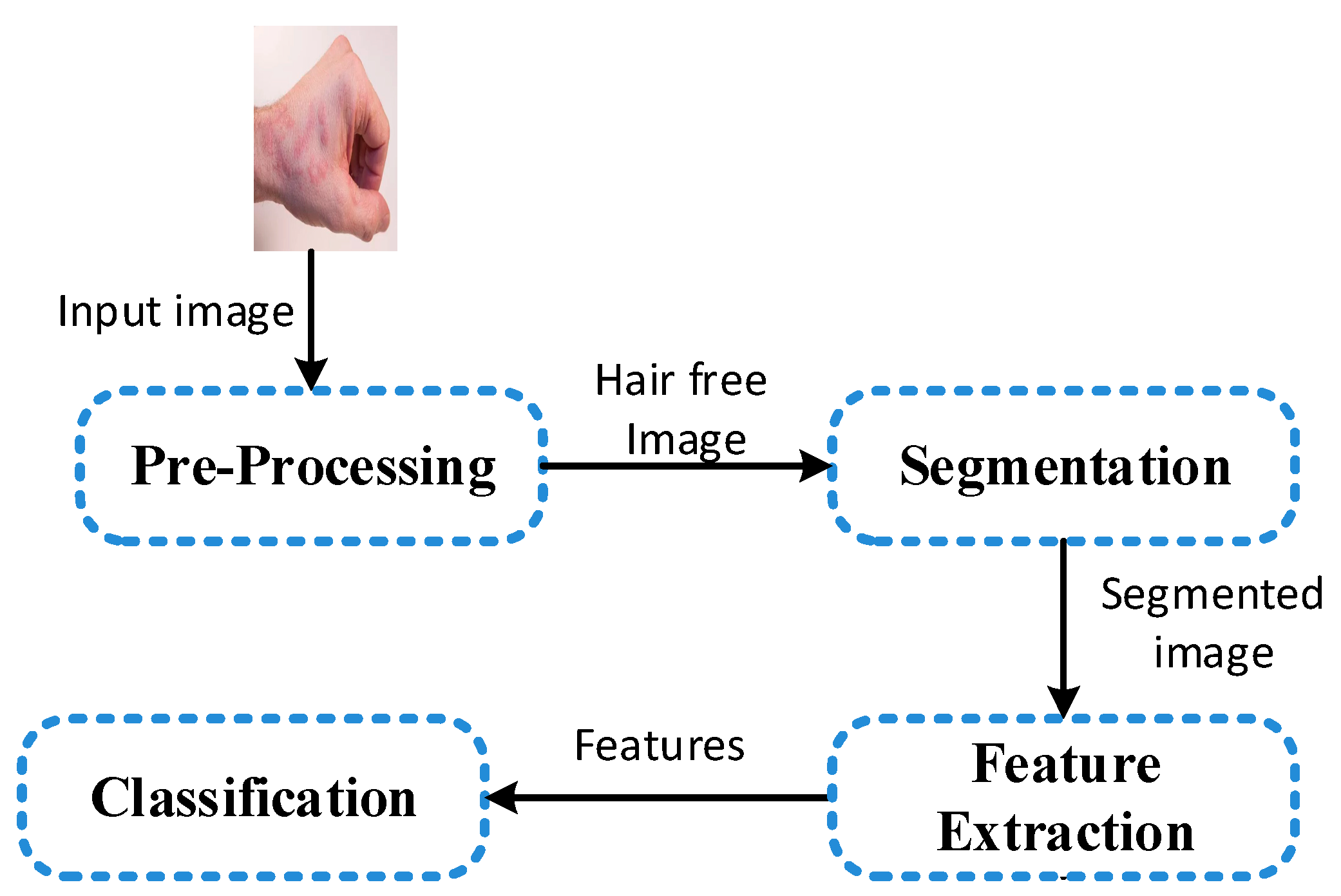
4. Method
4.1. Pre-Processing and Segmentation
4.2. Feature Extraction
4.2.1. Colour Features
4.2.2. Texture Features
4.3. Classification
5. Results and Discussion
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Seth, D.; Cheldize, K.; Brown, D.; Freeman, E.E. Global burden of skin disease: Inequities and innovations. Curr. Dermatol. Rep. 2017, 6, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Hay, R.J.; Augustin, M.; Griffiths, C.E. The global challenge for skin health. Br. J. Dermatol. 2015, 172, 1469–1472. [Google Scholar] [CrossRef] [PubMed]
- Picardi, A.; Lega, I.; Tarolla, E. Suicide risk in skin disorders. Clin. Dermatol. 2013, 31, 47–56. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. New Report Shows that 400 Million Do not Have Access to Essential Health Services; World Health Organization: Geneva, Switzerland, 2015. [Google Scholar]
- 5 Most Common Skin Disorders. Available online: http://www.foxnews.com/story/2009/12/15/5-most-common-skin-disorders.html (accessed on 1 June 2018).
- Hameed, N.; Shabut, A.; Hossain, M.A. A Computer-aided diagnosis system for classifying prominent skin lesions using machine learning. In Proceedings of the 10th Computer Science and Electronic Engineering (CEEC), Colchester, UK, 19–21 September 2018; pp. 186–191. [Google Scholar]
- Williams, H.C.; Dellavalle, R.P.; Garner, S. Acne vulgaris. Lancet 2012, 379, 361–372. [Google Scholar] [CrossRef]
- Karimkhani, C.; Dellavalle, R.P.; Coffeng, L.E.; Flohr, C.; Hay, R.J.; Langan, S.M.; Nsoesie, E.O.; Ferrari, A.J.; Erskine, H.E.; Silverberg, J.I.; et al. Global skin disease morbidity and mortality: An update from the global burden of disease study 2013. JAMA Dermatol. 2017, 153, 406–412. [Google Scholar] [CrossRef] [PubMed]
- Atopic Eczema. 2018. Available online: https://www.nhs.uk/conditions/atopic-eczema/ (accessed on 14 May 2018).
- Hameed, N.; Ruskin, A.; Hassan, K.A.; Hossain, M.A. A comprehensive survey on image-based computer aided diagnosis systems for skin cancer. In Proceedings of the 10th International Conference on Software, Knowledge, Information Management & Applications (SKIMA), Chengdu, China, 15–17 December 2016; pp. 205–214. [Google Scholar]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Al Abbadi, N.K.; Dahir, N.S.; Al-Dhalimi, M.A.; Restom, H. Psoriasis Detection Using Skin Color and Texture Features 1. J. Comput. Sci. 2010, 6, 648–652. [Google Scholar] [CrossRef]
- Umbaugh, S.E.; Moss, R.H.; Stoecker, W.V. Applying artificial intelligence to the identification of variegated coloring in skin tumors. IEEE Eng. Med. Biol. Mag. 1991, 10, 57–62. [Google Scholar] [CrossRef]
- Ercal, F.; Chawla, A.; Stoecker, W.V.; Lee, H.C.; Moss, R.H. Neural network diagnosis of malignant melanoma from color images. IEEE Trans. Biomed. Eng. 1994, 41, 837–845. [Google Scholar] [CrossRef]
- Nischik, M.; Forster, C. Analysis of skin erythema using true-color images. IEEE Trans. Med. Imaging 1997, 16, 711–716. [Google Scholar] [CrossRef]
- Vasconcelos, C.N.; Vasconcelos, B.N. Experiments using deep learning for dermoscopy image analysis. Pattern Recognit. Lett. 2017. [Google Scholar] [CrossRef]
- Zhang, Z.; Stoecker, W.V.; Moss, R.H. Border detection on digitized skin tumor images. IEEE Trans. Med. Imaging 2000, 19, 1128–1143. [Google Scholar] [CrossRef] [PubMed]
- Dorj, U.O.; Lee, K.K.; Choi, J.Y.; Lee, M. The skin cancer classification using deep convolutional neural network. Multimed. Tools Appl. 2018, 77, 9909–9924. [Google Scholar] [CrossRef]
- Taufiq, M.A.; Hameed, N.; Anjum, A.; Hameed, F. m-Skin Doctor: A mobile enabled system for early melanoma skin cancer detection using support vector machine. In Health 360°; Springer: Cham, Switzerland, 2017; pp. 468–475. [Google Scholar]
- Güvenir, H.A.; Emeksiz, N. An expert system for the differential diagnosis of erythemato-squamous diseases. Expert Syst. Appl. 2000, 18, 43–49. [Google Scholar] [CrossRef]
- Übeyli, E.D. Multiclass support vector machines for diagnosis of erythemato-squamous diseases. Expert Syst. Appl. 2008, 35, 1733–1740. [Google Scholar] [CrossRef]
- Chang, C.L.; Chen, C.H. Applying decision tree and neural network to increase quality of dermatologic diagnosis. Expert Syst. Appl. 2009, 36, 4035–4041. [Google Scholar] [CrossRef]
- Xie, J.; Wang, C. Using support vector machines with a novel hybrid feature selection method for diagnosis of erythemato-squamous diseases. Expert Syst. Appl. 2011, 38, 5809–5815. [Google Scholar] [CrossRef]
- Kumar, V.B.; Kumar, S.S.; Saboo, V. Dermatological disease detection using image processing and machine learning. In Proceedings of the 2016 Third International Conference on Artificial Intelligence and Pattern Recognition (AIPR), Lodz, Poland, 19–21 September 2016. [Google Scholar]
- Nanni, L. An ensemble of classifiers for the diagnosis of erythemato-squamous diseases. Neurocomputing 2006, 69, 842–845. [Google Scholar] [CrossRef]
- DermIS. Available online: http://www.dermis.net/dermisroot/en/home/index.htm (accessed on 29 June 2017).
- Derm101 Image Library. Available online: https://www.derm101.com/image-library/ (accessed on 12 January 2018).
- DermNZ-Image Library. Available online: https://www.dermnetnz.org/image-library/ (accessed on 13 January 2018).
- PH2 Database. Available online: http://www.fc.up.pt/addi/ph2 database.html (accessed on 12 January 2018).
- Lam, M.N.; Munia, T.T.; Tavakolian, K.; Vasefi, F.; MacKinnon, N.; Fazel-Rezai, R. Automatic detection and severity measurement of eczema using image processing. In Proceedings of the 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Orlando, FL, USA, 16–20 August 2016. [Google Scholar]
- Codella, N.C.; Gutman, D.; Celebi, M.E.; Helba, B.; Marchetti, M.A.; Dusza, S.W.; Kalloo, A.; Liopyris, K.; Mishra, N.; Kittler, H.; et al. Skin lesion analysis toward melanoma detection: A challenge at the 2017 international symposium on biomedical imaging (isbi), hosted by the international skin imaging collaboration (isic). In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 August 2018. [Google Scholar]
- Hameed, N.; Shabut, A.; Hameed, F.; Cirstea, S.; Hossain, M.A. An Intelligent Inflammatory Skin Lesions Classification Scheme for Mobile Devices. In Proceedings of the IEEE International Conference on Computing, Electronics & Communications Engineering, London, UK, 22–23 August 2019; pp. 83–88. [Google Scholar]
- Lee, T.; Ng, V.; Gallagher, R.; Coldman, A.; McLean, D. Dullrazor®: A software approach to hair removal from images. Comput. Biol. Med. 1997, 27, 533–543. [Google Scholar] [CrossRef]
- Phung, S.L.; Bouzerdoum, A.; Chai, D. Skin segmentation using color pixel classification: Analysis and comparison. IEEE Trans. Pattern Anal. Mach. Intell. 2005, 1, 148–154. [Google Scholar] [CrossRef]
- UCI Machine Learning Repository: Dermatology Data Set. Available online: https://archive.ics.uci.edu/ml/datasets/Dermatology (accessed on 27 April 2017).
- Erol, R.; Bayraktar, M.; Kockara, S.; Kaya, S.; Halic, T. Texture based skin lesion abruptness quantification to detect malignancy. BMC Bioinform. 2017, 18, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Schnürle, S.; Pouly, M.; vor der Brück, T.; Navarini, A.; Koller, T. On using Support Vector Machines for the Detection and Quantification of Hand Eczema. In Proceedings of the 9th International Conference on Agents and Artificial Intelligence (ICAART), Porto, Portugal, 24–26 February 2017; pp. 75–84. [Google Scholar]
- Hameed, N.; Shabut, A.M.; Hossain, M.A. Multi-Class Skin Diseases Classification Using Deep Convolutional Neural Network and Support Vector Machine. In Proceedings of the 12th International Conference on Software, Knowledge, Information Management & Applications (SKIMA), Phnom Penh, Cambodia, 3–5 December 2018. [Google Scholar]
- Texture Analysis Using the Gray-Level Co-Occurrence Matrix (GLCM). Available online: https://uk.mathworks.com/help/images/texture-analysis-using-the-gray-level-co-occurrence-matrix-glcm.html (accessed on 23 June 2018).
- Amadasun, M.; King, R. Textural features corresponding to textural properties. IEEE Trans. Syst. Man Cybern. 1989, 19, 1264–1274. [Google Scholar] [CrossRef]
- Choose Classifier Options. Available online: https://uk.mathworks.com/help/stats/choose-a-classifier.html (accessed on 12 January 2018).
- Sokolova, M.; Lapalme, G. A systematic analysis of performance measures for classification tasks. Inf. Process. Manag. 2009, 45, 427–437. [Google Scholar] [CrossRef]
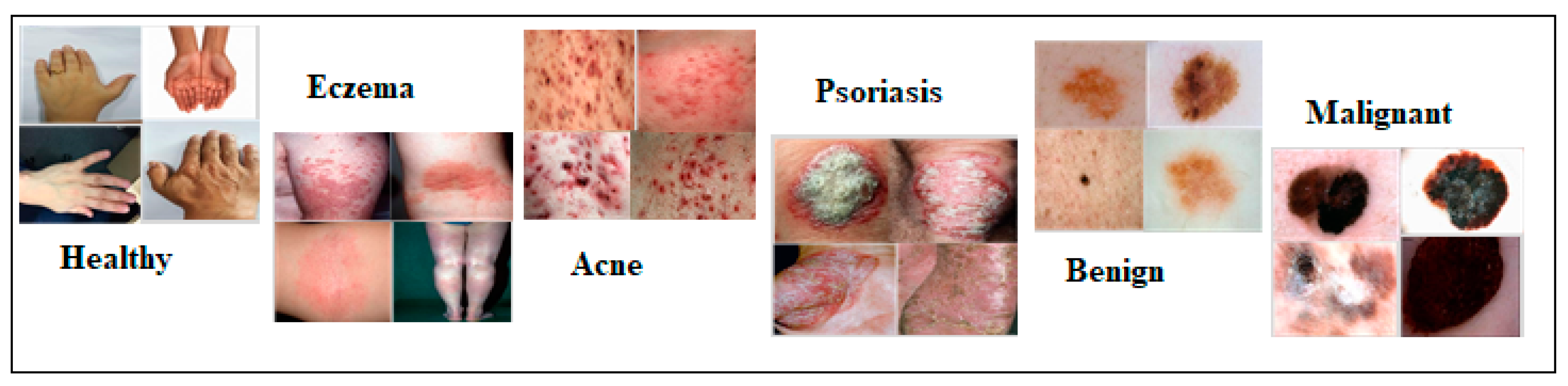
| Category | No. of Images |
|---|---|
| Healthy | 300 |
| Acne | 300 |
| Eczema | 300 |
| Psoriasis | 300 |
| Benign | 300 |
| Malignant | 300 |
| Total | 1800 |
| Feature Name | Description | Formula |
|---|---|---|
| Min | Minimum pixel value of R, G and B colour | Min(colour space) |
| Max | Maximum pixel value of R, G and B colour | Max(colour space) |
| Mean | Measures image overall intensity | |
| Mode | Gives information about the most occurring value | Mode(colour space) |
| Standard Deviation | Presents the spread of the data | |
| Skewness | Measure asymmetry of the probability distribution | |
| Energy | Gives information about the spread of the pixel values | |
| Entropy | Measure the required amount of information to code the image data | |
| Kurtosis | Measure of the peakness of the probability distribution of an image | |
| Legends*: is the number of intensity levels, is the intensity level, is the number of rows, is the number of columns in the image, is the mean, is the standard deviation | ||
| Name | Description | Formula |
|---|---|---|
| ContrastGLCM | Measure the local fluctuations of grey levels of neighbor pixels | |
| CorrelationGLCM | Measure the joint probability occurrence of specified pair pixels | |
| EnergyGLCM | Measure the sum of squared elements in the GLCM | |
| HomogeneityGLCM | Measures the local uniformity |
| Name | Description | Formula |
|---|---|---|
| Busyness | Measure changes in grey levels between neighboring voxels | |
| Complexity | Measure the non-uniformity and rapid changes in grey-levels | |
| Contrast | Measures the changes between voxels and their neighborhood | |
| Strength | Measure the primitives in an image |
| Classifier | Kernel |
|---|---|
| Tree | Fine Tree |
| Medium Tree | |
| Coarse Tree | |
| Support Vector Machine | Linear |
| Quadratic | |
| Cubic | |
| Fine Gaussian | |
| Coarse Gaussian | |
| k-Nearest Neighbors | Fine |
| Medium | |
| Coarse | |
| Cosine | |
| Cubic | |
| Weighted | |
| Ensemble | Boosted Trees |
| Bagged Trees | |
| Subspace Discriminant | |
| Subspace KNN | |
| RUSBoosted Trees |
| Measure | Formula | Description |
|---|---|---|
| Measure the number of correct classifications over the total number of examples evaluated | ||
| Measure the number of actual positive cases that are correctly identified | ||
| Measure the number of actual negative cases that are correctly identified | ||
| Legends: = Individual class i.e. Healthy, acne, eczema, psoriasis, benign and malignant = Total Number of classes = 6 | ||
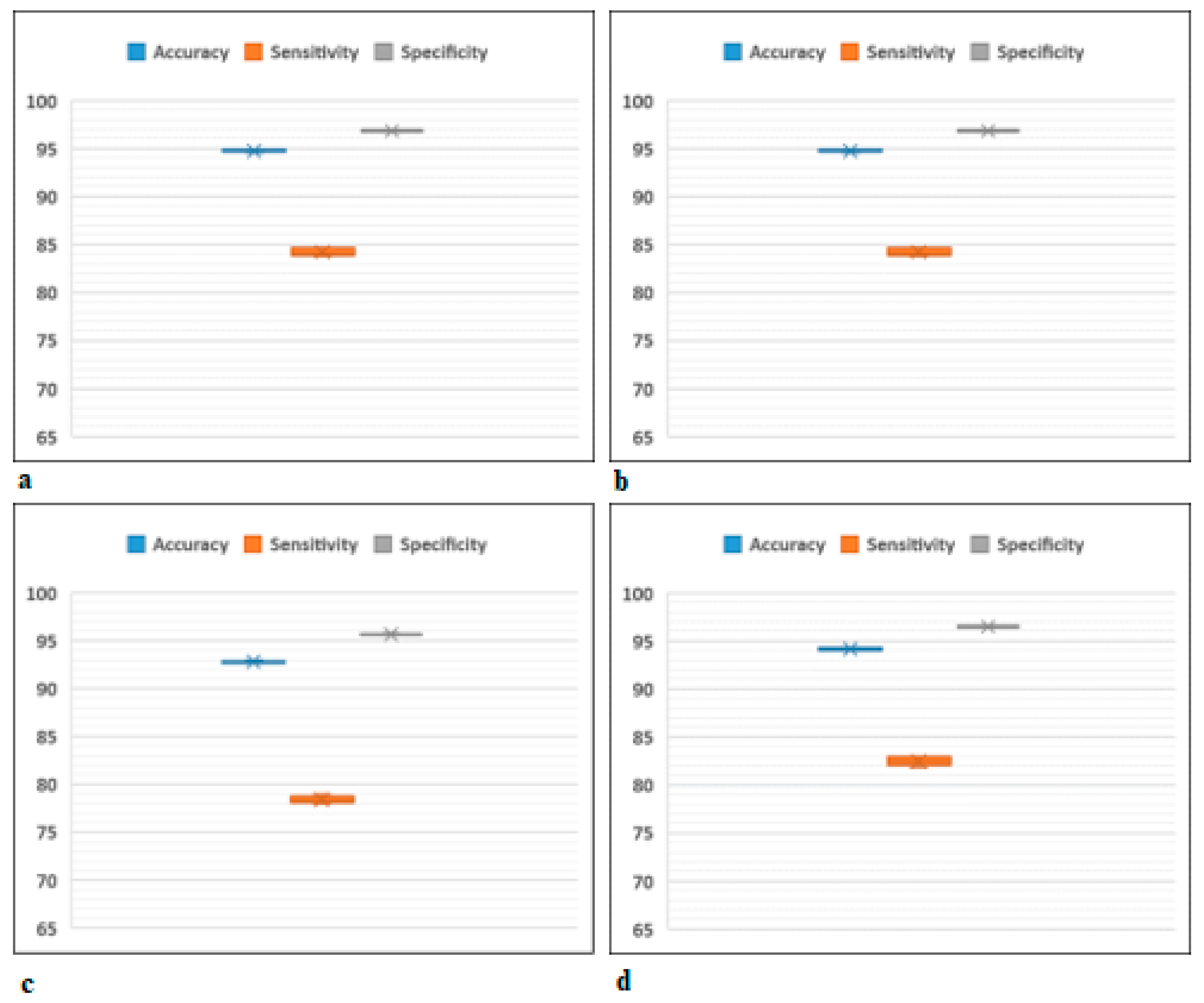
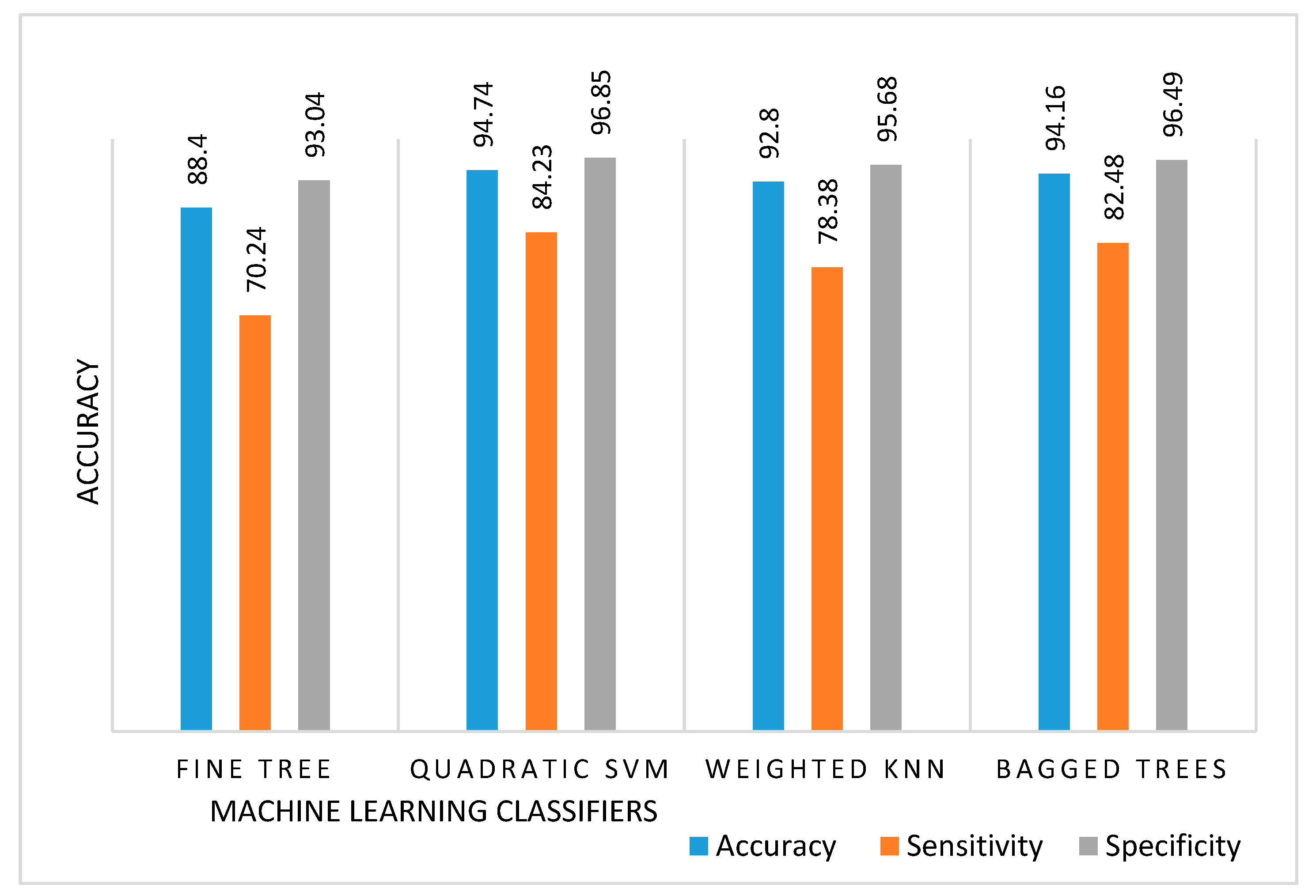
| Classifier | Accuracy (SD) | Sensitivity (SD) | Specificity (SD) |
|---|---|---|---|
| Fine Tree | 88.40 (0.27) | 70.24 (0.83) | 93.04 (0.17) |
| Quadratic SVM | 94.74 (0.11) | 84.23 (0.32) | 96.85 (0.06) |
| Weighted KNN | 92.80 (0.11) | 78.38 (0.33) | 95.68 (0.06) |
| Bagged Trees | 94.16 (0.13) | 82.48 (0.39) | 96.49 (0.07) |
| Reference | Accuracy | Sensitivity | Specificity |
|---|---|---|---|
| [1] | 83 | NA | NA |
| Proposed Work | 94.74 | 84.23 | 96.85 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hameed, N.; Hameed, F.; Shabut, A.; Khan, S.; Cirstea, S.; Hossain, A. An Intelligent Computer-Aided Scheme for Classifying Multiple Skin Lesions. Computers 2019, 8, 62. https://doi.org/10.3390/computers8030062
Hameed N, Hameed F, Shabut A, Khan S, Cirstea S, Hossain A. An Intelligent Computer-Aided Scheme for Classifying Multiple Skin Lesions. Computers. 2019; 8(3):62. https://doi.org/10.3390/computers8030062
Chicago/Turabian StyleHameed, Nazia, Fozia Hameed, Antesar Shabut, Sehresh Khan, Silvia Cirstea, and Alamgir Hossain. 2019. "An Intelligent Computer-Aided Scheme for Classifying Multiple Skin Lesions" Computers 8, no. 3: 62. https://doi.org/10.3390/computers8030062
APA StyleHameed, N., Hameed, F., Shabut, A., Khan, S., Cirstea, S., & Hossain, A. (2019). An Intelligent Computer-Aided Scheme for Classifying Multiple Skin Lesions. Computers, 8(3), 62. https://doi.org/10.3390/computers8030062





