An Embedded Convolutional Neural Network Model for Potato Plant Disease Classification
Abstract
1. Introduction
2. Related Work
3. Materials and Methods
3.1. Potato Plant Diseases
3.2. Convolutional Neural Networks
3.3. Dataset
3.4. Model Hyperparameters
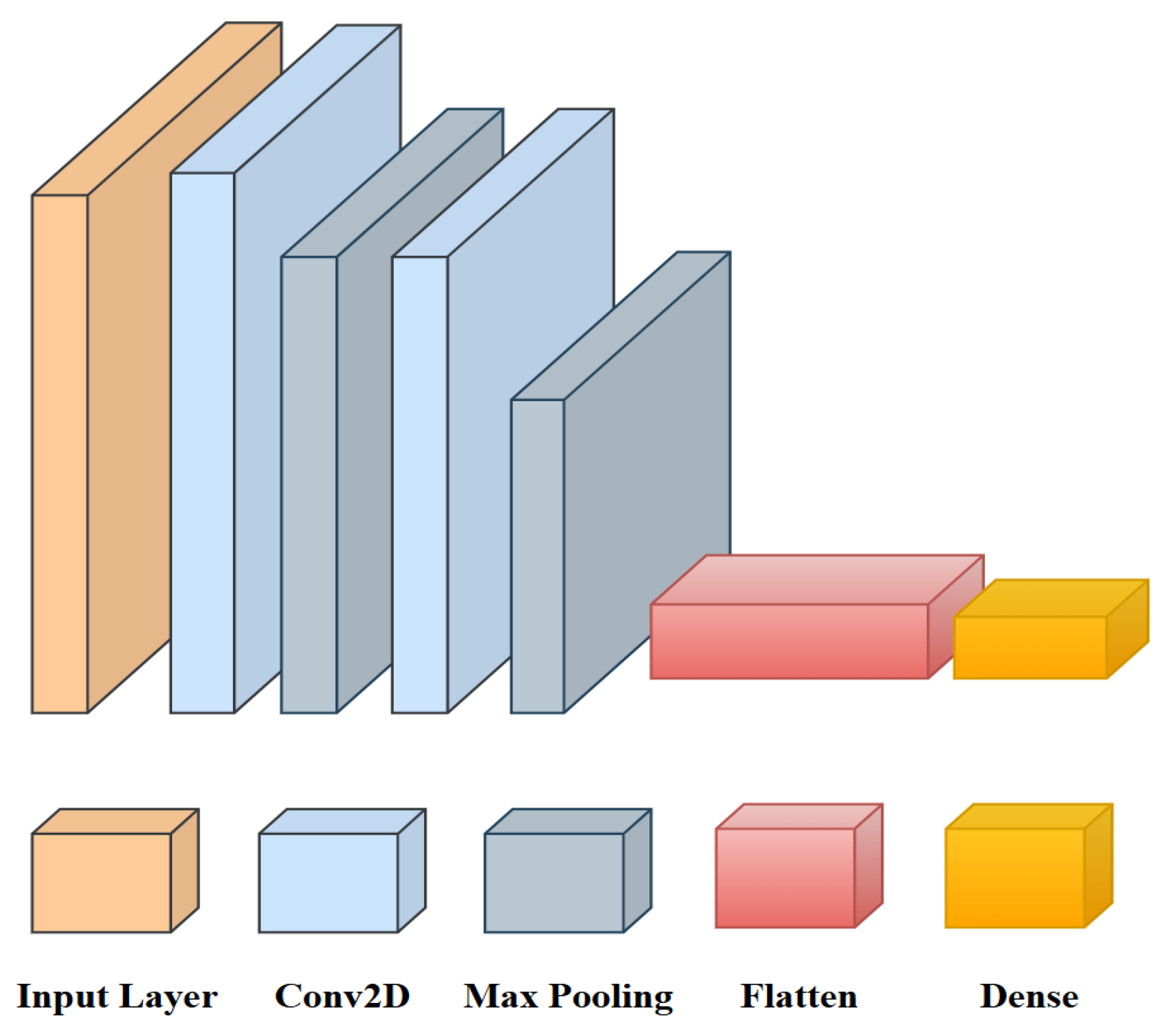
3.5. Proposed Model Architecture
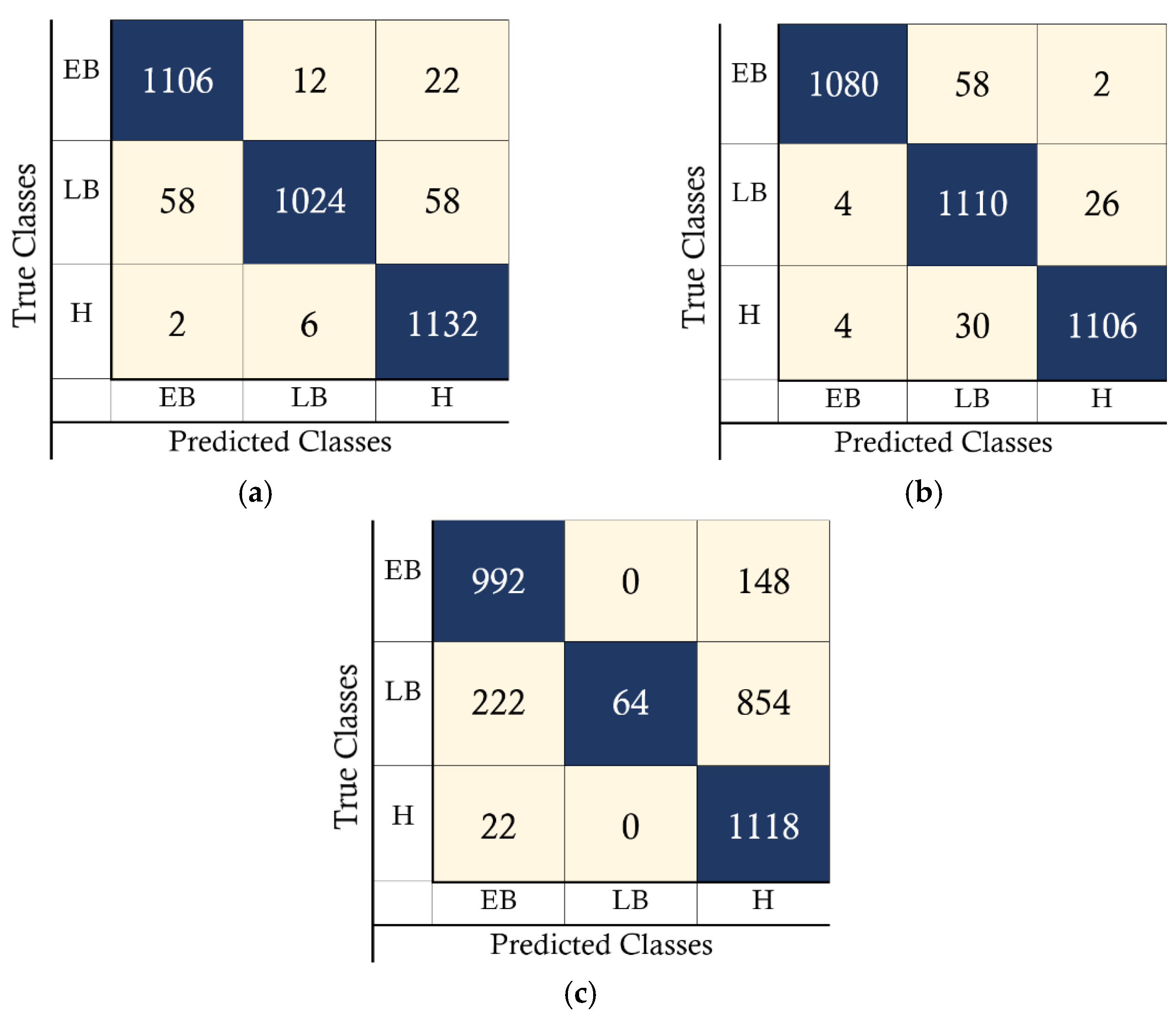
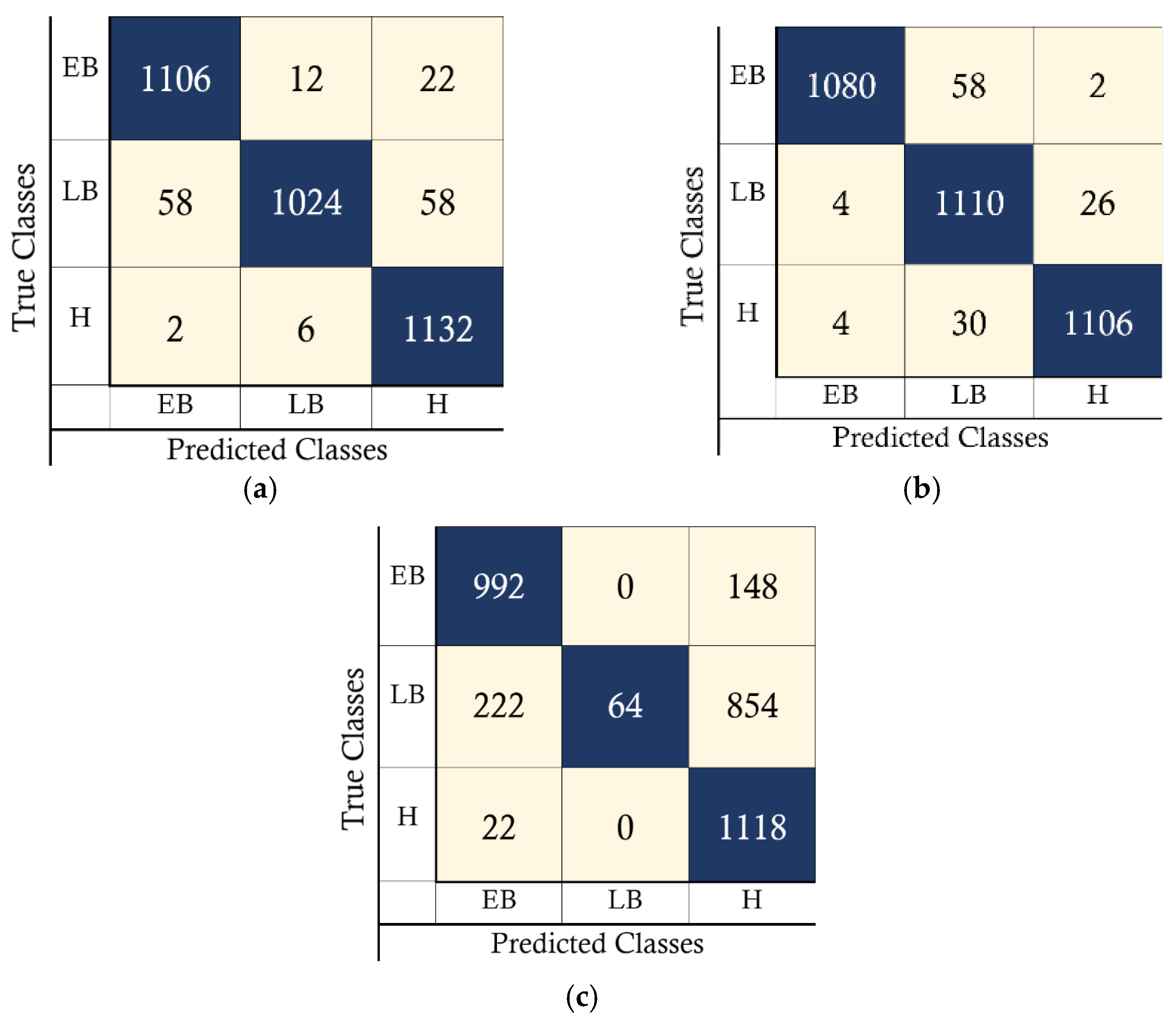
4. Results
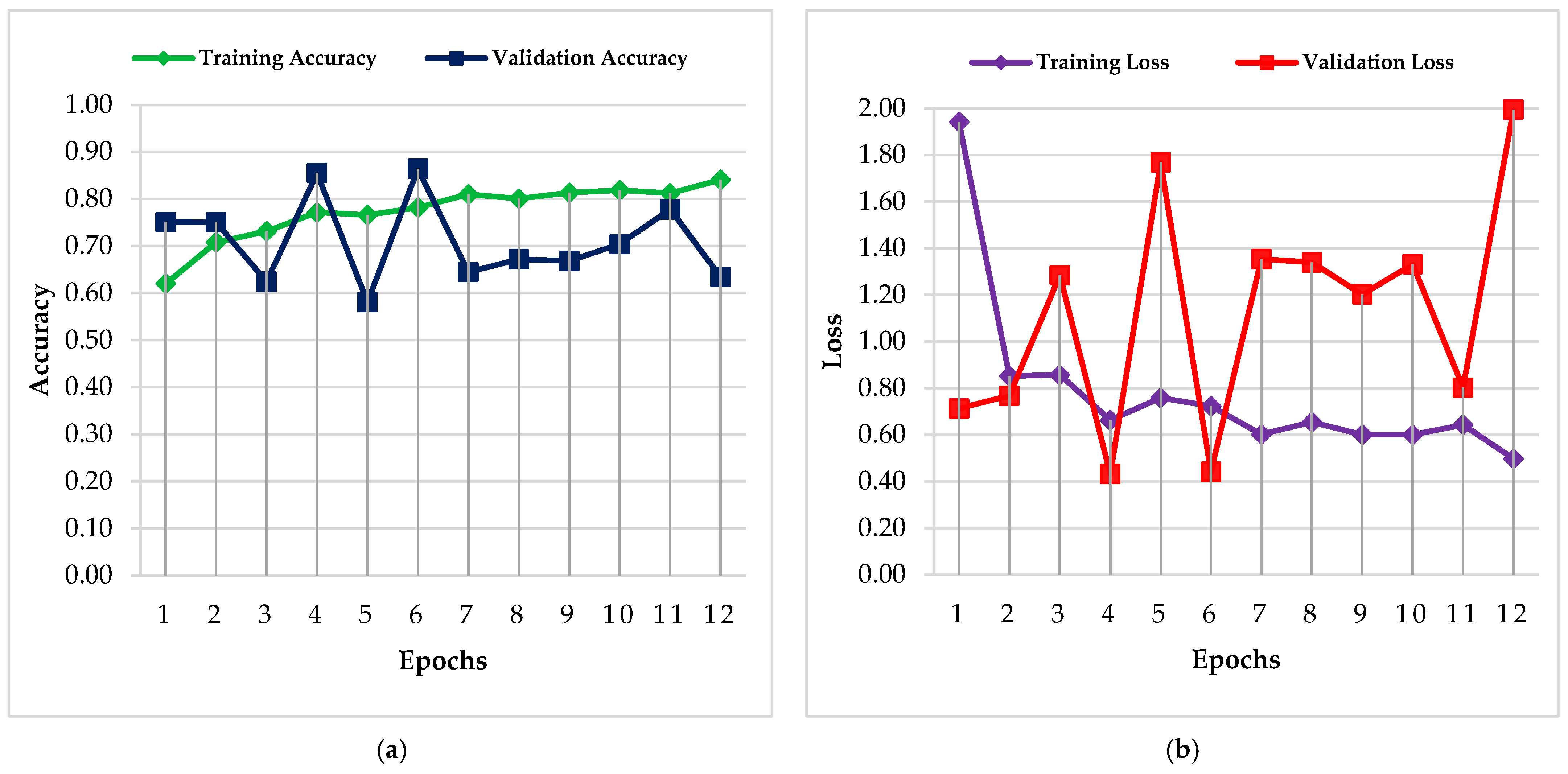
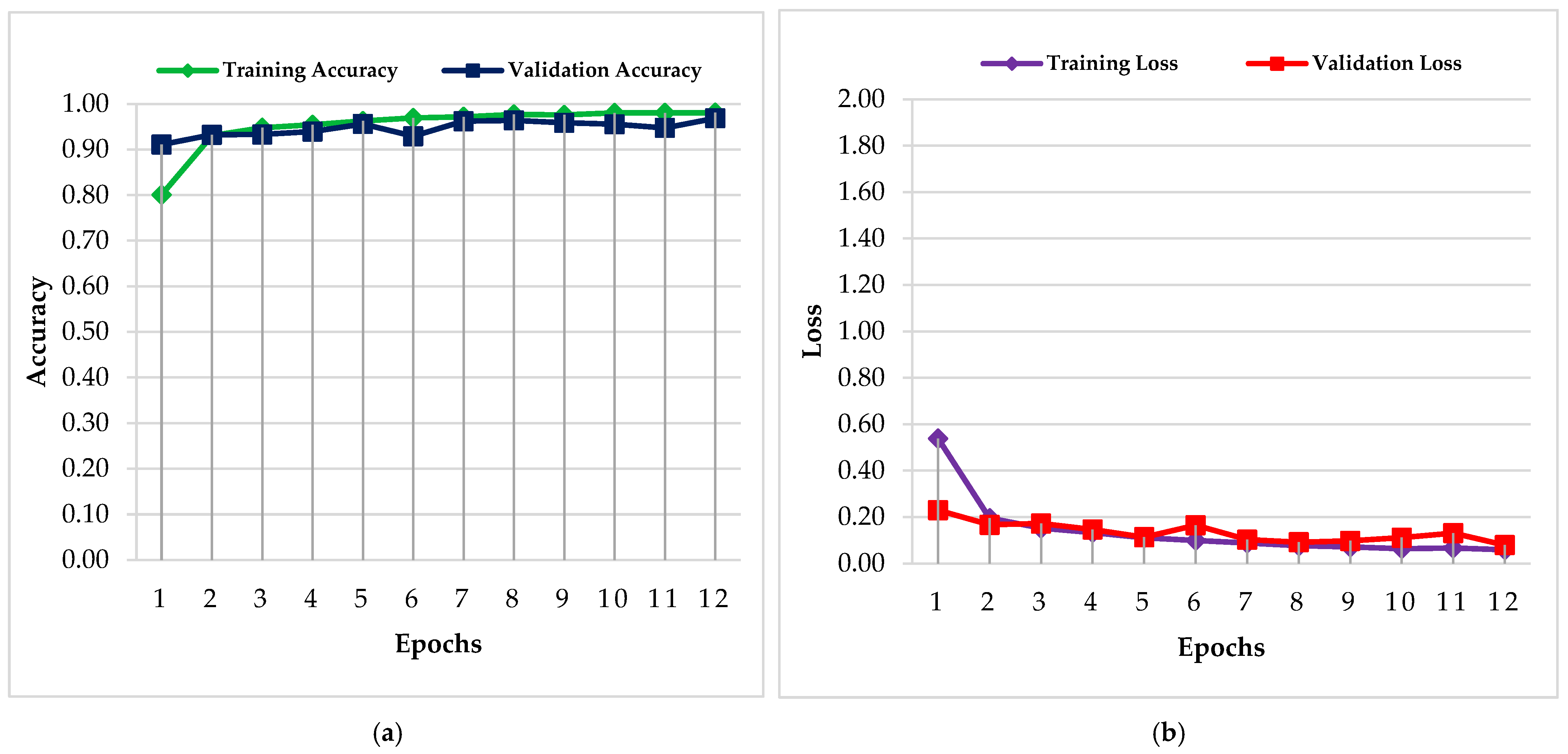
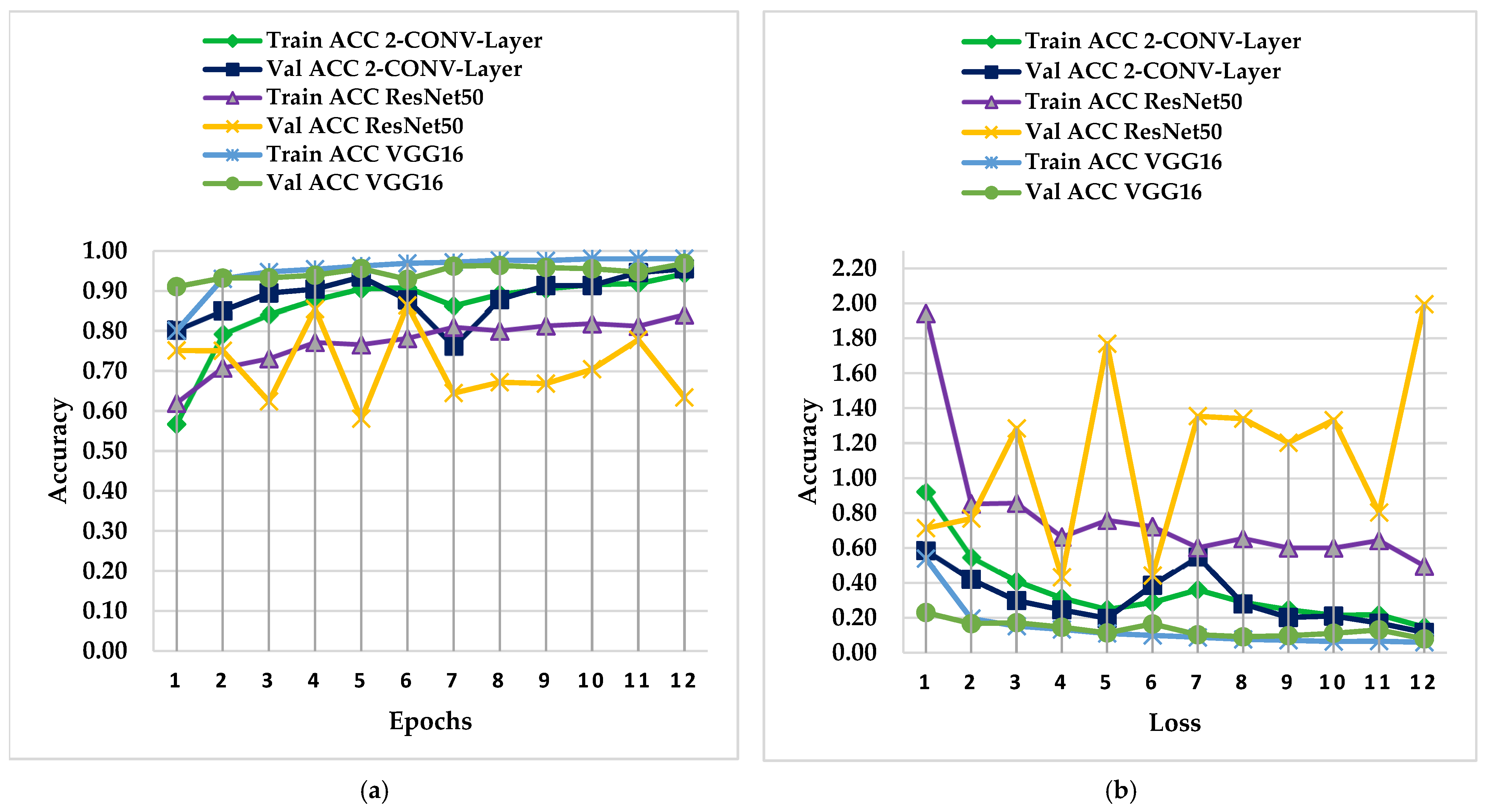
4.1. PC Simulation Implementation
4.2. Embedded Implementation
5. Discussions
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Gado, T.A.; El-Agha, D.E. Climate Change Impacts on Water Balance in Egypt and Opportunities for Adaptations. In Agro-Environmental Sustainability in MENA Regions; Springer: Berlin/Heidelberg, Germany, 2021; pp. 13–47. [Google Scholar]
- Lago-Olveira, S.; El-Areed, S.R.; Moreira, M.T.; González-García, S. Improving Environmental Sustainability of Agriculture in Egypt Through a Life-Cycle Perspective; Elsevier: Amsterdam, The Netherlands, 2023; Volume 890, pp. 1–11. [Google Scholar]
- Mansour, T.G.I.; Abdelazez, M.A.; Eleshmawiy, K.H. Challenges and Constraints Facing the Agricultural Extension System in Egypt. J. Agric. Sci. 2022, 17, 241–257. [Google Scholar] [CrossRef]
- Savary, S.; Ficke, A.; Aubertot, J.-N.; Hollier, C. Crop Losses Due to Diseases and Their Implications for Global Food Production Losses and Food Security. Food Secur. 2012, 4, 519–537. [Google Scholar] [CrossRef]
- John, M.A.; Bankole, I.; Ajayi-Moses, O.; Ijila, T.; Jeje, O.; Lalit, P. Relevance of Advanced Plant Disease Detection Techniques in Disease and Pest Management for Ensuring Food Security and Their Implication: A Review. Am. J. Plant Sci. 2023, 14, 1260–1295. [Google Scholar] [CrossRef]
- Shafik, W.; Tufail, A.; Namoun, A.; Silva, L.C.D.; Apong, R.A.A.H.M. A Systematic Literature Review on Plant Disease Detection: Motivations, Classification Techniques, Datasets, Challenges, and Future Trends. IEEE Access 2023, 11, 59174–59203. [Google Scholar] [CrossRef]
- Tugrul, B.; Elfatimi, E.; Eryigit, R. Convolutional Neural Networks in Detection of Plant Leaf Diseases: A Review. Agriculture 2022, 12, 1192. [Google Scholar] [CrossRef]
- Zhang, X.; Cao, Z.; Dong, W. Overview of Edge Computing in the Agricultural Internet of Things: Key Technologies, Applications, Challenges. IEEE Access 2020, 8, 141748–141761. [Google Scholar] [CrossRef]
- Garcia-Perez, A.; Miñón, R.; Torre-Bastida, A.I.; Zulueta-Guerrero, E. Analysing Edge Computing Devices for the Deployment of Embedded AI. Sensors 2023, 23, 9495. [Google Scholar] [CrossRef]
- Sayed, S.A.; Mahmoud, A.S.; Farg, E.; Mohamed, A.M.; Saleh, A.M.; AbdelRahman, M.A.E.; Moustafa, M.; AbdelSalam, H.M.; Arafat, S.M. A comparative study of big data use in Egyptian agriculture. J. Electr. Syst. Inf. Technol. 2023, 10, 2–20. [Google Scholar] [CrossRef]
- Shi, W.; Cao, J.; Zhang, Q.; Li, Y.; Xu, L. Edge Computing: Vision and Challenges. IEEE Internet Things J. 2016, 3, 637–646. [Google Scholar] [CrossRef]
- Mohanty, S.P.; Hughes, D.P.; Salathé, M. Using Deep Learning for Image-Based Plant Disease Detection. Front. Plant Sci. 2016, 7, 1419. [Google Scholar] [CrossRef]
- Ferentinos, K.P. Deep Learning Models for Plant Disease Detection and Diagnosis. Comput. Electron. Agric. 2018, 145, 311–318. [Google Scholar] [CrossRef]
- Jiang, Z.; Dong, Z.; Jiang, W.; Yang, Y. Recognition of Rice Leaf Diseases and Wheat Leaf Diseases Based on Multi-Task Deep Transfer Learning. Comput. Electron. Agric. 2021, 186, 106184. [Google Scholar] [CrossRef]
- Brahimi, M.; Boukhalfa, K.; Moussaoui, A. Deep Learning for Tomato Diseases: Classification and Symptoms Visualization. Appl. Artif. Intell. 2017, 31, 299–315. [Google Scholar] [CrossRef]
- Ha, J.G.; Moon, A.H.; Kwak, J.T.; Hassan, S.I.; Dang, M.; Lee, O.N.; Parkb, H.Y. Deep Convolutional Neural Network for Classifying Fusarium Wilt of Radish from Unmanned Aerial Vehicles. J. Appl. Remote Sens. 2017, 11, 042621. [Google Scholar] [CrossRef]
- Alatawi, A.; Alomani, S.M.; Alhawiti, N.I.; Ayaz, M. Plant Disease Detection using AI based VGG-16 Model. Int. J. Adv. Comput. Sci. Appl. 2022, 13, 718–727. [Google Scholar] [CrossRef]
- Costa, Z.D.; Figueroa, H.E.; Fracarolli, J.A. Computer Vision Based Detection of External Defects on Tomatoes Using Deep Learning. Biosyst. Eng. 2020, 190, 131–144. [Google Scholar] [CrossRef]
- Paul, S.G.; Biswas, A.A.; Saha, A.; Zulfiker, M.S.; Ritu, N.A.; Zahan, I.; Rahman, M.; Islam, M.A. A Real-Time Application-Based Convolutional Neural Network Approach for Tomato Leaf Disease Classification. Array 2023, 19, 100313. [Google Scholar] [CrossRef]
- Shin, J.; Chang, Y.K.; Heung, B.; Nguyen-Quang, T.; Price, G.W.; Al-Mallahi, A. A Deep Learning Approach for RGB Image-Based Powdery Mildew Disease Detection on Strawberry Leaves. Comput. Electron. Agric. 2021, 183, 106042. [Google Scholar] [CrossRef]
- Gonzalez-Huitron, V.; León-Borges, J.A.; Rodriguez-Mata, A.; Amabilis-Sosa, L.E.; Ramírez-Pereda, B.; Rodriguez, H. Disease Detection in Tomato Leaves via CNN with Lightweight Architectures Implemented in Raspberry Pi 4. Comput. Electron. Agric. 2021, 181, 105951. [Google Scholar] [CrossRef]
- Galstyan, D.; Harutyunyan, E.; Nikoghosyan, K. Usage of Nvidia Jetson Nano in Agriculture as an Example of Plant Leaves Illness Real-Time Detection and Classification. Agron. Agroecol. 2022, 2, 149–153. [Google Scholar] [CrossRef]
- Routis, G.; Michailidis, M.; Roussaki, I. Plant Disease Identification Using Machine Learning Algorithms on Single-Board Computers in IoT Environments. Electronics 2024, 13, 1010. [Google Scholar] [CrossRef]
- Dobnik, D.; Gruden, K.; Ramšak, Ž.; Coll, A. Importance of Potato as a Crop and Practical Approaches to Potato Breeding. In Solanum tuberosum; Springer: Berlin/Heidelberg, Germany, 2021; pp. 3–20. [Google Scholar]
- Chakrabart, S.K.; Sharma, S.; Shah, M.A. Potato Pests and Diseases: A Global Perspective. In Sustainable Management of Potato Pests and Diseases; Springer: Berlin/Heidelberg, Germany, 2022; pp. 1–23. [Google Scholar]
- El-Din, D.K.S.; Mostafa, D.Y.S.; ElShahed, D.M.A.A. The Current Situation of Egyptian Potatoes Exports in the shadow of Covid 19 Pandemic. J. Am. Sci. 2022, 18, 42–58. [Google Scholar]
- Kaur, S.; Mukerji, K.G. Potato Diseases and their Management. In Fruit and Vegetable Diseases; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2004; pp. 233–280. [Google Scholar]
- Javaid, M.; Haleem, A.; Khan, I.H.; Suman, R. Understanding the Potential Applications of Artificial Intelligence in Agriculture Sector. Adv. Agrochem 2023, 2, 15–30. [Google Scholar] [CrossRef]
- Lal, M.; Chaudhary, S.; Sharma, S.; Subhash, S.; Kumar, M. Bio-Intensive Management of Fungal Diseases of Potatoes. In Sustainable Management of Potato Pests and Diseases; Springer: Berlin/Heidelberg, Germany, 2022; pp. 453–493. [Google Scholar]
- Chaudhary, K.; Yadavb, J.; Guptac, A.K. Integrated Disease Management of Early Blight (Alternaria solani) of Potato. Trop. Agrobiodiversity (TRAB) 2021, 2, 77–81. [Google Scholar] [CrossRef]
- Fry, W.E.; Goodwin, S.B. Re-emergence of Potato and Tomato Late Blight in the United States. Plant Dis. 1997, 81, 1349–1357. [Google Scholar] [CrossRef] [PubMed]
- Arora, R.K.; Sharma, S. Late Blight Disease of Potato and its Management. Plant J. 2014, 41, 16–40. [Google Scholar]
- Kassaw, A.; Abera, M.; Eshetu, B. The Response of Potato Late Blight to Potato varieties and Fungicide Spraying Frequencies at Meket, Ethiopia. Cogent Food Agric. 2021, 7. [Google Scholar] [CrossRef]
- López, O.A.M.; López, A.M.; Crossa, J. Convolutional Neural Networks. In Multivariate Statistical Machine Learning Methods for Genomic Prediction; Springer: Berlin/Heidelberg, Germany, 2022; pp. 533–577. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. In 3rd International Conference on Learning Representations; ICLR: San Diego, CA, USA, 2015. [Google Scholar]
- Jia, X.; Jiang, X.; Li, Z.; Mu, J.; Wang, Y.; Niu, Y. Application of Deep Learning in Image Recognition of Citrus Pests. Agriculture 2023, 13, 1023. [Google Scholar] [CrossRef]
- Yang, L.; Xu, S.; Yu, X. A New Model Based on Improved VGG16 For Corn Weed Identification. Front. Plant Sci. 2023, 14, 1205151. [Google Scholar] [CrossRef]
- Tammina, S. Transfer Learning Using VGG-16 with Deep Convolutional Neural Network for Classifying Images. Int. J. Sci. Res. Publ. 2019, 9, 143–150. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Pranesh, M.; Atchaya, A.J.; Anitha, J.; Hemanth, D.J. Scene Classification in Enhanced Remote Sensing Images Using Pre-trained RESNET50 Architecture. In Electronic Governance with Emerging Technologies; Springer: Berlin/Heidelberg, Germany, 2023; pp. 78–88. [Google Scholar]
- Bhattarai, S. New Plant Diseases Dataset, Kaggle, 18 November 2018. Available online: https://www.kaggle.com/datasets/vipoooool/new-plant-diseases-dataset (accessed on 1 July 2025).
- Desai, C. Comparative Analysis of Optimizers in Deep Neural Networks. Int. J. Innov. Sci. Res. Technol. 2020, 5, 959–962. [Google Scholar]
- Zhou, P.; Feng, J.; Ma, C. Towards Theoretically Understanding Why SGD Generalizes Better Than ADAM in Deep Learning. In Proceedings of the 34th Conference on Neural Information Processing Systems (NeurIPS 2020), Vancouver, BC, Canada, 6–12 December 2020. [Google Scholar]
- Alzubaidi, L.; Zhang, J.; Humaidi, A.J.; Al-Dujaili, A.; Duan, Y.; Al-Shamma, O.; Santamaría, J.; Fadhel, M.A.; Al-Amidie, M.; Farhan, L. Review of Deep Learning: Concepts, CNN Architectures, Challenges, Applications, Future Directions. J. Big Data 2021, 8, 53. [Google Scholar] [CrossRef]
- Li, P.; He, X.; Song, D.; Ding, Z.; Qiao, M.; Cheng, X.; Li, R. Improved Categorical Cross-Entropy Loss for Training Deep Neural Networks with Noisy Labels. In Pattern Recognition and Computer Vision; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2021; pp. 78–89. [Google Scholar]
- López, O.A.M.; López, A.M.; Crossa, J. Fundamentals of Artificial Neural Networks and Deep Learning. In Multivariate Statistical Machine Learning Methods for Genomic Prediction; Springer: Cham, Switzerland, 2022; pp. 379–425. [Google Scholar]
- Kandel, I.; Castelli, M. The Effect of Batch Size on The Generalizability of the Convolutional Neural Networks on a Histopathology Dataset. ICT Express 2020, 6, 312–315. [Google Scholar] [CrossRef]
- Plant Disease Detection Using VGG16, Kaggle, 20 May 2020. Available online: https://www.kaggle.com/code/amitkrjha/plant-disease-detection-using-vgg16/notebook (accessed on 1 July 2025).
- Cotton Plant Disease Detection Using Resnet50, Kaggle, 25 November 2020. Available online: https://www.kaggle.com/code/sayooj98/cotton-plant-disease-detection-using-resnet50 (accessed on 1 July 2025).
- Raspberry Pi. Available online: https://www.raspberrypi.com/products/raspberry-pi-3-model-b/ (accessed on 3 July 2025).
- NVIDIA Jetson Nano. NVIDIA Corporation. 2024. Available online: https://www.nvidia.com/en-us/autonomous-machines/embedded-systems/jetson-nano/product-development/ (accessed on 3 July 2025).



| Hyperparameters | Assigned Value |
|---|---|
| Classes | 3 |
| Epochs | 12 |
| Batch size | 32 |
| Learning rate | 0.01 |
| Optimizer | SGD |
| Activation function in the output layer Activation function in the hidden layers Loss Function | SoftMax ReLU Categorical Cross-Entropy |
| Number of filters in each Conv layer | 32 |
| Kernel size | 3 × 3 |
| Classes | Accuracy | Precision | Recall | F1-Score | Macro-F1 |
|---|---|---|---|---|---|
| 2-Convolutional-Layer CNN | |||||
| EB | 0.973 | 0.95 | 0.970 | 0.959 | 0.952 |
| LB | 0.960 | 0.982 | 0.896 | 0.937 | |
| H | 0.973 | 0.930 | 0.992 | 0.960 | |
| VGG16 CNN | |||||
| EB | 0.983 | 0.994 | 0.954 | 0.973 | 0.966 |
| LB | 0.967 | 0.932 | 0.974 | 0.952 | |
| H | 0.982 | 0.975 | 0.971 | 0.973 | |
| ResNet50 CNN | |||||
| EB | 0.880 | 0.790 | 0.873 | 0.829 | 0.545 |
| LB | 0.687 | 1.000 | 0.061 | 0.115 | |
| H | 0.708 | 0.533 | 0.978 | 0.691 | |
| Classes | Accuracy | Precision | Recall | F1-Score | Macro-F1 |
|---|---|---|---|---|---|
| 2-Convolutional-Layer CNN | |||||
| EB | 0.972 | 0.948 | 0.970 | 0.959 | 0.953 |
| LB | 0.960 | 0.982 | 0.898 | 0.938 | |
| H | 0.974 | 0.933 | 0.992 | 0.962 | |
| VGG16 CNN | |||||
| EB | 0.980 | 0.992 | 0.947 | 0.969 | 0.963 |
| LB | 0.966 | 0.926 | 0.973 | 0.949 | |
| H | 0.981 | 0.975 | 0.970 | 0.972 | |
| ResNet50 CNN | |||||
| EB | 0.885 | 0.802 | 0.870 | 0.834 | 0.542 |
| LB | 0.685 | 1.000 | 0.056 | 0.106 | |
| H | 0.700 | 0.527 | 0.980 | 0.685 | |
| Classes | Accuracy | Precision | Recall | F1-Score | Macro-F1 |
|---|---|---|---|---|---|
| 2-Convolutional-Layer CNN | |||||
| EB | 0.972 | 0.948 | 0.970 | 0.959 | 0.953 |
| LB | 0.960 | 0.982 | 0.898 | 0.938 | |
| H | 0.974 | 0.933 | 0.992 | 0.962 | |
| VGG16 CNN | |||||
| EB | 0.980 | 0.992 | 0.947 | 0.969 | 0.963 |
| LB | 0.966 | 0.926 | 0.973 | 0.949 | |
| H | 0.981 | 0.975 | 0.970 | 0.972 | |
| ResNet50 CNN | |||||
| EB | 0.885 | 0.802 | 0.870 | 0.834 | 0.542 |
| LB | 0.685 | 1.000 | 0.056 | 0.106 | |
| H | 0.700 | 0.527 | 0.980 | 0.685 | |
| Metrics | 2-CONV-Layer Model | ResNet50 CNN | VGG16 CNN |
|---|---|---|---|
| Training Time | 68.46 min | 153.12 min | 415.28 min |
| Inference Time/image | 0.17 s | 0.38 s | 0.79 s |
| Code Execution Time | 3.35 min | 7.42 min | 15.17 min |
| Testing Accuracy | 95.32% | 63.80% | 96.67% |
| Total Parameters | 379,171 | 23,980,931 | 14,812,995 |
| Total FLOPs | 405,342,770 | 10,089,594,898 | 40,115,044,370 |
| CPU Consumption | 21.80% | 31.10% | 42.67% |
| Memory Utilization | 67.17% | 66.33% | 66.93% |
| Metrics | 2-CONV-Layer Model | ResNet50 CNN | VGG16 CNN |
|---|---|---|---|
| Inference Time | 0.79 s | 4.49 s | 8.91 s |
| Code Execution Time | 15.75 min | 86.95 min | 169.99 min |
| Testing Accuracy | 95.38% | 63.56% | 96.37% |
| CPU Consumption | 41.10% | 64.00% | 88.30% |
| Memory Utilization | 45.50% | 52.30% | 50.70% |
| Metrics | 2-CONV-Layer Model | ResNet50 CNN | VGG16 CNN |
|---|---|---|---|
| Inference Time | 0.23 s | 0.81 s | 1.92 s |
| Code Execution Time | 4.59 min | 16.09 min | 36.77 min |
| Testing Accuracy | 95.38% | 63.56% | 96.37% |
| CPU Consumption | 36.30% | 67.60% | 88.80% |
| Memory Utilization | 67.03% | 73.63% | 60.50% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hammam, L.; Bastawrous, H.A.; Ghali, H.; Ebrahim, G.A. An Embedded Convolutional Neural Network Model for Potato Plant Disease Classification. Computers 2025, 14, 498. https://doi.org/10.3390/computers14110498
Hammam L, Bastawrous HA, Ghali H, Ebrahim GA. An Embedded Convolutional Neural Network Model for Potato Plant Disease Classification. Computers. 2025; 14(11):498. https://doi.org/10.3390/computers14110498
Chicago/Turabian StyleHammam, Laila, Hany Ayad Bastawrous, Hani Ghali, and Gamal A. Ebrahim. 2025. "An Embedded Convolutional Neural Network Model for Potato Plant Disease Classification" Computers 14, no. 11: 498. https://doi.org/10.3390/computers14110498
APA StyleHammam, L., Bastawrous, H. A., Ghali, H., & Ebrahim, G. A. (2025). An Embedded Convolutional Neural Network Model for Potato Plant Disease Classification. Computers, 14(11), 498. https://doi.org/10.3390/computers14110498







