An Empirical Evaluation of Neural Network Architectures for 3D Spheroid Segmentation
Abstract
1. Introduction
- The analysis of ensemble images, where traditional imaging methods process images one by one [38].
- Generalizability and transferability, as DL-based models can learn and replicate shared properties of spheroids, enabling efficient segmentation [39].
- A reduction in the need for domain-specific experts to validate automated segmentation [40].
2. Related Works
3. Formulation of Deep Neural Network Models
3.1. UNet
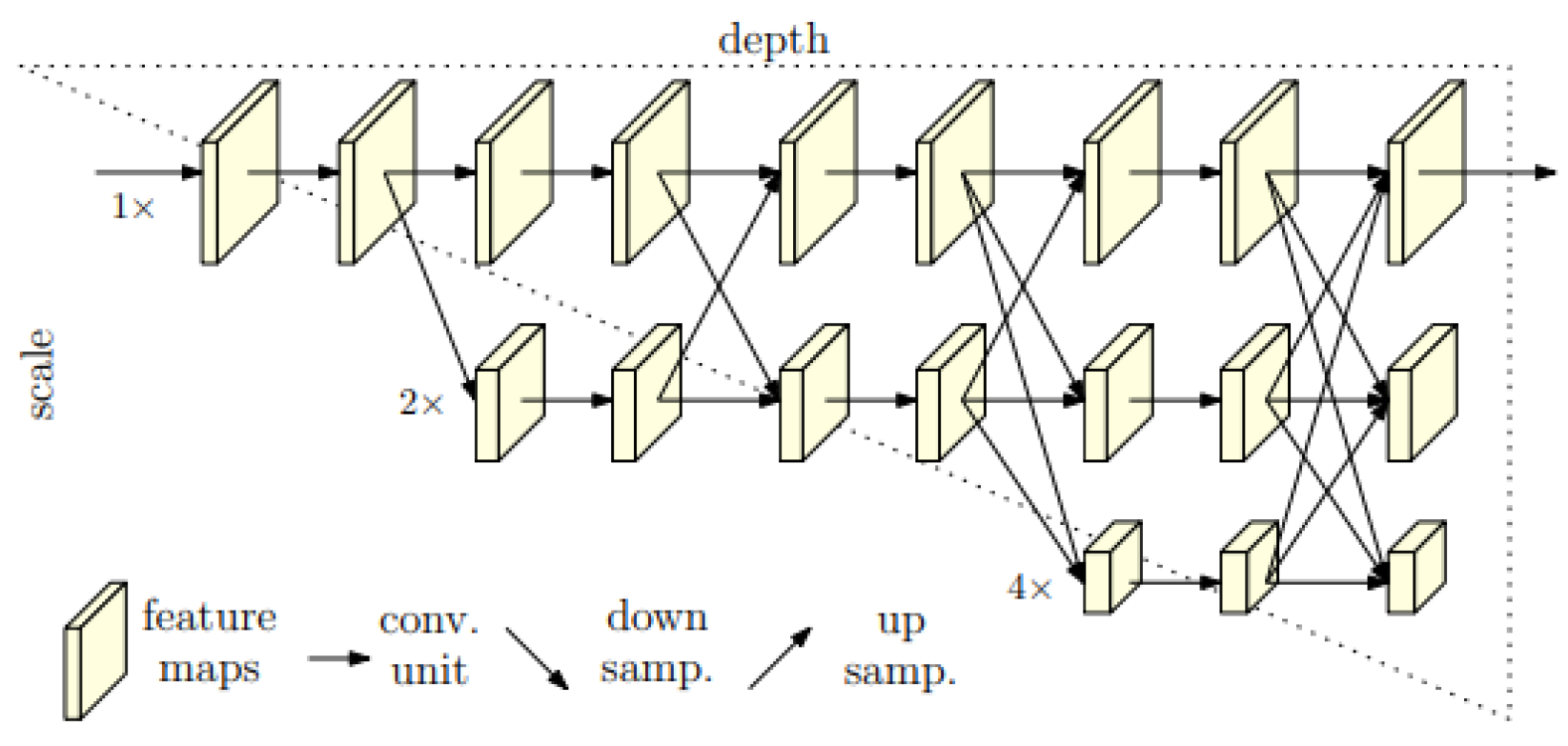
3.2. HRNet Architecture
3.3. DeepLabV3+
3.4. Optimizers
4. Proposed Methodology
| Algorithm 1: Image Segmentation Using UNet, HRNet, and DeepLabV3+ |
 |
5. Results and Discussion
5.1. Datasets
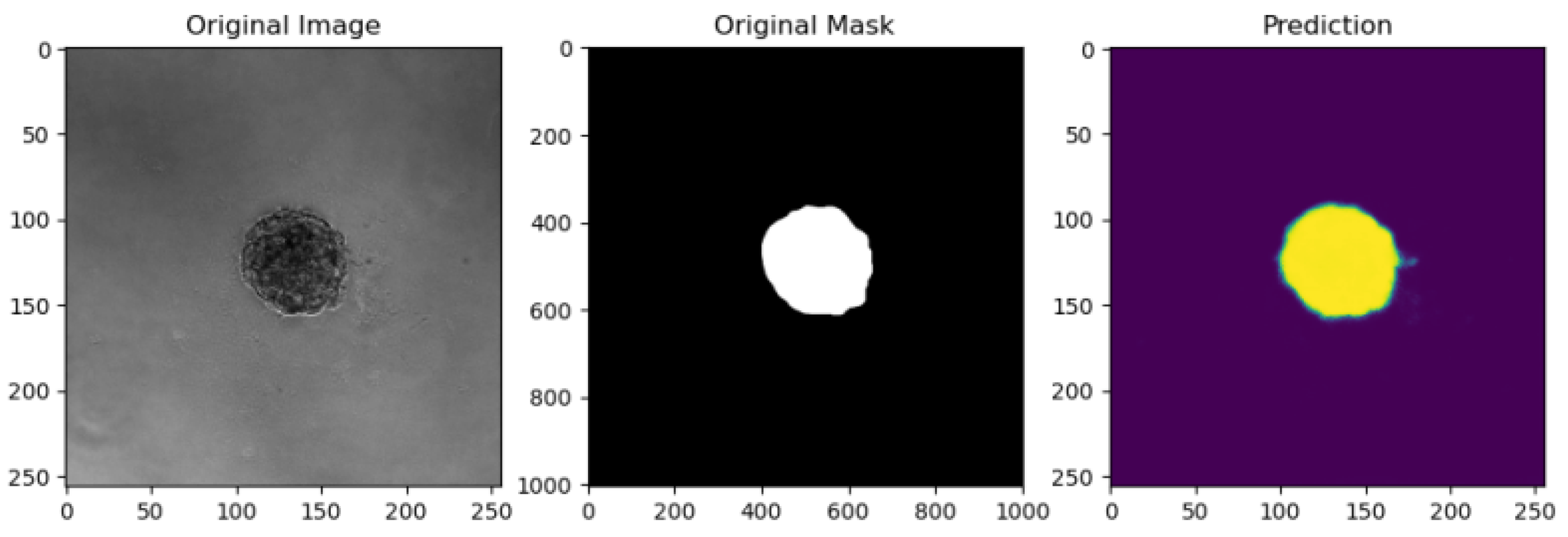
5.2. Evaluation Metrics
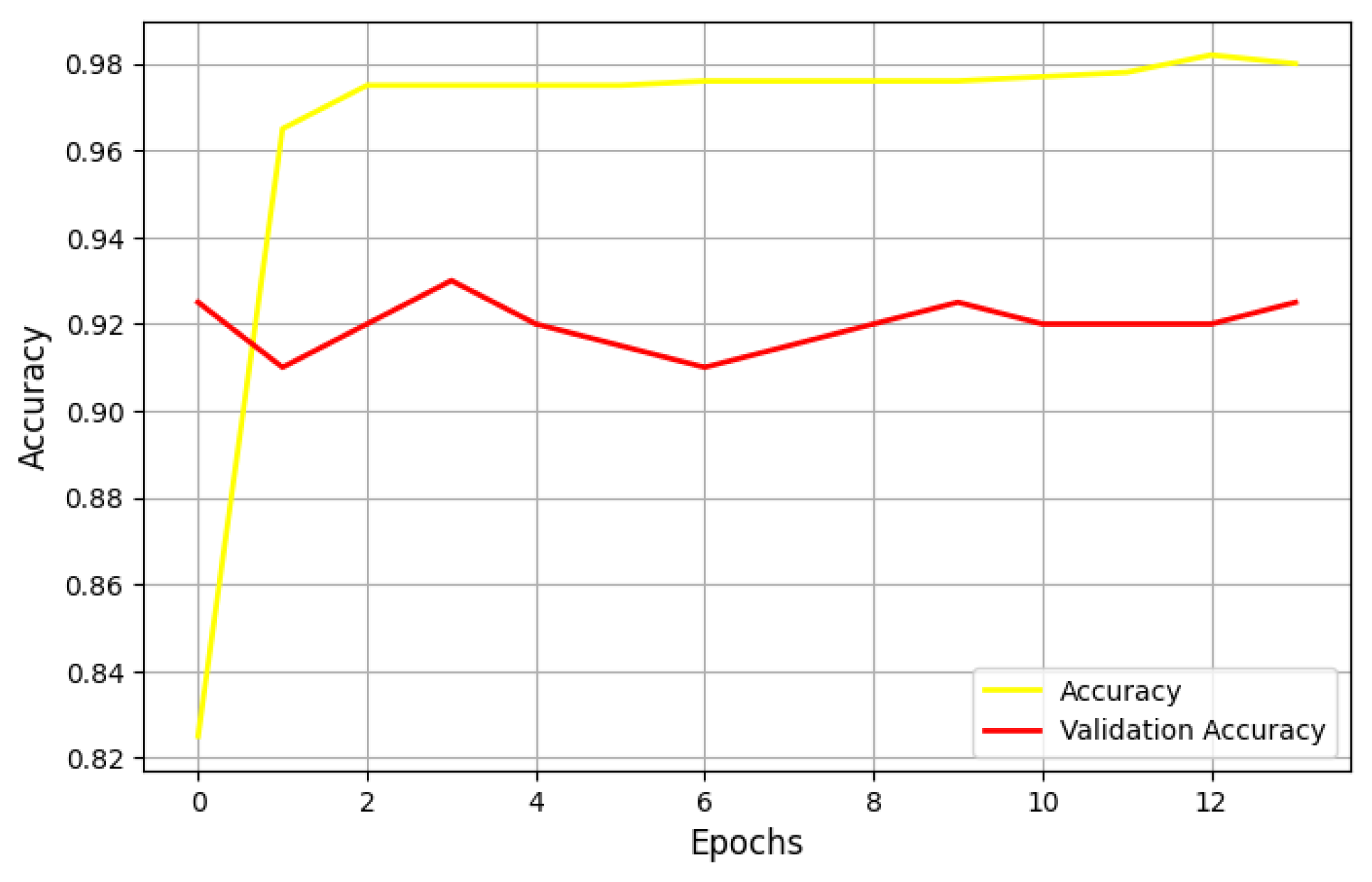
5.3. Performance of the U-Net Model
5.3.1. Impact of Learning Rates on Model Performance
5.3.2. Performance Using SGD and Adagrad Optimizers
5.4. Performance of the DeeplabV3+ Model
5.5. Performance of the HRNet Model
5.6. Comparison of Model Performance on Various Metrics
5.7. Independent Holdout Test Evaluation
- Training set (60% of the data): Used for model optimization during training.
- Validation set (20% of the data): Used for hyperparameter tuning and monitoring model performance during training.
- Test set (20% of the data): Reserved exclusively for final model evaluation on unseen data.
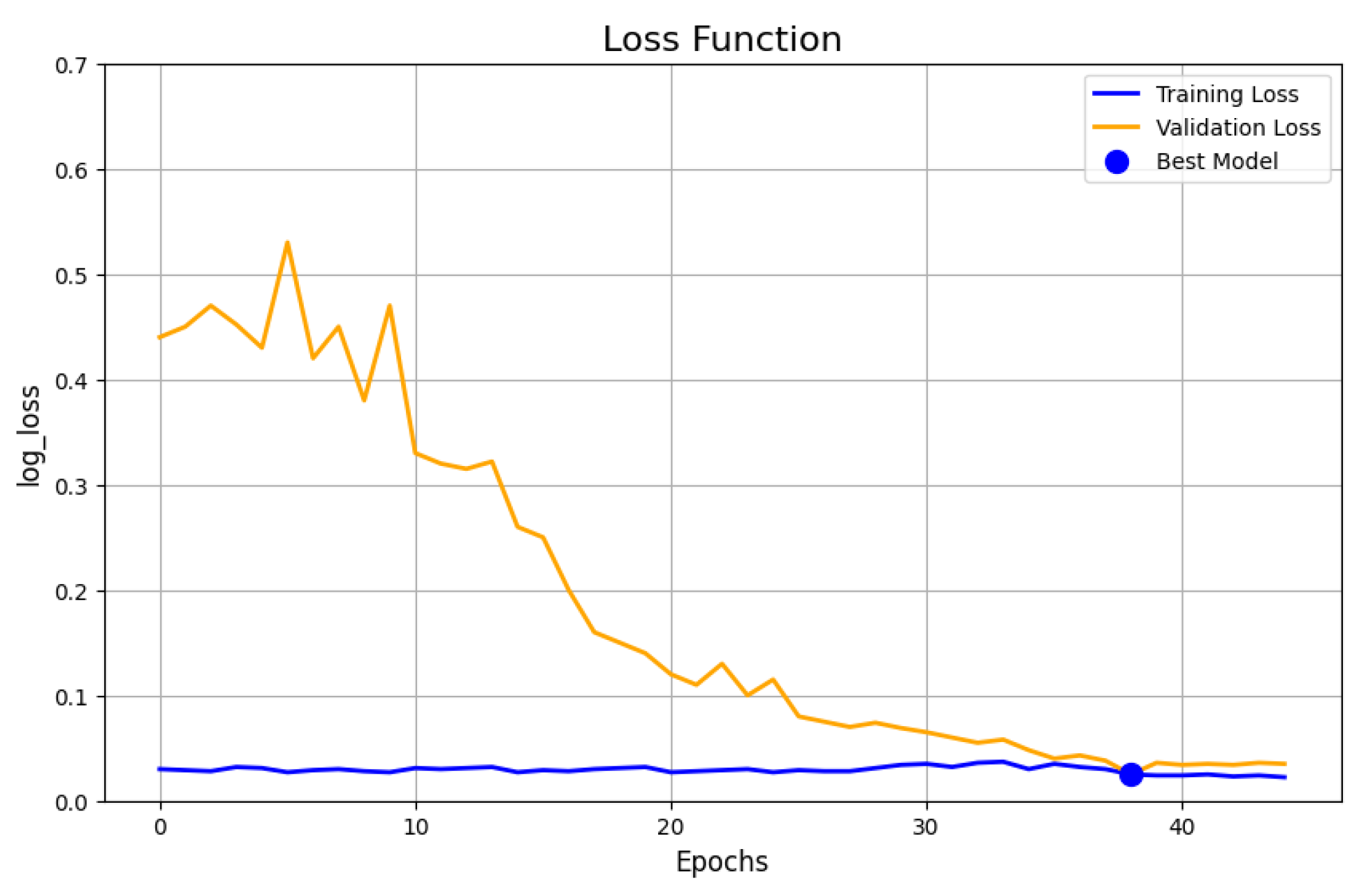
5.8. Addressing Overfitting Through Data Augmentation and Regularization
5.9. Dataset Limitations and Diversity Considerations
5.10. Evaluation Metrics and Their Relevance to Biomedical Segmentation
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Lee, S.Y.; Koo, I.S.; Hwang, H.J.; Lee, D.W. In Vitro three-dimensional (3D) cell culture tools for spheroid and organoid models. SLAS Discov. 2023, 28, 119–137. [Google Scholar] [CrossRef]
- Nguyen, T.H.; Ngo, T.K.N.; Vu, M.A.; Tu, T.Y. Blurry-Consistency Segmentation Framework with Selective Stacking on Differential Interference Contrast 3D Breast Cancer Spheroid. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 16–22 June 2024; pp. 5223–5230. [Google Scholar]
- Yun, C.; Kim, S.H.; Kim, K.M.; Yang, M.H.; Byun, M.R.; Kim, J.H.; Kwon, D.; Pham, H.T.; Kim, H.S.; Kim, J.H.; et al. Advantages of Using 3D Spheroid Culture Systems in Toxicological and Pharmacological Assessment for Osteogenesis Research. Int. J. Mol. Sci. 2024, 25, 2512. [Google Scholar] [CrossRef]
- Arora, S.; Singh, S.; Mittal, A.; Desai, N.; Khatri, D.K.; Gugulothu, D.; Lather, V.; Pandita, D.; Vora, L. Spheroids in cancer research: Recent advances and opportunities. J. Drug Deliv. Sci. Technol. 2024, 100, 106033. [Google Scholar] [CrossRef]
- Jadhav, S.; Rath, S.N.; Roopavath, U.K. A Review on Multicellular Spheroids and Organoids for Breast Cancer Diagnosis and Therapy. Biomed. Mater. Devices 2024, 1–23. [Google Scholar] [CrossRef]
- Feng, L.; Pan, R.; Ning, K.; Sun, W.; Chen, Y.; Xie, Y.; Wang, M.; Li, Y.; Yu, L. The impact of 3D tumor spheroid maturity on cell migration and invasion dynamics. Biochem. Eng. J. 2025, 213, 109567. [Google Scholar] [CrossRef]
- Ascione, F.; Ferraro, R.; Dogra, P.; Cristini, V.; Guido, S.; Caserta, S. Gradient-induced instability in tumour spheroids unveils the impact of microenvironmental nutrient changes. Sci. Rep. 2024, 14, 20837. [Google Scholar] [CrossRef]
- Zhu, Y.; Kang, E.; Wilson, M.; Basso, T.; Chen, E.; Yu, Y.; Li, Y.R. 3D tumor spheroid and organoid to model tumor microenvironment for cancer immunotherapy. Organoids 2022, 1, 149–167. [Google Scholar] [CrossRef]
- Piccinini, F.; Peirsman, A.; Stellato, M.; Pyun, J.c.; Tumedei, M.M.; Tazzari, M.; De Wever, O.; Tesei, A.; Martinelli, G.; Castellani, G. Deep learning-based tool for morphotypic analysis of 3D multicellular spheroids. J. Mech. Med. Biol. 2023, 23, 2340034. [Google Scholar] [CrossRef]
- Li, W.; Zhou, Z.; Zhou, X.; Khoo, B.L.; Gunawan, R.; Chin, Y.R.; Zhang, L.; Yi, C.; Guan, X.; Yang, M. 3D Biomimetic Models to Reconstitute Tumor Microenvironment In Vitro: Spheroids, Organoids, and Tumor-on-a-Chip. Adv. Healthc. Mater. 2023, 12, 2202609. [Google Scholar] [CrossRef]
- Rodrigues, C.N.; Nunes, I.M.; Pereira, M.B.; Oliveira, H.; dos Santos, J.A. From superpixels to foundational models: An overview of unsupervised and generalizable image segmentation. Comput. Graph. 2024, 123, 104014. [Google Scholar] [CrossRef]
- Koval, V.; Zahorodnia, D.; Adamiv, O. An Image Segmentation Method for Obstacle Detection in a Mobile Robot Environment. In Proceedings of the 2019 9th International Conference on Advanced Computer Information Technologies (ACIT), Ceske Budejovice, Czech Republic, 4–7 June 2019; IEEE: Piscataway, NJ, USA, 2019. [Google Scholar] [CrossRef]
- Vasuki, Y.; Holden, E.J.; Kovesi, P.; Micklethwaite, S. An interactive image segmentation method for lithological boundary detection: A rapid mapping tool for geologists. Comput. Geosci. 2017, 100, 27–40. [Google Scholar] [CrossRef]
- Ebert, L.; Dobay, A.; Franckenberg, S.; Thali, M.; Decker, S.; Ford, J. Image segmentation of post-mortem computed tomography data in forensic imaging: Methods and applications. Forensic Imaging 2022, 28, 200483. [Google Scholar] [CrossRef]
- Hoorali, F.; Khosravi, H.; Moradi, B. IRUNet for medical image segmentation. Expert Syst. Appl. 2022, 191, 116399. [Google Scholar] [CrossRef]
- Liu, M. Momentum Contrast Learning for Aerial Image Segmentation and Precision Agriculture Analysis. In Proceedings of the 2022 International Conference on Image Processing, Computer Vision and Machine Learning (ICICML), Xi’an, China, 28–30 October 2022; IEEE: Piscataway, NJ, USA, 2022. [Google Scholar] [CrossRef]
- Kaymak, Ç.; Uçar, A. Semantic Image Segmentation for Autonomous Driving Using Fully Convolutional Networks. In Proceedings of the 2019 International Artificial Intelligence and Data Processing Symposium (IDAP), Malatya, Turkey, 21–22 September 2019; IEEE: Piscataway, NJ, USA, 2019. [Google Scholar] [CrossRef]
- Grexa, I.; Diosdi, A.; Harmati, M.; Kriston, A.; Moshkov, N.; Buzas, K.; Pietiäinen, V.; Koos, K.; Horvath, P. SpheroidPicker for automated 3D cell culture manipulation using deep learning. Sci. Rep. 2021, 11, 14813. [Google Scholar] [CrossRef]
- Habchi, Y.; Himeur, Y.; Kheddar, H.; Boukabou, A.; Atalla, S.; Chouchane, A.; Ouamane, A.; Mansoor, W. Ai in thyroid cancer diagnosis: Techniques, trends, and future directions. Systems 2023, 11, 519. [Google Scholar] [CrossRef]
- Górriz, J.M.; Álvarez-Illán, I.; Álvarez-Marquina, A.; Arco, J.E.; Atzmueller, M.; Ballarini, F.; Barakova, E.; Bologna, G.; Bonomini, P.; Castellanos-Dominguez, G.; et al. Computational approaches to explainable artificial intelligence: Advances in theory, applications and trends. Inf. Fusion 2023, 100, 101945. [Google Scholar] [CrossRef]
- Gupta, M.; Mishra, A. A systematic review of deep learning based image segmentation to detect polyp. Artif. Intell. Rev. 2024, 57, 7. [Google Scholar] [CrossRef]
- Himeur, Y.; Aburaed, N.; Elharrouss, O.; Varlamis, I.; Atalla, S.; Mansoor, W.; Ahmad, H.A. Applications of Knowledge Distillation in Remote Sensing: A Survey. arXiv 2024, arXiv:2409.12111. [Google Scholar] [CrossRef]
- Li, Z.; Zheng, Y.; Shan, D.; Yang, S.; Li, Q.; Wang, B.; Zhang, Y.; Hong, Q.; Shen, D. Scribformer: Transformer makes cnn work better for scribble-based medical image segmentation. IEEE Trans. Med. Imaging 2024, 43, 2254–2265. [Google Scholar] [CrossRef]
- García-Domínguez, M.; Domínguez, C.; Heras, J.; Mata, E.; Pascual, V. Deep style transfer to deal with the domain shift problem on spheroid segmentation. Neurocomputing 2024, 569, 127105. [Google Scholar] [CrossRef]
- Shahbazi, A.S.; Irandoost, F.; Mahdavian, R.; Shojaeilangari, S.; Allahvardi, A.; Naderi-Manesh, H. A multi-stage weakly supervised design for spheroid segmentation to explore mesenchymal stem cell differentiation dynamics. BMC Bioinform. 2025, 26, 20. [Google Scholar] [CrossRef] [PubMed]
- Sampaio Da Silva, C.; Boos, J.A.; Goldowsky, J.; Blache, M.; Schmid, N.; Heinemann, T.; Netsch, C.; Luongo, F.; Boder-Pasche, S.; Weder, G.; et al. High-throughput platform for label-free sorting of 3D spheroids using deep learning. Front. Bioeng. Biotechnol. 2024, 12, 1432737. [Google Scholar] [CrossRef] [PubMed]
- Lacalle, D.; Castro-Abril, H.A.; Randelovic, T.; Domínguez, C.; Heras, J.; Mata, E.; Mata, G.; Méndez, Y.; Pascual, V.; Ochoa, I. SpheroidJ: An Open-Source Set of Tools for Spheroid Segmentation. Comput. Methods Programs Biomed. 2021, 200, 105837. [Google Scholar] [CrossRef] [PubMed]
- Olofsson, K.; Carannante, V.; Takai, M.; Önfelt, B.; Wiklund, M. Single cell organization and cell cycle characterization of DNA stained multicellular tumor spheroids. Sci. Rep. 2021, 11, 17076. [Google Scholar] [CrossRef]
- Cigla, C.; Alatan, A.A. Region-based image segmentation via graph cuts. In Proceedings of the 2008 15th IEEE International Conference on Image Processing, San Diego, CA, USA, 12–15 October 2008; IEEE: Piscataway, NJ, USA, 2008. [Google Scholar] [CrossRef]
- Sharma, M.; Bhattacharya, M. Segmentation of CA3 Hippocampal Region of Rat Brain Cells Images Based on Bio-inspired Clustering Technique. In Proceedings of the 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), San Diego, CA, USA, 18–21 November 2019; IEEE: Piscataway, NJ, USA, 2019. [Google Scholar] [CrossRef]
- Rao, R.V.; Savsani, V.J.; Vakharia, D.P. Teaching–learning-based optimization: A novel method for constrained mechanical design optimization problems. Comput.-Aided Des. 2011, 43, 303–315. [Google Scholar] [CrossRef]
- Hossain, M.D.; Chen, D. A hybrid image segmentation method for building extraction from high-resolution RGB images. ISPRS J. Photogramm. Remote Sens. 2022, 192, 299–314. [Google Scholar] [CrossRef]
- Fallah, F.; Yang, B.; Walter, S.S.; Bamberg, F. Hierarchical Feature-learning Graph-based Segmentation of Fat-Water MR Images. In Proceedings of the 2018 Signal Processing: Algorithms, Architectures, Arrangements, and Applications (SPA), Poznan, Poland, 19–21 September 2018; IEEE: Piscataway, NJ, USA, 2018. [Google Scholar] [CrossRef]
- Luo, D.; Zeng, W.; Chen, J.; Tang, W. Deep Learning for Automatic Image Segmentation in Stomatology and Its Clinical Application. Front. Med. Technol. 2021, 3, 767836. [Google Scholar] [CrossRef]
- Wang, J.; Sun, K.; Cheng, T.; Jiang, B.; Deng, C.; Zhao, Y.; Xiao, B. Deep High-Resolution Representation Learning for Visual Recognition. IEEE Trans. Pattern Anal. Mach. Intell. 2021, 43, 3349–3364. [Google Scholar] [CrossRef]
- Bhandari, A.K.; Singh, V.K.; Kumar, A.; Singh, G.K. Cuckoo search algorithm and wind driven optimization based study of satellite image segmentation for multilevel thresholding using Kapur’s entropy. Expert Syst. Appl. 2014, 41, 3538–3560. [Google Scholar] [CrossRef]
- Baydoun, M.; Al-Alaoui, M.A. Modified edge detection for segmentation. In Proceedings of the 2015 International Symposium on Signals, Circuits and Systems (ISSCS), Iasi, Romania, 9–10 July 2015; IEEE: Piscataway, NJ, USA, 2015. [Google Scholar] [CrossRef]
- Bechar, A.; Elmir, Y.; Himeur, Y.; Medjoudj, R.; Amira, A. Federated and Transfer Learning for Cancer Detection Based on Image Analysis. arXiv 2024, arXiv:2405.20126. [Google Scholar] [CrossRef]
- Bechar, A.; Elmir, Y.; Medjoudj, R.; Himeur, Y.; Amira, A. Transfer Learning for Cancer Detection based on Images Analysis. Procedia Comput. Sci. 2024, 239, 1903–1910. [Google Scholar] [CrossRef]
- Hamza, A.; Lekouaghet, B.; Himeur, Y. Hybrid whale-mud-ring optimization for precise color skin cancer image segmentation. In Proceedings of the 2023 6th International Conference on Signal Processing and Information Security (ICSPIS), Dubai, United Arab Emirates, 8–9 November 2023; IEEE: Piscataway, NJ, USA, 2023; pp. 87–92. [Google Scholar]
- Habchi, Y.; Kheddar, H.; Himeur, Y.; Boukabou, A.; Atalla, S.; Mansoor, W.; Al-Ahmad, H. Deep Transfer Learning for Kidney Cancer Diagnosis. arXiv 2024, arXiv:2408.04318. [Google Scholar]
- Khalifa, M.; Albadawy, M. Artificial Intelligence for Clinical Prediction: Exploring Key Domains and Essential Functions. Comput. Methods Programs Biomed. Update 2024, 5, 100148. [Google Scholar] [CrossRef]
- Wankhede, N.L.; Kale, M.B.; Shukla, M.; Nathiya, D.; Roopashree, R.; Kaur, P.; Goyanka, B.; Rahangdale, S.R.; Taksande, B.G.; Upaganlawar, A.B.; et al. Leveraging AI for the diagnosis and treatment of autism spectrum disorder: Current trends and future prospects. Asian J. Psychiatry 2024, 101, 104241. [Google Scholar] [CrossRef]
- Yaqoob, A.; Verma, N.K.; Aziz, R.M. Optimizing gene selection and cancer classification with hybrid sine cosine and cuckoo search algorithm. J. Med. Syst. 2024, 48, 10. [Google Scholar] [CrossRef]
- Alharbi, F.; Vakanski, A. Machine learning methods for cancer classification using gene expression data: A review. Bioengineering 2023, 10, 173. [Google Scholar] [CrossRef]
- Stephan, P.; Stephan, T.; Kannan, R.; Abraham, A. A hybrid artificial bee colony with whale optimization algorithm for improved breast cancer diagnosis. Neural Comput. Appl. 2021, 33, 13667–13691. [Google Scholar] [CrossRef]
- Thawkar, S.; Sharma, S.; Khanna, M.; kumar Singh, L. Breast cancer prediction using a hybrid method based on butterfly optimization algorithm and ant lion optimizer. Comput. Biol. Med. 2021, 139, 104968. [Google Scholar] [CrossRef]
- Bir-Jmel, A.; Douiri, S.M.; Bernoussi, S.E.; Maafiri, A.; Himeur, Y.; Atalla, S.; Mansoor, W.; Al-Ahmad, H. GFLASSO-LR: Logistic Regression with Generalized Fused LASSO for Gene Selection in High-Dimensional Cancer Classification. Computers 2024, 13, 93. [Google Scholar] [CrossRef]
- Li, J.; Liang, K.; Song, X. Logistic regression with adaptive sparse group lasso penalty and its application in acute leukemia diagnosis. Comput. Biol. Med. 2022, 141, 105154. [Google Scholar] [CrossRef]
- Tian, L.; Li, X.; Zheng, H.; Wang, L.; Qin, Y.; Cai, J. Fisher discriminant model based on LASSO logistic regression for computed tomography imaging diagnosis of pelvic rhabdomyosarcoma in children. Sci. Rep. 2022, 12, 15631. [Google Scholar] [CrossRef]
- Castillo, D.L. Deep-Tumour-Spheroid. GitHub. 2023. Available online: https://github.com/WaterKnight1998/Deep-Tumour-Spheroid (accessed on 21 October 2024).
- Michálek, J.; Štěpka, K.; Kozubek, M.; Navrátilová, J.; Pavlatovská, B.; Machálková, M.; Pruška, A. Quantitative Assessment of Anti-Cancer Drug Efficacy From Coregistered Mass Spectrometry and Fluorescence Microscopy Images of Multicellular Tumor Spheroids. Microsc. Microanal. 2019, 25, 1311–1322. [Google Scholar] [CrossRef]
- Tasnadi, E.A.; Toth, T.; Kovacs, M.; Diosdi, A.; Pampaloni, F.; Molnar, J.; Horvath, P. 3D-Cell-Annotator: An open-source active surface tool for single-cell segmentation in 3D microscopy images. Bioinformatics 2020, 36, 2948–2949. [Google Scholar] [CrossRef]
- Schmitz, A.; Fischer, S.C.; Mattheyer, C.; Pampaloni, F.; Stelzer, E.H.K. Multiscale image analysis reveals structural heterogeneity of the cell microenvironment in homotypic spheroids. Sci. Rep. 2017, 7, 43693. [Google Scholar] [CrossRef]
- Antoni, D.; Burckel, H.; Josset, E.; Noel, G. Three-Dimensional Cell Culture: A Breakthrough in Vivo. Int. J. Mol. Sci. 2015, 16, 5517–5527. [Google Scholar] [CrossRef]
- Barbosa, M.A.G.; Xavier, C.P.R.; Pereira, R.F.; Petrikaitė, V.; Vasconcelos, M.H. 3D Cell Culture Models as Recapitulators of the Tumor Microenvironment for the Screening of Anti-Cancer Drugs. Cancers 2021, 14, 190. [Google Scholar] [CrossRef]
- Wrzesinski, K.; Magnone, M.C.; Visby, H.L.; Kruse, M.E.; Bergauer, T.; Bobadilla, M.; Fey, S.J. HepG2/C3A 3D spheroids exhibit stable physiological functionality for at least 24 days after recovering from trypsinisation. Toxicol. Res. 2013, 2, 163. [Google Scholar] [CrossRef]
- Rousseau, D.; Huaman, R.; Rasti, P.; Riviere, C. Supervised machine learning for 3D light microscopy without manual annotation: Application to spheroids. In Proceedings of the Unconventional Optical Imaging, Strasbourg, France, 22–26 April 2018; SPIE: Bellingham, WA, USA, 2018. [Google Scholar] [CrossRef]
- Vaidyanathan, K.; Wang, C.; Krajnik, A.; Yu, Y.; Choi, M.; Lin, B.; Bae, Y. A machine learning pipeline revealing heterogeneous responses to drug perturbations on vascular smooth muscle cell spheroid morphology and formation. Sci. Rep. 2021, 11, 23285. [Google Scholar] [CrossRef]
- Kecheril Sadanandan, S.; Karlsson, J.; Wahlby, C. Spheroid segmentation using multiscale deep adversarial networks. In Proceedings of the IEEE International Conference on Computer Vision Workshops, Montreal, BC, Canada, 11–17 October 2017; pp. 36–41. [Google Scholar]
- Khoshdeli, M.; Winkelmaier, G.; Parvin, B. Multilayer encoder-decoder network for 3D nuclear segmentation in spheroid models of human mammary epithelial cell lines. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, Salt Lake City, UT, USA, 18–23 June 2018; pp. 2239–2245. [Google Scholar]
- Souadih, K.; Belaid, A.; Salem, D.B. Automatic segmentation of the sphenoid sinus in CT-scans volume with deepmedics 3D CNN architecture: Array. Med. Technol. J. 2019, 3, 334–346. [Google Scholar] [CrossRef]
- Gupta, A.; Harrison, P.J.; Wiesl, e.H.; Pielawski, N.; Kartasalo, K.; Partel, G.; Wählby, C. Deep Learning in Image Cytometry: A Review. Cytometry Part A 2018, 95, 366–380. [Google Scholar] [CrossRef]
- Wang, L. Deep Learning Techniques to Diagnose Lung Cancer. Cancers 2022, 14, 5569. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, A.; Goodarzi, S.; Frindel, C.; Recher, G.; Riviere, C.; Rousseau, D. Clearing spheroids for 3D fluorescent microscopy: Combining safe and soft chemicals with deep convolutional neural network. Cold Spring Harb. Lab. 2021. [Google Scholar] [CrossRef]
- Chen, Z.; Ma, N.; Sun, X.; Li, Q.; Zeng, Y.; Chen, F.; Gu, Z. Automated evaluation of tumor spheroid behavior in 3D culture using deep learning-based recognition. Biomaterials 2021, 272, 120770. [Google Scholar] [CrossRef] [PubMed]
- Rettenberger, L.; Schilling, M.; Reischl, M. Annotation Efforts in Image Segmentation can be Reduced by Neural Network Bootstrapping. Curr. Dir. Biomed. Eng. 2022, 8, 329–332. [Google Scholar] [CrossRef]
- Wen, C.; Miura, T.; Voleti, V.; Yamaguchi, K.; Tsutsumi, M.; Yamamoto, K.; Kimura, K.D. 3DeeCellTracker, a deep learning-based pipeline for segmenting and tracking cells in 3D time lapse images. eLife 2021, 10, 59187. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the Lecture Notes in Computer Science, Washington, DC, USA, 13–17 July 2015; Springer International Publishing: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar] [CrossRef]
- Yang, M.; Yu, K.; Zhang, C.; Li, Z.; Yang, K. DenseASPP for Semantic Segmentation in Street Scenes. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; IEEE: Piscataway, NJ, USA, 2018. [Google Scholar] [CrossRef]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Chintala, S. PyTorch: An Imperative Style, High-Performance Deep Learning Library. arXiv 2019. [Google Scholar] [CrossRef]
- Howard, J.; Gugger, S. Fastai: A Layered API for Deep Learning. Information 2020, 11, 108. [Google Scholar] [CrossRef]







| Ref. | Model Used | Dataset/Type of Data | Main Contribution | Best Performance Value | Limitation |
|---|---|---|---|---|---|
| [52] | Workflow integrating MALDI MSI and LSCM | Spheroid sections with matrix-assisted laser desorption/ionization imaging | Novel approach for coregistering low-resolution MALDI MS with high-resolution LSCM images, correlating drug and cell distribution in spheroids | Differentiates therapy-induced from natural cell death using new peeling algorithms | Limited scalability and generalizability for diverse datasets |
| [53] | 3D-Cell-Annotator | 3D microscopy images of biological structures (e.g., spheroids, organoids) | Semi-automated segmentation tool with precision comparable to human experts | Annotation time reduced by factor of 3; Jaccard Index not specified but superior to state-of-the-art tools | Precision limited to tested biological structures |
| [54] | Automated nuclei segmentation using graph and computational topology | Light sheet-based fluorescence microscopy of spheroids | High-resolution, 3D image analysis pipeline providing structural data for tissues | F-Score for Nuclei Segmentation: 0.88 | Computational complexity and potential challenges for very large spheroids |
| [57] | HepG2/C3A spheroid culture model | 3D culture of human hepatocytes for long-term metabolic function analysis | Stable homeostatic model for medium- to long-term studies of drug metabolism and biomarkers | Long-term stability of metabolic functions for 24 days | Limited scope for applications beyond liver-related studies |
| [58] | Shallow and deep learning detection models | Synthetic and real 3D microscopy images | Use of synthetic images to train supervised machine learning models for real data | Over 90% accurate cell detection on real images | Dependency on synthetic data for training |
| [59] | VGG16-U-Net | VSMC spheroid microscopy images | Identification of heterogeneous drug responses using deep-learning-based morphological clustering | Dice coefficient: 0.96 for segmentation | Focus limited to vascular smooth muscle cells |
| [60] | Multiscale deep adversarial network | 2D bright-field microscopy images | Novel adversarial network for stable segmentation of spheroids under varying conditions | maF: 77.09% (easy), 64.21% (hard) | Relatively lower performance on challenging datasets |
| [61] | 3D CNN with encoder–decoder architecture | 3D nuclear segmentation of human mammary epithelial cells | Superior pixel-based segmentation for non-uniform and heterogeneous cell morphologies | F1-Score: 0.95 | Limited to specific cell lines |
| [62] | 3D CNN with preprocessing and postprocessing | CT volumes of the sphenoid sinus | Fully automated segmentation with high accuracy and reliability | Dice: 0.92, Precision: 0.94, Recall: 0.90 | Small dataset used for validation |
| [65] | Deep CNN with segmentation pipeline | Cleared and non-cleared 3D spheroid images | Combined clearing techniques with digital segmentation for nuclei and transfer of segmentation knowledge across clearing protocols | F1-Score: 0.91 ± 0.01, AJI: 0.71 ± 0.02 (Glycerol clearing method) | Challenges with scalability and generalized application |
| [66] | PSP-U-Net | Tumor spheroid boundary detection in 3D cultures | Developed AI-based indices (EPI, MSEI) for tumor invasiveness and segmentation | F1-Score: >0.95 (after 125,000 training rounds) | High computational cost for extensive training |
| [67] | U-Net with unsupervised pre-annotations | Biomedical datasets (Droplet Microarray, ISIC Melanoma) | Reduced annotation effort using automated segmentation mask generation methods | DSC: 0.76 (Droplet dataset), 0.64 (ISIC dataset) | Limited performance improvement in complex datasets |
| [27] | HRNet-Seg | Spheroid images from diverse experimental conditions | Developed open-source tools for generalizable segmentation across conditions (SpheroidJ) | Jaccard Index: 0.9512 (Validation), 0.97 ± 0.01 (BN10S dataset) | Generic algorithms struggle with unseen scenarios |
| [18] | Mask R-CNN, U-Net | 3D cell culture images for spheroid manipulation | Automated AI-guided system for spheroid selection and transfer in precision medicine workflows | Spheroid transfer success: 89% (semi-auto), 80% (fully auto) | Lower success rate in fully automated mode |
| [68] | 3DeeCellTracker (deep learning-based pipeline) | 3D + T images of tumor spheroids, zebrafish heart, worm brain | Integrated segmentation and tracking for dynamic cell activity analysis in diverse datasets | 90–100% tracking accuracy for cells in most datasets | Requires parameter tuning for diverse optical systems |
| Model | Val. Loss (%) | Val. Acc (%) | Dice Coeff. (%) | Jaccard Coeff. (%) |
|---|---|---|---|---|
| HRNet | 1.99 | 99.72 | 96.70 | 93.62 |
| DeeplabV3+ | 2.42 | 99.72 | 96.45 | 93.14 |
| UNET (Adam) | 41.43 | 92.07 | 82.55 | 70.48 |
| UNET (Adagrad) | 5.94 | 97.24 | 80.44 | 67.29 |
| UNET (SGD) | 6.09 | 97.23 | 80.82 | 67.89 |
| Model | Val. Acc. (%) | Test Acc. (%) | Dice Coeff. (%) | Jaccard Coeff. (%) |
|---|---|---|---|---|
| HRNet | 99.72 | 99.54 | 96.08 | 92.67 |
| DeepLabV3+ | 99.72 | 99.43 | 95.62 | 91.73 |
| U-Net (Adam) | 92.07 | 90.34 | 82.55 | 70.48 |
| U-Net (SGD) | 97.23 | 96.87 | 80.82 | 67.89 |
| U-Net (Adagrad) | 97.24 | 96.76 | 80.44 | 67.29 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oudouar, F.; Bir-Jmel, A.; Grissette, H.; Douiri, S.M.; Himeur, Y.; Miniaoui, S.; Atalla, S.; Mansoor, W. An Empirical Evaluation of Neural Network Architectures for 3D Spheroid Segmentation. Computers 2025, 14, 86. https://doi.org/10.3390/computers14030086
Oudouar F, Bir-Jmel A, Grissette H, Douiri SM, Himeur Y, Miniaoui S, Atalla S, Mansoor W. An Empirical Evaluation of Neural Network Architectures for 3D Spheroid Segmentation. Computers. 2025; 14(3):86. https://doi.org/10.3390/computers14030086
Chicago/Turabian StyleOudouar, Fadoua, Ahmed Bir-Jmel, Hanane Grissette, Sidi Mohamed Douiri, Yassine Himeur, Sami Miniaoui, Shadi Atalla, and Wathiq Mansoor. 2025. "An Empirical Evaluation of Neural Network Architectures for 3D Spheroid Segmentation" Computers 14, no. 3: 86. https://doi.org/10.3390/computers14030086
APA StyleOudouar, F., Bir-Jmel, A., Grissette, H., Douiri, S. M., Himeur, Y., Miniaoui, S., Atalla, S., & Mansoor, W. (2025). An Empirical Evaluation of Neural Network Architectures for 3D Spheroid Segmentation. Computers, 14(3), 86. https://doi.org/10.3390/computers14030086






