Abstract
Antiviral peptides (AVPs) are promising therapeutic candidates, yet the rapid growth of sequence data and the field’s emphasis on predictors have left a gap: the lack of an integrated view linking peptide chemistry with biological context. Here, we map the AVP landscape through interactive data mining using Half-Space Proximal Networks (HSPNs) and Metadata Networks (MNs) in the StarPep toolbox. HSPNs minimize edges and avoid fixed thresholds, reducing computational cost while enabling high-resolution analysis. A threshold-free HSPN resolved eight chemically and biologically distinct communities, while MNs contextualized AVPs by source, function, and target, revealing structural–functional relationships. To capture diversity compactly, we applied centrality-guided scaffold extraction with redundancy removal (90–50% identity), producing four representative subsets suitable for modeling and similarity searches. Alignment-free motif discovery yielded 33 validated motifs, including 10 overlapping with reported AVP signatures and 23 apparently novel. Motifs displayed category-specific enrichment across antimicrobial classes, and sequences carrying multiple motifs (≥4–5) consistently showed higher predicted antiviral probabilities. Beyond computational insights, scaffolds provide representative “entry points” into AVP chemical space, while motifs serve as modular building blocks for rational design. Together, these resources provide an integrated framework that may inform AVP discovery and support scaffold- and motif-guided therapeutic design.
1. Introduction
Viruses comprise a diverse group of pathogens responsible for major infectious disease outbreaks throughout human history. From smallpox to the recent SARS-CoV-2 pandemic, viral diseases have remained a central focus of scientific, agricultural, and medical research [1]. Many viruses rapidly adapt to new hosts and environments through frequent mutations [2], driving the continual emergence of viral threats and underscoring the urgent need for effective therapeutic strategies [3]. Given this persistent risk, the development of antiviral therapeutics has been a long-standing scientific priority; in recent decades, numerous agents have been designed to target either viral proteins or host pathways, providing effective options across multiple infections [4,5].
Antiviral drugs can be broadly classified as small molecules or peptide-based agents. Historically, development favored small molecules due to simpler design and manufacturing [6]. Recent advances in chemistry, formulation, and delivery have mitigated key peptide limitations (instability, low permeability), renewing interest in their therapeutic potential [6]. Therapeutic peptides—typically <50 amino acids—offer close biomimicry, high specificity, low toxicity, and strong efficacy [7].
Despite the remaining challenges [8], peptides have significantly impacted the modern pharmaceutical industry [9]. As of 2022, more than 60 peptide drugs had marketing approval across the United States, Europe, and Japan [10,11], and over 400 peptides were in clinical trials, with 150 in active development and 260 completed human trials [10]. Within this context, antiviral peptides (AVPs) are increasingly important, with notable examples targeting HIV [12], SARS-CoV-2 [13], influenza [14], herpes simplex virus [15], dengue [16], tobacco mosaic virus [17], HSV [18], and Zika virus [19].
The growing interest in AVPs has led to numerous studies on their structures, mechanisms of action, and the development of dedicated databases [20]. AVPs are often considered a subclass of antimicrobial peptides (AMPs), and major AMP repositories such as dbAMP, LAMP, and CAMPR4 [21,22,23] contain thousands of peptides organized by functional class. For a targeted resource, the AVPpd database provides 2,683 experimentally validated AVPs [24,25]. However, the rapid expansion of AVP collections has outpaced manual interpretation, calling for systematic integration [26]. In cheminformatics, the concept of chemical space, though not uniquely defined, is generally tied to molecular similarity and diversity [27]. Mapping chemical space has proven highly valuable in drug discovery, helping relate structure to properties and activities and bridging chemical and biological domains to guide design [27].
Network-based methodologies efficiently handle large chemical datasets. Constructing similarity networks yields intuitive, analyzable maps of molecular relationships that support information extraction, connectivity analyses, and interactive data mining [28,29,30]. Although most applications have focused on small molecules, similar strategies have been applied to peptide classes such as tumor-homing [31], antibiofilm [32], haemolytic [33], and antiparasitic peptides [34]. Building on these methodologies, we investigate the AVP landscape in chemical and biological terms. While prior work has focused on activity predictors and machine-learning tools [35], a comprehensive network-level view that connects structural diversity with source, function, origin, and viral targets is still missing. To fill this gap, we combine Half-Space Proximal Networks (HSPNs) with Metadata Networks (MNs) to map, simplify, and contextualize the AVP space [36].
In this study, we used StarPepDB [37] and StarPep toolbox [36], available at https://github.com/Grupo-Medicina-Molecular-y-Traslacional/StarPep/ (accessed on 15 January 2025). StarPepDB is a graph-based repository integrating multiple peptide databases and currently represents one of the largest compilation of AMPs [37]. The StarPep toolbox supports visual network analysis [36,38] and enables the construction of two types of similarity complex networks: Chemical Space Networks (CSNs) and Half-Space Proximal Networks (HSPNs). CSNs model peptide similarity using alignment-free (AF) metrics derived from sequence descriptors, whereas HSPNs apply the Half-Space Proximal Test [38,39], which considers only a subset of pairwise relationships between nodes rather than all possible connections [34,38]. In parallel, the toolbox also builds Metadata Networks (MNs), modeled as bipartite graphs, that link peptides to their metadata—database source, function, origin, and viral target—thereby supplying the biological context needed to interpret chemical communities [36]. This chemical–biological coupling is central to our approach.
HSPNs offer several advantages over CSNs, including a substantially reduced number of edges, which lowers computational demands and processing time. In this work, we apply HSPNs in a novel way to explore and characterize the structural diversity of AVPs within chemical space. Notably, HSPNs can operate without a predefined similarity threshold, although thresholds can still be applied if desired [31,32]. Here, we examine the influence of applying a similarity threshold by comparing an HSPN without a cut-off to one constructed with a threshold, using the latter as a reference. Together with MNs, this design lets us connect chemically derived communities with their biological attributes (sources, functions, targets), supporting interpretation and downstream selection.
We evaluated the sequence diversity of representative AVP sets (scaffolds), extracted from HSPN communities, using Dover Analyzer [40], which supported the selection of the most representative subsets. HSPN analysis combined with alignment-free motif discovery (MEME suite v5.5.2) [41] enabled the identification of previously unreported motifs associated with antiviral activity. To assess their significance, we performed motif enrichment analysis with SEA v5.5.2 [42] and validated results against publicly available external datasets. MNs further contextualized these findings by revealing source/target linkages across peptide sets. This network-based strategy not only revealed novel antiviral motifs but also provided insights into the potential of representative AVPs across the mapped chemical space. Collectively, these findings advance our understanding of AVP structural and functional diversity and their prospects as antiviral therapeutics.
In summary, this study contributes (i) the construction and analysis of HSPNs to map the AVP chemical space without arbitrary thresholds, (ii) the integration of MNs to couple the AVP chemical space with biological attributes (sources, functions, viral targets), (iii) the extraction of representative scaffolds and enriched motifs capturing both known and novel antiviral patterns, (iv) the systematic validation of motif significance against external datasets to support rational AVP design, and (v) the demonstration of interactive data mining with the StarPep toolbox as a practical framework for AVP discovery and prioritization.
2. Materials and Methods
2.1. Basic Concepts
There are numerous concepts that it would be helpful to address previously to discuss further sections.
A Half-Space Proximal Network (HSPN) is a type of complex network designed to handle large datasets with lower memory requirements for storing (dis)similarity matrices. It reduces the number of links between nodes while preserving the metric space properties of (dis)similarity. By applying the Half-Space Proximal Test, HSPNs generate sparser graphs than conventional Chemical Space Networks (CSNs), making them efficient for exploring and analyzing large chemical datasets, including the structural diversity of AVPs [38,39].
The Euclidean (dis)similarity metric is widely used in data mining to measure the distance between data points. It is valued for its effectiveness with compact clusters, ease of computation, and sensitivity to outliers [43]. It is calculated as:
Modularity, as defined by Newman, measures the difference between the number of edges within groups in a network and the expected number of such edges in a random network with similar characteristics. Higher modularity values indicate a stronger community structure, and maximizing modularity reveals distinct communities within the graph [44].
The Louvain algorithm is a community detection method that optimizes modularity through two iterative phases: (1) local movement of nodes and (2) network aggregation [45]. initially, each node is assigned to its own community. For each node, the algorithm evaluates the modularity gain from moving it to a neighbor’s community, selecting the move with the highest gain. If no gain is possible, the node remains in its current community. This process repeats until a local maximum of modularity is reached. In the second phase, communities are aggregated into new nodes, and the weights of links between them are recalculated as the sum of all connections between their member nodes [46].
The Average Clustering Coefficient (ACC) summarizes the overall tendency of nodes in a network to form tightly connected groups; a property associated with the “small-world” effect. The local Clustering Coefficient (CC) for a node is the ratio of actual connections among its neighbors to the total possible connections. The ACC is the mean of CC values across all nodes, providing a global view of clustering tendencies and network efficiency in information transfer [47,48].
A bipartite graph is a graph whose vertices can be divided into two disjoint sets, V1 and V2, such that each edge connects a vertex from V1 to one from V2 [49]. In this study, metadata networks are modeled as bipartite graphs, where one set represents peptides and the other set represents metadata attributes, capturing relationships between these two types of entities.
Community Hub-Bridge Centrality (HB) identifies nodes that play dual roles as both hubs within their own community and bridges to other communities. In this measure, intra-community links are weighted by the size of the community, and inter-community links are weighted by the number of neighboring communities [50,51]. For a node i, is its internal strength, its external strength, card(ci) the size of its community, and nnc(i) the number of neighboring communities. The HB centrality is computed as [38]:
This metric highlights nodes that are key for maintaining intra-community cohesion and inter-community connectivity.
Harmonic Centrality (HC) is a global centrality measure based on the harmonic mean of the shortest path lengths in a network. For each pair of distinct nodes x and y, the reciprocal of the shortest path length 1/d(x,y) is summed across all reachable pairs:
Nodes with higher HC values are more centrally positioned, being closer on average to other nodes in the network and thus potentially more influential [52,53].
Betweenness Centrality (BC) measures the importance of a node in connecting other nodes via the shortest paths. It is calculated as the fraction of all shortest paths in the network that pass through a given node. Nodes with high BC act as key intermediaries, facilitating efficient communication between different parts of the network and often controlling information flow [54].
See Supplementary Materials S1.1 for a more detailed explanation of these and other concepts used in the manuscript.
2.2. Half-Space Proximal Network
The HSPNs were constructed following methodologies described in previous studies [31,32,34,38]. Data were obtained from StarPepDB [37], which contains 45,120 AMPs, including 4663 classified as antiviral peptides (AVPs). To reduce redundancy, sequences with >95% identity were removed using the Smith–Waterman algorithm [55], resulting in 3494 AVPs. Pairwise similarity was computed with the Euclidean metric and Min–Max normalization. Feature extraction employed multiple molecular descriptors, including peptide length, net charge, isoelectric point, molecular weight, Boman index, hydrophobic moment, average hydrophilicity, hydrophobic periodicity, aliphatic index, instability index, and aggregation-operator-based indices [38].
To evaluate the effect of the similarity threshold (t) in the HSPNs, various values of t from 0.3 to 0.9 were tested, and a network with t = 0 (parameter-free complex network) was included (Table S1.1). Clustering was performed with the Louvain algorithm (48). Network centrality was characterized mainly with HB centrality, and HC was used for smaller networks [52,53]. All these analyses were conducted using the StarPep Toolbox (https://github.com/Grupo-Medicina-Molecular-y-Traslacional/StarPep/) (GrapML files available in Supplementary Materials S2) [36].
2.3. Metadata Complex Network
The StarPep toolbox [38] was also used to generate Metadata Networks (MNs), represented as bipartite graphs. One node set corresponded to the 3494 AVPs, and the other to metadata categories: Database, Origin, Function, and Target. To preserve the bipartite structure, edges between peptides within the same set were removed from the initial similarity network. This arrangement highlights hierarchical relationships; whereby multiple peptides may connect to the same origin node but not directly to each other. Unlike the HSPNs, MN centrality was assessed using Betweenness centrality [54]. The resulting GrapML files are provided in Supplementary Materials S3.
2.4. Network Visualization and Characterization
HSPNs were visualized using Gephi, version 0.10 (https://gephi.org/, accessed on 30 January 2025) [56] using the Fruchterman–Reingold layout [57] with adjusted area and speed parameters. Clusters were color-coded, and node size was scaled by HB centrality. Network metrics—average degree, density, modularity, ACC, and average path length—were calculated in Gephi for each HSPN generated at similarity thresholds (t = 0.3–0.9). The number of singletons (atypical sequences) was determined from the giant component subgraph and/or nodes with degree zero. These metrics guided the selection of the optimal t, which served as the primary reference for comparison with the t = 0 network to assess the effect of the similarity threshold on AVP representation. From each HSPN, the five most central AVPs were extracted for further characterization. Their molecular properties—aliphatic index, Boman index, hydrophobicity, isoelectric point, net charge, and length—were calculated with the Peptides R package version 4.3.2 [58]. Biological activity data were added by cross-referencing StarPepDB metadata [37]. These sequences were visualized as a 10-node HSPN, and similarity overlap was measured using Dover Analyzer [40].
2.5. Exploration of Scaffold and Selection of Most Representative Subset
From the selected HSPNs (t = 0.75 and t = 0), scaffolds were extracted using the StarPep toolbox [36]. This procedure involves several parameters, such as the centrality measure HC or HB [52,53], the alignment algorithm type (Needleman-Wunsch [59] or Smith-Waterman [55]), and the sequence identity threshold (90–50%) to control redundancy during the scaffold extraction.
This exploration generated 20 distinct sets of scaffolds from each selected HSPN. Subsets were compared with Dover Analyzer [40] to evaluate the impact of centrality, alignment, and similarity threshold. For each identity level, subsets were grouped and compared by percentage of identical and similar sequences, enabling the identification of the most diverse and least redundant sets.
The optimal subsets for each similarity threshold were compiled into a consolidated dataset, representing the most informative scaffold collections. These were visualized in Gephi [56] following the methodology described in Section 2.4.
2.6. Motif Discovery
Motif discovery was performed using the alignment-free STREME algorithm [41], from the MEME Suite 5.5.2 [60]. The analysis used the eight communities identified in the HSPN (t = 0) generated with the StarPep toolbox [36]. Each community was exported as an individual FASTA file (S5) and processed in STREME with a motif width range of 3–6 amino acids and a significance threshold of p < 0.05.
In parallel, the three most central nodes from each cluster were chemically characterized using the Peptides R package, version 4.3.2 [58], as in Section 2.4. A literature review provided additional biological context, and the sequences were screened to verify the presence of discovered motifs within their respective clusters.
2.7. Alignment Free Motif Enrichment
The motifs identified by STREME [41] were validated using the Sequence Enrichment Analysis (SEA) tool from the MEME Suite 5.5.2 [60], assessing their enrichment in external datasets [61] (Supplementary Materials S6):
- B-TS_StarPepAVP (272 positives + 623 negatives)
- Ex_StarPepAVP (1230 positives + 10,771 negatives)
- TR_StarPepAVP (2321 positives + 2321 negatives)
- TS_StarPepAVP (623 positives + 623 negatives)
These datasets were built from StarPepDB [37] by selecting AVPs of 5–100 AAs and excluding peptides with multiple antimicrobial activities. The Ex_StarPepAVP set included 10,771 non-AMP sequences from [62], while negative sequences for the remaining sets were retrieved from UniProt [63]. Redundancy within and across datasets was minimized using Dover Analyzer [40].
In the SEA validation process, the E-value threshold was set to 10 or lower. The enrichment E-value of a motif was calculated by multiplying the adjusted p-value with the number of motifs in the input. The adjusted p-value represents the probability of the motif distinguishing the primary sequences from the control sequences [42].
An “inverse-validation” step was carried out using a negative dataset (13,715 unique sequences) compiled from the negatives of the four external datasets, to confirm that motifs were not equally probable in positives and negatives. Validated motifs were then searched within the most central sequences of each source cluster, followed by a literature review to identify previously reported associations or novel biological relevance. This dual validation ensured both statistical robustness and biological interpretability. The overall workflow is summarized in Scheme 1.
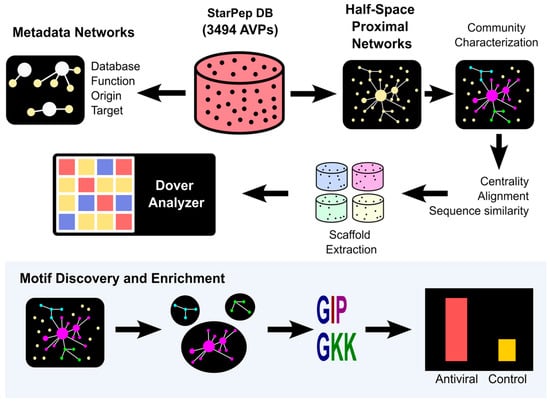
Scheme 1.
Methodology Review. Workflow illustrating the sequential analysis of representative scaffolds of the antiviral peptide space and the characterization of antiviral peptide motifs through complex network approaches. Arrows represent consecutive steps, beginning with metadata and half-space proximal networks, followed by scaffold extraction and motif enrichment, and culminating in computational validation.
2.8. Motif Scanning on Non-Antiviral Sequences
To assess the occurrence and potential relevance of the discovered motifs in peptides without reported antiviral activity, seven non-overlapping datasets were curated from StarPepDB (1–6) [37] and CPPsite 2.0 (7) [64].
- Antibacterial (12,936 seqs)
- Antifungal (4882)
- Antiparasitic (530)
- AMP dataset (13,107): antibacterial, antifungal, and antiparasitic peptides, excluding any overlap among these categories.
- Other dataset (8440): peptides classified as anticancer, antidiabetic, antihypertensive, enzymatic inhibitors, insecticidal, neuropeptides, and spermicidal.
- Toxic dataset (4653): peptides annotated as venom/toxic or toxic to mammals.
- CPP dataset (1171): cell-penetrating peptides from CPPsite 2.0.
All datasets excluded any sequence labeled as “Antiviral” in StarPepDB [37]. Motif enrichment in these datasets was evaluated using the SEA tool [42] from MEME Suite, following the same methodology applied for motif validation against external datasets, with an E-value threshold ≤ 10.
Additionally, the AMP dataset (13,107 sequences) was analyzed with FIMO (https://meme-suite.org/meme/tools/fimo, accessed on 12 February 2025) to detect individual motif occurrences, and with three AVP prediction tools—Meta-iAVP [65], AI4AVP [66], and seqprops [67]—selected for their combination of shallow and deep learning architectures. Each predictor generated a probability score per sequence, which was averaged to obtain a unified AVP-likelihood score. This score was then integrated with motif occurrence data for comparative assessment.
3. Results and Discussion
3.1. Metadata Complex Networks
Metadata Networks (MNs) provide a structured overview of AVP-related information in StarPepDB [37], enabling the analysis of sequence distribution and interconnections based on attributes such as databases, functions, origins, and targets. Constructed as bipartite graphs, MNs reveal hierarchical relationships between “peptide” and “metadata” nodes, as well as among “metadata” categories when redundant classification exists.
Database MN—The databases most contributing to the AVPs population in StarPepDB include SATPdb [68], AVPdb [25], DBAASP [69], DRAMP_General [70] and, LAMP_Experimental [22] (Figure 1A). These databases represent the top 5 central nodes in the network according to the BC [54] and exhibit high connectivity since they share a significant number of sequences. SATPdb stands out with 3,106 peptide connections (88.9% of the subset), far exceeding AVPdb. The least connected databases are DRAMP_Clinical [70] and MilkAMP [71], with node degrees of 4 and 6, respectively. While most AVPs are linked to multiple databases, peripheral nodes connect to only a single source, such as AVPdb (70 unique sequences) or CyBase_Cyclotides [72], with 81 cyclic backbone peptides. Although not representative for AVPs, the latter illustrates the methodological scope of the approach.
Function MN—As expected, the “Antiviral” subclass is the most central node, followed by Antimicrobial, Antibacterial, Antifungal, Anti Gram+, Anti Gram–, and Anti-HIV (Figure 1B). The prominence of “anti-HIV” reflects the targeted interest in therapeutics against this virus. Outside the antimicrobial category, “toxic to mammals” and “hemolytic” appear as highly connected functions, suggesting a possible link between antiviral activity and toxicity, in agreement with observations in [33,73]. The least connected function is “tumor-homing activity,” associated with a single peptide.
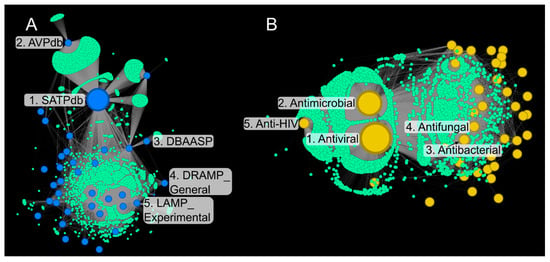
Figure 1.
Metadata Networks (MN). (A) “Database” MN, found in green, are the peptide nodes, and in blue database nodes, numbered nodes are: 1, SATPdb [68]; 2, AVPdb [25]; 3, DBAASP; 4, DRAMP General [70]; and 5, LAMP_ Experimental [22]. (B) “Function” MN, found in green are the peptide nodes, and in yellow are function nodes, numbered nodes are: 1, Antimicrobial; 2, Antibacterial; 3, Antifungal; 4, Anti Gram +; 5, Anti Gram -; 6, Anti-HIV; and 7, Toxic to mammals. Layout: Force Atlas2 [74].
Origin MN—The “produced by” category reveals two main spheres (Figure 2A): an inner sphere dominated by synthetic constructs (13.5% of sources) and an outer sphere of naturally isolated AVPs, primarily from Homo genus and Homininae family. Some peptides, such as StarPep_01104 and StarPep_00155, appear in both synthetic and natural origins, with 15 and 41 PubMed cross-references in StarPepDB, respectively. These are classified as a member of the mammalian tachykinin peptide family [75] and a transferase found in Homo sapien and Saccharomyces cerevisiae (PDB entry—4XNH (pdb_00004xnh)). The “is a” classification highlights small taxonomically related clusters (Figure S1.1) and peripheral unique sources such as Macaca mulatta, Rana temporaria, and Odorrana andersonii. These peripheral nodes may represent unique sources of AVPs with distinct biological activities, making them promising candidates for future investigations in the field of antiviral peptide research.
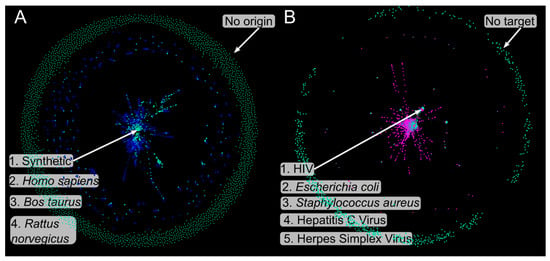
Figure 2.
Metadata Networks (MNs). (A) “Origin” MN “produced by” edges, found in green are the peptide nodes, and in blue origin nodes, numbered nodes are: 1, Synthetic; 2, Homo sapiens; 3, Bos taurus; 4, Rattus norvegicus. (B) “Target” MN “assessed against” nodes, found in green are the peptide nodes, and in fuchsia are target nodes. Numbered nodes are the following: 1, HIV; 2, Escherichia coli; 3, Staphylococcus aureus; 4, Hepatitis C Virus; and 5, Herpes Simplex Virus. Layout: Force Atlas2 [74].
Target MN—This network incorporates both “is a” and “assessed against” relationships to examine antiviral activity against different targets. The inner circle contains the most frequent targets, including HIV, Escherichia coli, Staphylococcus aureus, hepatitis C virus, and herpes simplex virus (Figure 2B). HIV, connected to 666 peptides (19% of targets), highlights its prominence in antiviral research. The occurrence of bacterial targets further supports the functional overlap between AVPs and AMPs. A small intermediate cluster connects to Andes virus, while the outer ring contains peptides linked to broad taxonomic groups without explicit targets. Taxonomic relationships and relative proximities among targets are depicted in Figure S1.2.
3.2. Half Space Proximal Networks
In exploring HSPNs for AVPs, the Euclidean metric was employed as the most intuitive similarity measure, widely used in cheminformatics to compute distances in multi-dimensional descriptor spaces. Beyond its interpretability, Euclidean distance was selected for three reasons: (i) in exploratory analyses, it consistently yielded stable networks with good topological resolution [38]; (ii) it enables straightforward methodological continuity with our prior studies on peptide similarity networks [32,33]; and (iii) in a previous comparative evaluation across haemolytic peptides, Euclidean, Bhattacharyya, and Soergel distances produced highly similar behaviors in terms of degree distributions, cutoff-dependent modularity, and scaffold subsets, supporting its robustness as a representative metric [33]. Nevertheless, we acknowledge that alternative alignment-free distances such as Angular Separation or Chebyshev may project peptide relatedness differently and thus affect community structure and motif detection, offering promising avenues for future network-based analyses.
Here, the Euclidean distances between nodes—defined by n molecular descriptors—were transformed into similarity values and aggregated into a similarity matrix. While such a matrix can be directly visualized as a similarity network, its conversion requires applying a threshold matrix. The choice of similarity threshold (t) directly impacts the network topology, influencing the preservation or removal of edges between peptides/nodes [76]. In this study, t was systematically varied from 0.3 to 0.9, and an additional threshold-free HSPN was constructed to enable direct comparison between networks with and without cutoffs. For readers seeking further technical details on HSPN construction, we refer to our earlier work [38], where Figure 1 presents a detailed schematic of the process.
Although the number of nodes remains constant, edge counts vary considerably across similarity thresholds. This variation, seemingly minor, has a marked effect on community formation and on the number of singletons—nodes lacking connections to others. After constructing all HSPNs at the selected thresholds (Figure S1.3), network topology was characterized in Gephi [56] using parameters including the number of edges, communities, and singletons, as well as density, modularity, average clustering coefficient (ACC), average path length, and average degree (Table S1.1). Density, ACC, and modularity trends across thresholds are shown in Figure 3.
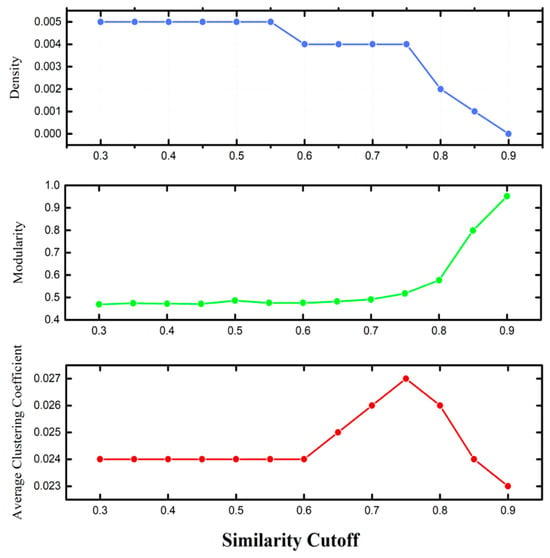
Figure 3.
HSPN’s characterization using different parameters. The similarity cut-off (t) is varied from 0.3 to 0.9.
These metrics guided the identification of the optimal t for AVP representation. Importantly, t selection is not an automated process but requires researcher judgment: a single overly cohesive community obscures heterogeneity, while excessive singletons dilute similarity insights. The aim is to balance connectivity and resolution for meaningful AVP representation [76].
A distinctive feature of HSPNs, setting them apart from conventional similarity networks, is that they can remain sparse even without a threshold—making them parameter-free. This property eliminates the need for optimal t selection, offering flexibility in analyses. Here, both the optimal-threshold network and the threshold-free network were examined in parallel to compare their densities and structures. This dual approach enabled a more complete view of network dynamics and relationships among AVPs.
As expected, increasing t reduced network density. Modularity, which the Louvain clustering algorithm seeks to maximize [46], increased with t, indicating fewer but more cohesive communities—particularly after t = 0.8. In contrast, ACC showed a non-linear relationship with t, peaking around t = 0.75, reflecting the highest connectivity. Based on these observations, t = 0.75 was chosen as optimal.
Figure 4A compares the HSPN at t = 0.75 (HSPN_OP) with the threshold-free version (t = 0; HSPN_NC). In HSPN_NC, Louvain clustering yielded eight communities (Figure 4B), ranging from 100 nodes (Cluster 4) to 692 nodes (Cluster 5). Node sizes reflect centrality, and these communities can be analyzed similarly to whole-space networks despite their smaller size. In contrast, HSPN_OP produced 74 clusters and 75 singletons, making community-level interpretation more fragmented and less tractable.

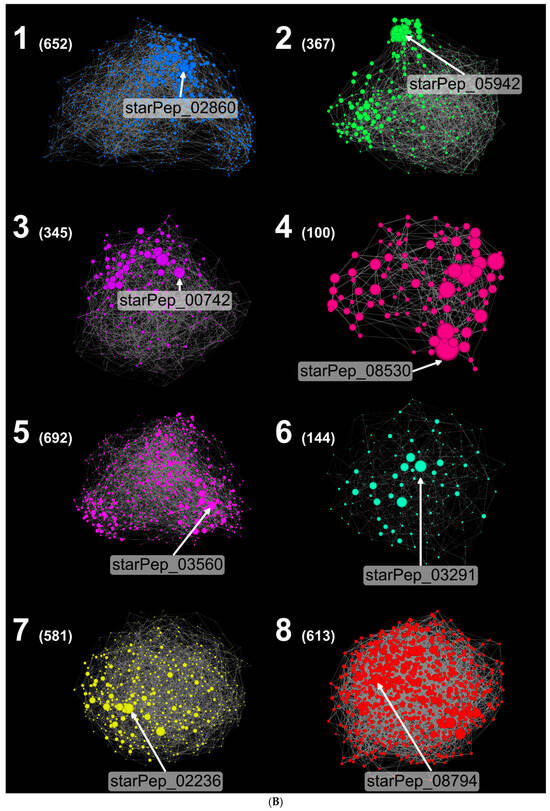
Figure 4.
(A) HSPNs. (I) t = 0 and (II) t = 0.75. A different color is assigned in each HSPN to show communities. Additionally, the most central nodes using HB centrality [38] are labeled, and the label size is proportional to the centrality value. Layout: Fruchterman Reingold [57] and NoOverlap. (B) HSPNs (t = 0) for every eight communities were obtained by using the Louvain algorithm [46]. The most central AVP using HC [52,53] are labeled in each cluster. The number in parenthesis represents the number of peptides in the cluster. Layout: Force Atlas2 [74].
Degree distributions (Figure 5) reveal that HSPN_NC exhibits a more balanced connectivity profile, whereas HSPN_OP contains many singletons and low-degree nodes.
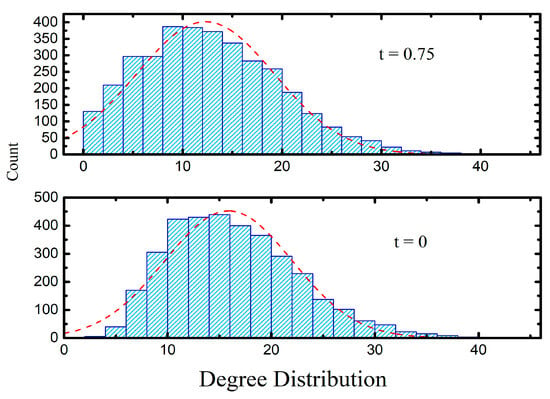
Figure 5.
HSPNs’ Degree distribution. t = 0.75, HSPN with optimal similarity cut-off. t = 0, HSPN with no cutoff (free parameter). The dashed red line indicates the standard fit for the respective distribution.
The most central nodes (by HB centrality) in both networks were further examined using physicochemical descriptors (Table 1) and amino acid (AA) composition generated with the “Peptides” R package [58]. While the central node sets differ between networks (Figure 4A), overlap exists: HSPN_OP’s most central nodes from clusters 19 and 9 correspond to clusters 1 and 3 in HSPN_NC. In HSPN_NC, central peptides are distributed across clusters 1, 5, and 8, which map to clusters 9, 18, and 31 in HSPN_OP. These results confirm that identical sequences consistently cluster together, regardless of network topology, while HSPN_OP tends to fragment chemical space more, making inter-cluster relationships harder to interpret.

Table 1.
Molecular Descriptors for the Most Central AVPs in each HSPNs.
Chemically, the most central peptides mapped in HSPN_OP display lower aliphatic index and hydrophobicity and are on average ~13 residues shorter than those in HSPN_NC. Differences in Boman Index, isoelectric point, and net charge remain below 10%. In AA composition, HSPN_OP’s central peptides show greater homogeneity, likely due to dominance by a single large cluster. HSPN_NC has higher mole percentages of aromatic, charged, and basic residues, while HSPN_OP has relatively more tiny, aliphatic, and acidic residues. Both networks share the predominance of non-polar residues (Figure 6).
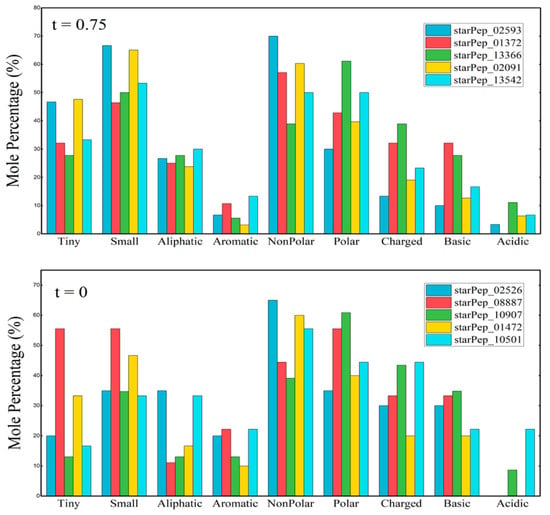
Figure 6.
Occurrence of different types of AAs corresponding to the five most central nodes (Harmonic Centrality [52,53] of each HSPNs selected.
Representative literature references for each central peptide are listed to illustrate biological relevance.
HSPN_OP central nodes:
- StarPep_02593—Cycloviolacin-O17 from Viola odorata; anti-HIV, antibacterial, hemolytic. [77]. Seq: “GIPCGESCVWIPGISAAIGCSCKNKVCYRN”.
- StarPep_01372—Kenojeinin I from fermented skate skin; cationic residues aid bacterial membrane binding; hydrophobic residues disrupt membranes [78]. Seq: “GKQYFPKVGGRLSGKAPLAAKTHRRLKP”.
- StarPep_13366—Hepatitis C virus genome polyprotein fragment (E1/E2 envelope region), implicated in viral entry [79]. Seq: “VATRDGKLPTTQLRRHID”.
- StarPep_02091—Ascaris suum antibacterial factor abf-2; active against Gram-positive/negative bacteria and yeast [80]. Seq: “DIDFSTCARMDVPILKKAAQGLCITSCSMQNCGTGSCKKRSGRPTCVCYRCANGGGDIPLGAL”.
- StarPep_13542—Classical swine fever virus genome polyprotein fragment, envelope glycoprotein-associated [81]. Seq: “VSRRYLASLHKKALPTSVTFELLFDGTNPS”.
HSPN_NC central nodes:
- StarPep_02526—D51 synthetic AMP designed via linguistic model, amphipathic, active against Gram-positive/negative bacteria [82]. Seq: “FLFRVASKVFPALIGKFKKK”.
- StarPep_08887—Andes virus inhibitor, identified via cysteine-constrained phage display [83]. Seq: “CSLHSHKGC”.
- StarPep_10907—Feline immunodeficiency virus gp150-derived peptide, likely interfering with viral entry [84]. Seq: “KQRNRWEWRPDFKSKKVKISLPC”.
- StarPep_01472—Deer (Cervus elaphus) blood-derived AMP, especially active against Gram-negative bacteria [85]. Seq: “IRNSLTCRFNFGICLPKRCPGRMRQIGTCF”
- StarPep_10501—Amphipathic helix peptide targeting HIV envelope glycoprotein to inhibit membrane fusion [86]. Seq: “KAFEEVLAKKFYDKALWD”.
As in the Metadata Network analysis, antiviral activity shows strong overlap with broader AMP classifications, with three of the ten central peptides also targeting bacteria. Notably, each network contains at least one HIV-targeting peptide, reflecting HIV’s prominence in AVP research.
A similarity network built from these ten sequences (Figure 7A) shows sparse connectivity—most nodes link to only two others—with StarPep_02091 being the most connected and bigger, likely due to its longer length. Local alignment-based similarity analysis (Dover Analyzer) produced a heatmap (Figure 7B) confirming low compositional similarity among the sequences. This heterogeneity suggests that the chemical space of AVPs is diverse even among the most central representatives and motivates future intracommunity analyses to identify distinctive features and functional relationships.
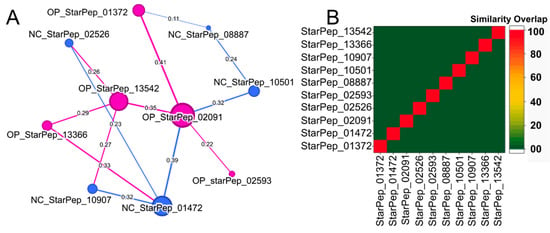
Figure 7.
(A) Similarity Network between the most central nodes from HSPN_OP and HSPN_NC. The node size is proportional to the centrality measure (HB) applied to the network. Layout: Fruchterman Reingold [57] (B) Similarity Overlaps between the 10 top sequences of each HSPN done by Dover Analyzer [40] with a local type of alignment algorithm [55].
Compared with earlier applications of peptide similarity networks (e.g., tumor-homing, antibiofilm, haemolytic, and antiparasitic peptides [31,32,34,73]), our AVP-focused framework introduces three differentiating elements. First, we use HSPNs that can operate without arbitrary similarity cut-offs, yielding high-resolution yet computationally tractable maps of AVP chemical space. Second, we integrate Metadata Networks (MNs) to couple the AVP chemical space with biological attributes (sources, functions, viral targets), enabling functional interpretation beyond purely structural views. Third, we combine two HSPN topologies (HSPN_OP and HSPN_NC) with a two-stage motif pipeline (external enrichment and inverse validation) and community-aware scaffold extraction, strengthening interpretability and facilitating reuse (scaffold subsets and validated motifs) in downstream discovery. Together, these aspects position our approach as complementary to prior peptide network methodologies while being specifically tailored to antiviral peptides.
3.3. Scaffold Extraction
The primary goal of this analysis was to identify the subset of peptides that best captures the diversity and representativeness of the AVP chemical space. Using the scaffold extraction tool available in the Subnetwork Mining module of the StarPep toolbox [36], a total of 20 scaffolds were obtained from both HSPN_OP and HSPN_NC by systematically varying the alignment algorithm, centrality measure, and similarity threshold. Scaffold extraction reduces network complexity while preserving its topological and chemical representativeness, thus enabling a condensed yet meaningful view of the AVP landscape.
It is important to note that the purpose of scaffold extraction in this study was not to provide exhaustive biological annotation of the scaffolds themselves, but rather to generate representative, non-redundant subsets of AVPs that preserve chemical diversity. These subsets are intended as reusable network resources for downstream applications—such as training datasets for ML models, benchmarking of predictive pipelines, or multi-query similarity searches—rather than as final biological classifications [35]. Scaffold-level biological interpretation will therefore be addressed in future integrative studies, where chemical diversity can be coupled with experimental or functional annotations.
The comparison between scaffold sets was conducted using Dover Analyzer [40], focusing on pairwise similarity overlaps represented as percentages. These values indicate the proportion of redundant sequences between two scaffold sets. The results were visualized as heatmaps, enabling the simultaneous comparison of eight scaffolds under different conditions. Due to differences in scaffold sizes, the heatmaps are inherently asymmetric—two scaffolds may share the same absolute number of overlapping sequences, but the relative redundancy percentage will differ if one set is substantially smaller.
Across all comparisons (Figure 8), a consistent pattern emerged: scaffold sets derived from different parent HSPNs showed a predominantly red section in the heatmaps, reflecting a high degree of overlap and suggesting similar representativeness and diversity. This finding indicates that, at least for AVPs, exhaustive optimization of the similarity threshold (t) in HSPN construction may not be necessary for scaffold extraction purposes.
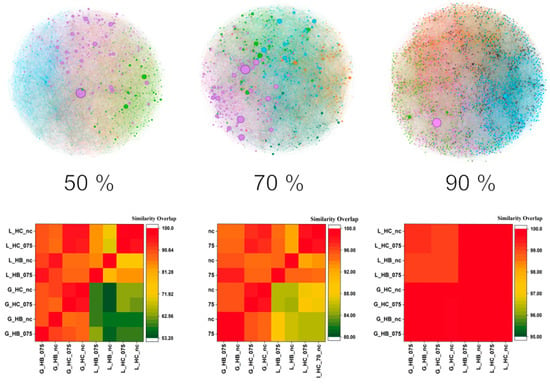
Figure 8.
HSPNs were obtained from the subsets with different % identities (top). Clusters are colored, and the node size is configured to represent the HB’s centrality [38] within the Fruchterman Reingold layout [57]. Similarity overlap heatmaps (bottom) of scaffolds. From left to right, the lowest similarity for each heatmap is: 53.20%, 80%, and 95%. L/G = alignment used in scaffold extraction—L: local (Smith–Waterman), G: global (Needleman–Wunsch); HC/HB = centrality metric—HC: harmonic, HB: hub–bridge; NC = threshold-free HSPN (no similarity cut-off); 0.75 = optimal similarity cut-off (t = 0.75) applied to the HSPN.
This insight has direct implications for streamlining future AVP chemoinformatic workflows.
By contrast, the alignment algorithm exerted the strongest influence on scaffold composition. This effect was especially pronounced in scaffolds generated using different centrality measures. HB centrality, being more localized and community-oriented, produced scaffold sets more sensitive to alignment method variations, whereas HC centrality, based on shortest-path distances, yielded scaffolds less affected by alignment changes. However, even with HC, differences became noticeable at lower sequence identity thresholds (≤60%). Detailed results for all scaffold sets are presented in Table 2 and Table 3.

Table 2.
Characterization of Scaffolds from HSPN_NC varying the alignment algorithm and Centrality Measure.

Table 3.
Characterization of Scaffolds from HSPN_OP varying the alignment algorithm and Centrality Measure.
From these analyses, four merged scaffold subsets were created at decreasing pairwise sequence identity thresholds (80%, 70%, 60%, and 50%), containing 2703, 2445, 2152, and 1872 positive sequences, respectively. These curated subsets represent highly diverse, non-redundant fractions of the AVP chemical space and are provided as a community resource for downstream applications such as training datasets for machine learning models or multi-query similarity searches [35] (subsets available in Supplementary Materials S4).
The principal advantage of these scaffold-derived subsets lies in their ability to prevent overfitting and family overrepresentation in predictive modeling. They therefore provide a compact and chemically diverse representation of AVPs, suitable for computational modeling, similarity searching, and other chemoinformatic analyses. In parallel to scaffold extraction, a separate motif discovery analysis was performed on the communities identified in the threshold-free HSPN, aiming to detect short amino acid patterns enriched within AVP clusters and potentially linked to antiviral activity.
Looking ahead, scaffold subsets may also offer a rational entry point for experimental exploration. Their non-redundant composition can facilitate the prioritization of diverse peptide panels for synthesis and initial screening, serving as chemically representative “templates” rather than final therapeutic candidates. In this prospective role, scaffolds could help select distinct sequence chemotypes for in vitro assays (e.g., envelope destabilization, pseudovirus entry), while reducing family bias and redundancy. We stress, however, that this translational perspective is exploratory, and that experimental validation will be essential to determine whether scaffold-derived representatives indeed display a promising antiviral activity.
3.4. Motif Discovery
Although peptide sequences can undergo vast diversification, sequence variation must remain within functional limits. Certain amino acids (AAs) remain conserved because of their crucial role in peptide activity. These conserved residues, found across multiple sequences, are called motifs, signifying their orthologous relationship [87]. In biological contexts, motifs are recurrent sequence patterns; here, they are defined as short AA segments of 3–6 residues. Peptide motifs act as compact building blocks with functional autonomy, potentially yielding more effective AVPs [88]. Their experimental discovery is challenging and often requires labor-intensive methodologies [89].
Before reporting motif results, it is important to note two potential sources of bias. (i) Database bias: AVP annotations and the external validation sets may vary in their experimental support and assay conditions, which can influence enrichment outcomes. Although STREME’s shuffled controls and SEA’s inverse validation help to mitigate spurious patterns, they cannot fully eliminate biases introduced during dataset curation or uneven sampling. (ii) Descriptor/graph dependence: the communities used for motif mining derive from HSPN_NC, built on global physicochemical descriptors and sequence-based molecular descriptors; however, alternative descriptor sets, or similarity metrics could yield different partitions and, consequently, different candidate motifs. Accordingly, motif specificity should be regarded as hypothesis-generating and confirmed experimentally.
Previous results showed no significant difference between HSPNs with and without an optimal cut-off; therefore, motif searches were conducted exclusively on HSPN_NC. The search targeted the eight clusters/communities identified from HSPN_NC (Figure 4B). As a first step, each cluster’s chemical space was characterized. Table 4.

Table 4.
List of Most Central AVPs corresponding to each Community in HSPN.
The search targeted the eight clusters/communities identified from HSPN_NC (Figure 4B). As a first step, each cluster’s chemical space was characterized. Table 4 lists the three most central peptides per cluster, including their sequences and relevant external information. Notably, clusters 1 and 2 contain plant-derived cyclotides, clusters 3 and 6 contain amphibian-derived peptides and cluster 4 contains HIV-interacting peptides (Table 4). The remaining clusters lack clear biological relationships among their central nodes.
Distinct chemical profiles were generated for each cluster (Figure S1.4), and differences in global peptide descriptors are summarized in Figure 9. Key observations include:
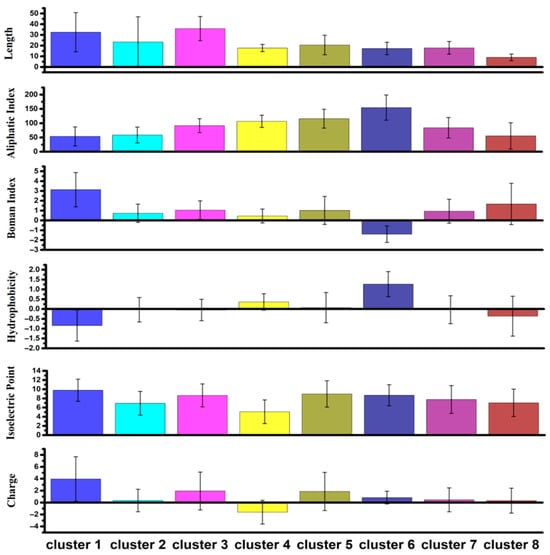
Figure 9.
Representation of the mean of several molecular descriptors calculated for the peptides in each of the clusters of HSPN_NC using the ‘Peptides’ Library for R [58]. Each color represents a different cluster.
- Cluster 4: more acidic AAs, average negative charge, lowest isoelectric point.
- Cluster 1: highest positive charge and Boman index, lowest hydrophobicity.
- Clusters 2, 7, 8: near-neutral charges.
- Cluster 6: highest aliphatic index, high hydrophobicity, negative Boman index.
- Clusters 1 and 3: longest sequences; Cluster 8: shortest sequences.
These profiles align with known AVP characteristics, where cationic and hydrophobic residues are critical for activity against enveloped viruses [107]. Importantly, chemical profiling also clarified why certain clusters formed separately—for example, clusters 1 and 2 share biological sources but differ significantly in chemical composition, justifying their division.
Motif discovery was performed using the STREME algorithm from MEME Suite [60], as described in Materials and Methods—Motif Discovery. STREME generated shuffled control datasets to filter irrelevant patterns, providing statistical significance by comparing motif occurrence in primary vs. control sequences.
3.4.1. Motif Enrichment
The STREME search identified 42 motifs (Table S1.2). Validation proceeded in two stages, following the approach in Materials and Methods—Motif Enrichment.
Stage 1—Positive Dataset Validation
Using the SEA algorithm [42] and four positive external datasets (Tables S1.3 and S1.4), we assessed motif enrichment. To ensure the reliability of the validation process, a similarity overlap analysis was performed on the external datasets used. Figure S1.5 shows that most overlaps are below 60%, except for complete overlap between the B-TS and TS datasets. Using non-redundant, diverse datasets increased the reliability and generalizability of the motif selection.
Stage 2—Inverse Validation
A unique negative dataset was built from the combined negatives of the four positive datasets, with redundancies removed. SEA confirmed motifs that appeared significantly more in positives than in negatives, filtering out chance patterns.
After both stages, 33 motifs remained, which are listed with their respective significance in Table 5.

Table 5.
Full list of validated motifs by SEA software [42], after removing motifs occurring in negative datasets.
These motifs were also mapped in the central peptides of each cluster (see Table 4, for red-marked entries). Clusters 2 and 4 contained multiple motifs—suggesting greater sequence diversity and functional variation—whereas cluster 1 initially showed none.
Comparison with literature: A comparison was conducted with the existing state of the art literature to gain further insights into these motifs. Table 6 summarize the AVP motifs previously reported and the convergence with them are discussed as follows:
- Position-specific glycine: Machine-learning models for SARS-CoV-2 peptide prediction reported an elevated frequency of glycine at position 1 [108]. In our validated motifs, 18% begin with glycine and 21% contain glycine at other positions, mirroring that positional signal [108].
- Anti-coronavirus peptide motifs: A second study reporting anti-coronavirus peptides provided functional AVP motifs in its Supplementary Information [109]. We observe higher motif-level similarity to our set (see Table 6), including enrichment of arginine, leucine, and valine, consistent with other studies [109].
- GKK motif and residue composition (ENNAVIA-D): The GKK motif appears among positive samples in the ENNAVIA-D model [110]. Consistently with [110], our motifs frequently include lysine, leucine, asparagine, glutamic acid, and valine; specifically, lysine, leucine, and valine occur in 36%, 30%, and 27% of validated motifs, respectively.
- Specificity caveat for lysine: While lysine is prevalent among AVPs, its occurrence is even higher in non-AVPs, an important consideration when interpreting motif specificity and when using lysine-rich motifs for design [110].
- Residue concordance across other studies: Other reports highlight frequent leucine, glutamic acid, valine, and tryptophan in AVPs [107,108]. This residue-level convergence further supports the biological relevance of the identified motifs herein.

Table 6.
Overlap between network-derived AVP motifs and literature-reported motifs.
Table 6.
Overlap between network-derived AVP motifs and literature-reported motifs.
| MOTIF | Cluster | Reference Sequence | Reference |
|---|---|---|---|
| CYCR | 1 | CYCRTGRCATRERRSGTCIIQGRL | [111] |
| RRRRRH | RRRRRRRRHPAEPGSTVTTQNTASQTMS | [109] | |
| CGES | 2 | IPCGESCVWIPCITA | [109] |
| VWIPCI | IPCGESCVWIPCITA | [109] | |
| QAVG | VYSRCGFAQTLYYDYGVTDMNTLANWVCLVQYESSFNDQAVGAINYNGTQDFGLFQINNKYWCQGAVSSSDSCGIACTSLLGNLSASWSCAQLVYQQQGFSAWYGWLNNCNGTAPSVADCF | [112] | |
| FNK | 3 | GVTQNVLYENQKQIANQFNKAISQIQESLTTTSTALGKLQ | [13] |
| NGIGVTQNVLYENQKQIANQFNKAISQIQESLTTTSTA | |||
| AASFNKAMTNIVDAFTGVNDAITQTSQALQTVATALNKIQDVVNQQGNSLNHLTSQ | |||
| QIANQFNKAISQIQE | [109] | ||
| KQFNKCSLATELSRLGVPKSELPDWVCLVQHESNFKTNWINKKNSNGSWDFGLFQINDKWWCEGHIRSHNTCNVKCEELVTEDIEKALECAKVIKRERGYKAWYGWLNNCQNKKPSVDECF | [112] | ||
| KKKKVV | 5 | KKKKVVAATYV | [109] |
| WLRDI | SWLRDIWDWICEVLS | [109] | |
| WDWIC | SWLRDIWDWICEVLS | [109] | |
| SGSWLRDVWDWICTVLTDFKTWLQSKL | [19] | ||
| GKK | 6 | ILPLLKKFGKKFGKKVWKAL | [113] |
| AFAFDVTRKINPETSAVERPEVSEYPEIPKGTKLQEFVMMDIEIEEEGADNRAETIQRIKCVPSQCNQICRVLGKKCGYCKNASTCVCLG | [112] |
Note: The table highlights in red validated motifs from this study that align with previously reported computationally predicted or experimentally characterized AVP motifs.
The subset of validated motifs that overlap with previously reported patterns (Table 6) can also be mechanistically linked to established antiviral principles. For instance, the GKK motif represents a glycine-initiated pattern, associated with structural flexibility and enhanced binding to viral proteins, as reported for SARS-CoV-2 inhibitory peptides [108]. Likewise, lysine- and arginine-rich signatures such as RRRRH and KKKKVV are consistent with electrostatic destabilization of viral envelopes and interactions with negatively charged membranes [107,110]. In addition, motifs like WDWIC and WLRDI combine hydrophobic and cationic residues, reflecting the amphipathic balance required for membrane insertion and disruption, a well-established mechanism of AVPs [109]. These convergences underscore that the motifs discovered here do not only carry statistical enrichment but also recapitulate functional signatures known to underpin antiviral activity. Although our validation pipeline was primarily computational, this convergence with experimentally and computationally characterized motifs reported in the literature (Table 6) provides an additional layer of biological plausibility. Taken together, these findings underscore that our network-derived motifs are not only statistically enriched but also aligned with functional signatures known to underpin antiviral activity.
Out of the 33 computationally validated motifs, only 10 could be cross-referenced with state-of-the-art literature, including recent reviews and computational studies that integrate curated AVP datasets and predictive frameworks (e.g., ENNAVIA-D and anti-coronavirus peptide models), as summarized in Table 6. The remaining 23 motifs appear to be novel, as they could not be traced in recent literature sources and showed no matches in PROSITE’s pattern and profile databases when queried through the Motif Search engine of the GenomeNet suite [114]. Taken together, these negative results reinforce the interpretation that the 23 validated motifs represent previously undescribed antiviral sequence signatures, although their functional roles will require subsequent experimental validation.
3.4.2. Mapping Antiviral Motifs Against Non-AVPs
To assess how antiviral motifs distribute beyond explicitly annotated AVPs, we mapped the 33 validated motifs against non-antiviral datasets (see Materials and Methods—Motif Scanning): Antibacterial, Antifungal, Antiparasitic, AMP, Toxic, Others, and CPP.
Overall, motif occurrence varied by group (Table 7). The most ubiquitous pattern was “GKK”, which appeared in every category but peaked in antibacterials (56.9%) and was least frequent in toxic peptides (24.5%). Although several motifs were comparatively rare overall, e.g., “RRRRH” and “LLDK”, they were enriched in antifungal peptides. Among the antimicrobial classes, “CYCR” was far more common in antifungals (23%) than antibacterials (2.1%) or antiparasitic (0.7%).

Table 7.
Relative Enrichment of Motifs in different Peptides Datasets measured by SEA.
“GCSCK” showed a high prevalence in AMP (16.5%) and was also present in antibacterials (8.1%) and antifungals (5.4%). “GTCNTP” was detected across a range of datasets: antibacterial (7.4%), antifungal (9.1%) and antiparasitic (4.9%). In the Others dataset, “GLPV” (33.20%) and “SAAJ” (19.90%) were highly prevalent, suggesting these motifs may mark bioactive peptides more generally. By contrast, the CPP dataset carried the lowest burden of antiviral motifs among all groups (Table 7).
Taken together, these distributions suggest that specific motifs are linked to particular peptide classes and that the presence of a motif can be used to make preliminary, informed assessments about potential activity profiles, even if this is not definitive (Table 7). This supports the utility of motif screening as a lightweight, early-stage filter in broader discovery pipelines.
We further examined the link between motif burden and predicted antiviral activity within the AMP dataset. As shown in Figure 10, the presence of motifs consistently elevates the predicted AVP probability, and peptides carrying four or five motifs achieve the highest predicted scores.
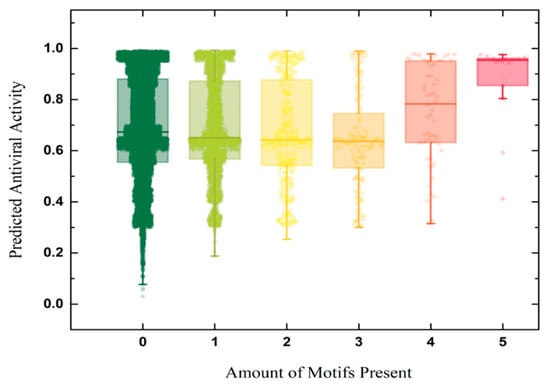
Figure 10.
Distribution of peptides antiviral activity score and the corresponding amount of motifs scanned by FIMO [115] within the AMP dataset.
This finding implies that designing sequences from validated motifs—rather than arranging residues at random—substantially increases the likelihood of antiviral activity. Guided by this principle, we highlight several candidate sequences for experimental follow-up in Table 8.

Table 8.
Example sequences from StarPepDB containing many motifs and have high probability of antiviral activity. The mapped motifs are highlighted in red.
Overall, the combination of network-based clustering, detailed chemical profiling, and rigorous motif validation yielded novel and biologically relevant patterns. By confirming overlaps with known AVPs and revealing unique, previously undescribed motifs, our results highlight the potential of motif analytics as an effective route to expand the antiviral sequence space. These findings indicate that motif occurrence is not only informative of organismal source and target categories, but also predictive of antiviral potential, reinforcing the value of integrating chemical-space analysis with motif discovery in AVP research.
Beyond these computational insights, the 33 validated motifs may also serve as modular sequence elements to guide therapeutic peptide design. By embedding such short, functionally enriched motifs into synthetic constructs or grafting them onto privileged scaffolds, it becomes possible to generate candidate sequences tailored to relevant viral targets (e.g., HIV, influenza, SARS-CoV-2). This translational perspective remains prospective, as these motifs have only been computationally validated; nevertheless, they underscore how network-derived motifs can bridge large-scale in silico analyses with experimentally testable hypotheses, providing a rational basis for future antiviral development.
While these motifs provide a rational basis for peptide design, it must be stressed that their validation was entirely computational. Database biases and in silico enrichment cannot substitute for biological testing; thus, the 33 motifs should be viewed as hypothesis-generating elements. Experimental follow-up will be required to confirm their antiviral functionality and therapeutic potential.
4. Conclusions
We present an integrated, network-driven map of the AVP landscape. Using HSPNs and MNs in StarPep, we show that AVP space can be explored at high resolution without a fixed similarity threshold: a threshold-free HSPN remains sparse and, with Louvain clustering, resolves eight chemically distinct, biologically coherent communities. MNs link chemistry to AVP context, highlighting the prevalence of synthetic entries, overlap with broader AMP functions, and the centrality of HIV as a tested target. A systematic centrality-guided scaffold extraction yields four non-redundant, diversity-preserving subsets for modeling, benchmarking, and multi-query searching. Alignment-free motif analytics produce 33 enriched motifs—23 apparently novel—that carry source/target information across antimicrobial classes; cross-AMP classess motif mapping revealed category-specific enrichment patterns, and within AMPs, motif load (≥4–5 mapped motifs per sequence) consistently associated with higher predicted antiviral probability across three independent predictors.
Identifying HB/HC central peptides yields privileged scaffolds that capture intra-community cores and inter-community bridges. Prioritizing these scaffolds produces representative, low redundancy chemotypes for antiviral design, screening and benchmarking, while mitigating family bias and overfitting.
Strengths and contributions: Unified chemical–biological mapping (HSPN/MN); centrality-informed scaffold selection; reusable graphs/subsets; and short, portable motifs as actionable design elements for AVPs. Limitations: Computational study depending on StarPepDB coverage (e.g., HIV testing bias) and on descriptor/alignment choices; motif specificity ultimately requires experiments. Future directions: Next steps include (i) experimental testing of motif-based candidates, (ii) benchmark AVP predictors using our released non-redundant scaffold subsets, (iii) tailor selected scaffolds/motifs to virus-specific panels (e.g., HIV, influenza, SARS-CoV-2); and (iv) integrating network features with structural/biophysical data and modern sequence embeddings to build mechanism-aware models.
In sum, coupling HSPNs, MNs, centrality-guided scaffolds, and motif analytics provides a practical route to navigate AVP diversity and accelerate rational, motif- and scaffold-driven design of next-generation antiviral peptides.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/computers14100423/s1. File S1 includes Content S1.1. Basic Concepts and Terms. Table S1.1. Full centrality characterization of each of the HSPNs constructed. Table S1.2. Motifs discovered using STREME and their significance statistics. Table S1.3. Motifs enriched with SEA and significance statistics. Table S1.4. Motifs tested against negative datasets. Figure S1.1. Metadata Network –“Origin” of AVPs. Figure S1.2. Metadata Network –“Targets” of AVPs. Figure S1.3. Effect of the similarity threshold (t) on HSPN topology. Figure S1.4. Chemical profiles of the eight clusters and Figure S1.5. Similarity overlaps among the four external datasets used. S2. GraphML files of HSPNs built at different similarity threshold. S3. GraphML files of Metadata Networks. S4. Scaffold subsets in FASTA obtained under different extraction conditions—50–90% identity redundancy removal—Dover Analyzer comparisons, and the four final FASTA subsets best representing the AVP chemical space. S5. Physicochemical descriptors for the eight clusters resolved in the threshold-free HSPN with corresponding FASTA sequences and S6. The four external datasets in FASTA described in Section 2.7.
Author Contributions
Y.M.-P. and G.A.-C. contributed to the conceptualization, methodology, supervision, drafting/reviewing of the manuscript and funding acquisition. D.d.L.G. was responsible for data curation, networks building, scaffolds extraction, statistical analysis and data/result visualization. H.R., F.J.F. and E.A.M. participated in sequence comparative analyses, motif discovery/enrichment. F.M.-R., J.R.M. and Y.P.-C. participated in programming tasks, data visualization and review/editing. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the USFQ MED Grant 2024-25 (Project ID: 30509) and the Portuguese Foundation for Science and Technology (FCT) under the scope of UIDB/04423/2020 and UIDP/04423/2020.
Data Availability Statement
The original data presented in the study are openly available at the journal site while the StarPep toolbox software and the respective user manual are freely available online at http://mobiosd-hub.com/starpep/. All underlying code and installation files are accessible through GitHub (https://github.com/Grupo-Medicina-Molecular-y-Traslacional/StarPep) under the Apache 2.0.
Acknowledgments
Yovani Marrero-Ponce thanks the program Profesor convitado for a fellowship to work at Valencia University in 2024. We thanks Kevin Castillo-Mendieta for his assistance with artwork.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Miranda, M.N.S.; Pingarilho, M.; Pimentel, V.; Torneri, A.; Seabra, S.G.; Libin, P.J.K.; Abecasis, A.B. A Tale of Three Recent Pandemics: Influenza, HIV and SARS-CoV-2. Front. Microbiol. 2022, 13, 889643. [Google Scholar] [CrossRef] [PubMed]
- Markov, P.V.; Ghafari, M.; Beer, M.; Lythgoe, K.; Simmonds, P.; Stilianakis, N.I.; Katzourakis, A. The Evolution of SARS-CoV-2. Nat. Rev. Microbiol. 2023, 21, 361–379. [Google Scholar] [CrossRef]
- Meganck, R.M.; Baric, R.S. Developing Therapeutic Approaches for Twenty-First-Century Emerging Infectious Viral Diseases. Nat. Med. 2021, 27, 401–410. [Google Scholar] [CrossRef]
- Von Delft, A.; Hall, M.D.; Kwong, A.D.; Purcell, L.A.; Saikatendu, K.S.; Schmitz, U.; Tallarico, J.A.; Lee, A.A. Accelerating Antiviral Drug Discovery: Lessons from COVID-19. Nat. Rev. Drug Discov. 2023, 22, 585–603. [Google Scholar] [CrossRef]
- Wallis, R.S.; O’Garra, A.; Sher, A.; Wack, A. Host-Directed Immunotherapy of Viral and Bacterial Infections: Past, Present and Future. Nat. Rev. Immunol. 2023, 23, 121–133. [Google Scholar] [CrossRef]
- Wang, L.; Wang, N.; Zhang, W.; Cheng, X.; Yan, Z.; Shao, G.; Wang, X.; Wang, R.; Fu, C. Therapeutic Peptides: Current Applications and Future Directions. Signal Transduct. Target. Ther. 2022, 7, 48. [Google Scholar] [CrossRef] [PubMed]
- Lau, J.L.; Dunn, M.K. Therapeutic Peptides: Historical Perspectives, Current Development Trends, and Future Directions. Bioorganic Med. Chem. 2018, 26, 2700–2707. [Google Scholar] [CrossRef]
- Lamers, C. Overcoming the Shortcomings of Peptide-Based Therapeutics. Future Drug Discov. 2022, 4, FDD75. [Google Scholar] [CrossRef]
- Henninot, A.; Collins, J.C.; Nuss, J.M. The Current State of Peptide Drug Discovery: Back to the Future? J. Med. Chem. 2018, 61, 1382–1414. [Google Scholar] [CrossRef] [PubMed]
- Mousavi Maleki, M.S.; Sardari, S.; Ghandehari Alavijeh, A.; Madanchi, H. Recent Patents and FDA-Approved Drugs Based on Antiviral Peptides and Other Peptide-Related Antivirals. Int. J. Pept. Res. Ther. 2022, 29, 5. [Google Scholar] [CrossRef]
- Usmani, S.S.; Bedi, G.; Samuel, J.S.; Singh, S.; Kalra, S.; Kumar, P.; Ahuja, A.A.; Sharma, M.; Gautam, A.; Raghava, G.P.S. THPdb: Database of FDA-Approved Peptide and Protein Therapeutics. PLoS ONE 2017, 12, e0181748. [Google Scholar] [CrossRef]
- Carter, E.P.; Ang, C.G.; Chaiken, I.M. Peptide Triazole Inhibitors of HIV-1: Hijackers of Env Metastability. Curr. Protein Pept. Sci. 2023, 24, 59–77. [Google Scholar] [CrossRef]
- Heydari, H.; Golmohammadi, R.; Mirnejad, R.; Tebyanian, H.; Fasihi-Ramandi, M.; Moosazadeh Moghaddam, M. Antiviral Peptides against Coronaviridae Family: A Review. Peptides 2021, 139, 170526. [Google Scholar] [CrossRef]
- Agamennone, M.; Fantacuzzi, M.; Vivenzio, G.; Scala, M.C.; Campiglia, P.; Superti, F.; Sala, M. Antiviral Peptides as Anti-Influenza Agents. Int. J. Mol. Sci. 2022, 23, 11433. [Google Scholar] [CrossRef]
- Jenssen, H. Anti Herpes Simplex Virus Activity of Lactoferrin/Lactoferricin—An Example of Antiviral Activity of Antimicrobial Protein/Peptide. Cell. Mol. Life Sci. CMLS 2005, 62, 3002–3013. [Google Scholar] [CrossRef]
- Becker, Y. Dengue Fever Virus and Japanese Encephalitis Virus Synthetic Peptides, with Motifs to Fit HLA Class I Haplotypes Prevalent in Human Populations in Endemic Regions, Can Be Used for Application to Skin Langerhans Cells to Prime Antiviral CD8+ Cytotoxic T Cells (CTLs)—A Novel Approach to the Protection of Humans. Virus Genes 1994, 9, 33–45. [Google Scholar] [CrossRef]
- Park, J.Y.; Yang, S.Y.; Kim, Y.C.; Kim, J.-C.; Dang, Q.L.; Kim, J.J.; Kim, I.S. Antiviral Peptide from Pseudomonas Chlororaphis O6 against Tobacco Mosaic Virus (TMV). J. Korean Soc. Appl. Biol. Chem. 2012, 55, 89–94. [Google Scholar] [CrossRef]
- Jenssen, H.; Gutteberg, T.J.; Rekdal, O.; Lejon, T. Prediction of Activity, Synthesis and Biological Testing of Anti-HSV Active Peptides. Chem. Biol. Drug Des. 2006, 68, 58–66. [Google Scholar] [CrossRef]
- Jackman, J.A.; Costa, V.V.; Park, S.; Real, A.L.C.V.; Park, J.H.; Cardozo, P.L.; Ferhan, A.R.; Olmo, I.G.; Moreira, T.P.; Bambirra, J.L.; et al. Therapeutic Treatment of Zika Virus Infection Using a Brain-Penetrating Antiviral Peptide. Nat. Mater. 2018, 17, 971–977. [Google Scholar] [CrossRef]
- Agarwal, G.; Gabrani, R. Antiviral Peptides: Identification and Validation. Int. J. Pept. Res. Ther. 2021, 27, 149–168. [Google Scholar] [CrossRef]
- Jhong, J.-H.; Yao, L.; Pang, Y.; Li, Z.; Chung, C.-R.; Wang, R.; Li, S.; Li, W.; Luo, M.; Ma, R.; et al. dbAMP 2.0: Updated Resource for Antimicrobial Peptides with an Enhanced Scanning Method for Genomic and Proteomic Data. Nucleic Acids Res. 2022, 50, D460–D470. [Google Scholar] [CrossRef]
- Zhao, X.; Wu, H.; Lu, H.; Li, G.; Huang, Q. LAMP: A Database Linking Antimicrobial Peptides. PLoS ONE 2013, 8, e66557. [Google Scholar] [CrossRef]
- Gawde, U.; Chakraborty, S.; Waghu, F.H.; Barai, R.S.; Khanderkar, A.; Indraguru, R.; Shirsat, T.; Idicula-Thomas, S. CAMPR4: A Database of Natural and Synthetic Antimicrobial Peptides. Nucleic Acids Res. 2023, 51, D377–D383. [Google Scholar] [CrossRef]
- Ramazi, S.; Mohammadi, N.; Allahverdi, A.; Khalili, E.; Abdolmaleki, P. A Review on Antimicrobial Peptides Databases and the Computational Tools. Database 2022, 2022, baac011. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, A.; Thakur, N.; Tandon, H.; Kumar, M. AVPdb: A Database of Experimentally Validated Antiviral Peptides Targeting Medically Important Viruses. Nucleic Acids Res. 2014, 42, D1147–D1153. [Google Scholar] [CrossRef] [PubMed]
- Cook, D.J.; Holder, L.B. Graph-Based Data Mining. IEEE Intell. Syst. 2000, 15, 32–41. [Google Scholar] [CrossRef]
- Medina-Franco, J.L.; Sánchez-Cruz, N.; López-López, E.; Díaz-Eufracio, B.I. Progress on Open Chemoinformatic Tools for Expanding and Exploring the Chemical Space. J. Comput. Aided Mol. Des. 2022, 36, 341–354. [Google Scholar] [CrossRef]
- Maggiora, G.M.; Bajorath, J. Chemical Space Networks: A Powerful New Paradigm for the Description of Chemical Space. J. Comput. Aided Mol. Des. 2014, 28, 795–802. [Google Scholar] [CrossRef]
- Holzinger, A.; Dehmer, M.; Jurisica, I. Knowledge Discovery and Interactive Data Mining in Bioinformatics—State-of-the-Art, Future Challenges and Research Directions. BMC Bioinform. 2014, 15, I1. [Google Scholar] [CrossRef]
- Wang, F.; Jin, R.; Agrawal, G.; Piontkivska, H. Graph and Topological Structure Mining on Scientific Articles. In Proceedings of the 2007 IEEE 7th International Symposium on BioInformatics and BioEngineering, Boston, MA, USA, 14–17 October 2007; IEEE: Piscataway, NJ, USA, 2007; pp. 1318–1322. [Google Scholar]
- Romero, M.; Marrero-Ponce, Y.; Rodríguez, H.; Agüero-Chapin, G.; Antunes, A.; Aguilera-Mendoza, L.; Martinez-Rios, F. A Novel Network Science and Similarity-Searching-Based Approach for Discovering Potential Tumor-Homing Peptides from Antimicrobials. Antibiotics 2022, 11, 401. [Google Scholar] [CrossRef]
- Agüero-Chapin, G.; Antunes, A.; Mora, J.R.; Pérez, N.; Contreras-Torres, E.; Valdes-Martini, J.R.; Martinez-Rios, F.; Zambrano, C.H.; Marrero-Ponce, Y. Complex Networks Analyses of Antibiofilm Peptides: An Emerging Tool for Next-Generation Antimicrobials’ Discovery. Antibiotics 2023, 12, 747. [Google Scholar] [CrossRef]
- Castillo-Mendieta, K.; Agüero-Chapin, G.; Marquez, E.A.; Perez-Castillo, Y.; Barigye, S.J.; Vispo, N.S.; García-Jacas, C.R.; Marrero-Ponce, Y. Peptide Hemolytic Activity Analysis Using Visual Data Mining of Similarity-Based Complex Networks. Npj Syst. Biol. Appl. 2024, 10, 115. [Google Scholar] [CrossRef]
- Ayala-Ruano, S.; Marrero-Ponce, Y.; Aguilera-Mendoza, L.; Pérez, N.; Agüero-Chapin, G.; Antunes, A.; Aguilar, A.C. Network Science and Group Fusion Similarity-Based Searching to Explore the Chemical Space of Antiparasitic Peptides. ACS Omega 2022, 7, 46012–46036. [Google Scholar] [CrossRef]
- Agüero-Chapin, G.; Galpert-Cañizares, D.; Domínguez-Pérez, D.; Marrero-Ponce, Y.; Pérez-Machado, G.; Teijeira, M.; Antunes, A. Emerging Computational Approaches for Antimicrobial Peptide Discovery. Antibiotics 2022, 11, 936. [Google Scholar] [CrossRef]
- Aguilera-Mendoza, L.; Ayala-Ruano, S.; Martinez-Rios, F.; Chavez, E.; García-Jacas, C.R.; Brizuela, C.A.; Marrero-Ponce, Y. StarPep Toolbox: An Open-Source Software to Assist Chemical Space Analysis of Bioactive Peptides and Their Functions Using Complex Networks. Bioinformatics 2023, 39, btad506. [Google Scholar] [CrossRef]
- Aguilera-Mendoza, L.; Marrero-Ponce, Y.; Beltran, J.A.; Tellez Ibarra, R.; Guillen-Ramirez, H.A.; Brizuela, C.A. Graph-Based Data Integration from Bioactive Peptide Databases of Pharmaceutical Interest: Toward an Organized Collection Enabling Visual Network Analysis. Bioinformatics 2019, 35, 4739–4747. [Google Scholar] [CrossRef]
- Aguilera-Mendoza, L.; Marrero-Ponce, Y.; García-Jacas, C.R.; Chavez, E.; Beltran, J.A.; Guillen-Ramirez, H.A.; Brizuela, C.A. Automatic Construction of Molecular Similarity Networks for Visual Graph Mining in Chemical Space of Bioactive Peptides: An Unsupervised Learning Approach. Sci. Rep. 2020, 10, 18074. [Google Scholar] [CrossRef]
- Chavez, E.; Dobrev, S.; Kranakis, E.; Opatrny, J.; Stacho, L.; Tejeda, H.; Urrutia, J. Half-Space Proximal: A New Local Test for Extracting a Bounded Dilation Spanner of a Unit Disk Graph. In Principles of Distributed Systems; Anderson, J.H., Prencipe, G., Wattenhofer, R., Eds.; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2006; Volume 3974, pp. 235–245. ISBN 978-3-540-36321-7. [Google Scholar]
- Aguilera-Mendoza, L.; Marrero-Ponce, Y.; Tellez-Ibarra, R.; Llorente-Quesada, M.T.; Salgado, J.; Barigye, S.J.; Liu, J. Overlap and Diversity in Antimicrobial Peptide Databases: Compiling a Non-Redundant Set of Sequences. Bioinformatics 2015, 31, 2553–2559. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bailey, T.L. STREME: Accurate and Versatile Sequence Motif Discovery. Bioinformatics 2021, 37, 2834–2840. [Google Scholar] [CrossRef]
- Bailey, T.L.; Grant, C.E. SEA: Simple Enrichment Analysis of Motifs. bioRxiv 2021. [Google Scholar] [CrossRef]
- Shirkhorshidi, A.S.; Aghabozorgi, S.; Wah, T.Y. A Comparison Study on Similarity and Dissimilarity Measures in Clustering Continuous Data. PLoS ONE 2015, 10, e0144059. [Google Scholar] [CrossRef] [PubMed]
- Newman, M.E.J. Modularity and Community Structure in Networks. Proc. Natl. Acad. Sci. USA 2006, 103, 8577–8582. [Google Scholar] [CrossRef]
- Traag, V.A.; Waltman, L.; Van Eck, N.J. From Louvain to Leiden: Guaranteeing Well-Connected Communities. Sci. Rep. 2019, 9, 5233. [Google Scholar] [CrossRef]
- Blondel, V.D.; Guillaume, J.-L.; Lambiotte, R.; Lefebvre, E. Fast Unfolding of Communities in Large Networks. J. Stat. Mech. Theory Exp. 2008, 2008, P10008. [Google Scholar] [CrossRef]
- Zahoránszky, L.A.; Katona, G.Y.; Hári, P.; Málnási-Csizmadia, A.; Zweig, K.A.; Zahoránszky-Köhalmi, G. Breaking the Hierarchy—A New Cluster Selection Mechanism for Hierarchical Clustering Methods. Algorithms Mol. Biol. 2009, 4, 12. [Google Scholar] [CrossRef] [PubMed]
- Watts, D.J.; Strogatz, S.H. Collective Dynamics of ‘Small-World’ Networks. Nature 1998, 393, 440–442. [Google Scholar] [CrossRef] [PubMed]
- Zepeda-Mendoza, M.L.; Resendis-Antonio, O. Bipartite Graph. In Encyclopedia of Systems Biology; Dubitzky, W., Wolkenhauer, O., Cho, K.-H., Yokota, H., Eds.; Springer: New York, NY, USA, 2013; pp. 147–148. ISBN 978-1-4419-9862-0. [Google Scholar]
- Ghalmane, Z.; Hassouni, M.E.; Cherifi, H. Immunization of Networks with Non-Overlapping Community Structure. Soc. Netw. Anal. Min. 2019, 9, 45. [Google Scholar] [CrossRef]
- Rajeh, S.; Savonnet, M.; Leclercq, E.; Cherifi, H. Investigating Centrality Measures in Social Networks with Community Structure. In Complex Networks & Their Applications IX.; Benito, R.M., Cherifi, C., Cherifi, H., Moro, E., Rocha, L.M., Sales-Pardo, M., Eds.; Studies in Computational Intelligence; Springer International Publishing: Cham, Switzerland, 2021; Volume 943, pp. 211–222. ISBN 978-3-030-65346-0. [Google Scholar]
- Marchiori, M.; Latora, V. Harmony in the Small-World. Phys. Stat. Mech. Its Appl. 2000, 285, 539–546. [Google Scholar] [CrossRef]
- Ruan, Y.; Tang, J.; Hu, Y.; Wang, H.; Bai, L. Efficient Algorithm for the Identification of Node Significance in Complex Network. IEEE Access 2020, 8, 28947–28955. [Google Scholar] [CrossRef]
- Brandes, U.; Pich, C. CENTRALITY ESTIMATION IN LARGE NETWORKS. Int. J. Bifurc. Chaos 2007, 17, 2303–2318. [Google Scholar] [CrossRef]
- Xia, Z.; Cui, Y.; Zhang, A.; Tang, T.; Peng, L.; Huang, C.; Yang, C.; Liao, X. A Review of Parallel Implementations for the Smith–Waterman Algorithm. Interdiscip. Sci. Comput. Life Sci. 2022, 14, 1–14. [Google Scholar] [CrossRef]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An Open Source Software for Exploring and Manipulating Networks. In Proceedings of the Third International Conference on Weblogs and Social Media, ICWSM 2009, San Jose, CA, USA, 17–20 May 2009. [Google Scholar] [CrossRef]
- Fruchterman, T.M.J.; Reingold, E.M. Graph Drawing by Force-Directed Placement. Softw. Pract. Exp. 1991, 21, 1129–1164. [Google Scholar] [CrossRef]
- Osorio, D.; Rondón-Villarreal, P.; Torres, R. Peptides: A Package for Data Mining of Antimicrobial Peptides. R J. 2015, 7, 4. [Google Scholar] [CrossRef]
- Needleman, S.B.; Wunsch, C.D. A General Method Applicable to the Search for Similarities in the Amino Acid Sequence of Two Proteins. J. Mol. Biol. 1970, 48, 443–453. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed]
- Pinacho-Castellanos, S.A.; García-Jacas, C.R.; Gilson, M.K.; Brizuela, C.A. Alignment-Free Antimicrobial Peptide Predictors: Improving Performance by a Thorough Analysis of the Largest Available Data Set. J. Chem. Inf. Model. 2021, 61, 3141–3157. [Google Scholar] [CrossRef] [PubMed]
- Gabere, M.N.; Noble, W.S. Empirical Comparison of Web-Based Antimicrobial Peptide Prediction Tools. Bioinformatics 2017, 33, 1921–1929. [Google Scholar] [CrossRef] [PubMed]
- The UniProt Consortium. UniProt: A Worldwide Hub of Protein Knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef]
- Agrawal, P.; Bhalla, S.; Usmani, S.S.; Singh, S.; Chaudhary, K.; Raghava, G.P.S.; Gautam, A. CPPsite 2.0: A Repository of Experimentally Validated Cell-Penetrating Peptides. Nucleic Acids Res. 2016, 44, D1098–D1103. [Google Scholar] [CrossRef] [PubMed]
- Schaduangrat, N.; Nantasenamat, C.; Prachayasittikul, V.; Shoombuatong, W. Meta-iAVP: A Sequence-Based Meta-Predictor for Improving the Prediction of Antiviral Peptides Using Effective Feature Representation. Int. J. Mol. Sci. 2019, 20, 5743. [Google Scholar] [CrossRef]
- Lin, T.-T.; Sun, Y.-Y.; Wang, C.-T.; Cheng, W.-C.; Lu, I.-H.; Lin, C.-Y.; Chen, S.-H. AI4AVP: An Antiviral Peptides Predictor in Deep Learning Approach with Generative Adversarial Network Data Augmentation. Bioinform. Adv. 2022, 2, vbac080. [Google Scholar] [CrossRef]
- Otović, E.; Njirjak, M.; Kalafatovic, D.; Mauša, G. Sequential Properties Representation Scheme for Recurrent Neural Network-Based Prediction of Therapeutic Peptides. J. Chem. Inf. Model. 2022, 62, 2961–2972. [Google Scholar] [CrossRef]
- Singh, S.; Chaudhary, K.; Dhanda, S.K.; Bhalla, S.; Usmani, S.S.; Gautam, A.; Tuknait, A.; Agrawal, P.; Mathur, D.; Raghava, G.P.S. SATPdb: A Database of Structurally Annotated Therapeutic Peptides. Nucleic Acids Res. 2016, 44, D1119–D1126. [Google Scholar] [CrossRef]
- Pirtskhalava, M.; Gabrielian, A.; Cruz, P.; Griggs, H.L.; Squires, R.B.; Hurt, D.E.; Grigolava, M.; Chubinidze, M.; Gogoladze, G.; Vishnepolsky, B.; et al. DBAASP v.2: An Enhanced Database of Structure and Antimicrobial/Cytotoxic Activity of Natural and Synthetic Peptides. Nucleic Acids Res. 2016, 44, D1104–D1112. [Google Scholar] [CrossRef] [PubMed]
- Shi, G.; Kang, X.; Dong, F.; Liu, Y.; Zhu, N.; Hu, Y.; Xu, H.; Lao, X.; Zheng, H. DRAMP 3.0: An Enhanced Comprehensive Data Repository of Antimicrobial Peptides. Nucleic Acids Res. 2022, 50, D488–D496. [Google Scholar] [CrossRef]
- Théolier, J.; Fliss, I.; Jean, J.; Hammami, R. MilkAMP: A Comprehensive Database of Antimicrobial Peptides of Dairy Origin. Dairy Sci. Technol. 2014, 94, 181–193. [Google Scholar] [CrossRef]
- Wang, C.K.L.; Kaas, Q.; Chiche, L.; Craik, D.J. CyBase: A Database of Cyclic Protein Sequences and Structures, with Applications in Protein Discovery and Engineering. Nucleic Acids Res. 2008, 36, D206–D210. [Google Scholar] [CrossRef] [PubMed]
- Castillo-Mendieta, K.; Agüero-Chapin, G.; Mora, J.R.; Pérez, N.; Contreras-Torres, E.; Valdes-Martini, J.R.; Martinez-Rios, F.; Marrero-Ponce, Y. Unraveling the Hemolytic Toxicity Tapestry of Peptides Using Chemical Space Complex Networks. Toxicol. Sci. 2024, 202, 236–249. [Google Scholar] [CrossRef] [PubMed]
- Jacomy, M.; Venturini, T.; Heymann, S.; Bastian, M. ForceAtlas2, a Continuous Graph Layout Algorithm for Handy Network Visualization Designed for the Gephi Software. PLoS ONE 2014, 9, e98679. [Google Scholar] [CrossRef]
- Chandrashekar, I.R.; Cowsik, S.M. Three-Dimensional Structure of the Mammalian Tachykinin Peptide Neurokinin A Bound to Lipid Micelles. Biophys. J. 2003, 85, 4002–4011. [Google Scholar] [CrossRef]
- Zahoránszky-Kőhalmi, G.; Bologa, C.G.; Oprea, T.I. Impact of Similarity Threshold on the Topology of Molecular Similarity Networks and Clustering Outcomes. J. Cheminform. 2016, 8, 16. [Google Scholar] [CrossRef] [PubMed]
- Ireland, D.C.; Colgrave, M.L.; Craik, D.J. A Novel Suite of Cyclotides from Viola odorata: Sequence Variation and the Implications for Structure, Function and Stability. Biochem. J. 2006, 400, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.-H.; Lee, B.-D.; An, H.; Eun, J.-B. Kenojeinin I, Antimicrobial Peptide Isolated from the Skin of the Fermented Skate, Raja Kenojei. Peptides 2005, 26, 581–587. [Google Scholar] [CrossRef]
- Op De Beeck, A.; Voisset, C.; Bartosch, B.; Ciczora, Y.; Cocquerel, L.; Keck, Z.; Foung, S.; Cosset, F.-L.; Dubuisson, J. Characterization of Functional Hepatitis C Virus Envelope Glycoproteins. J. Virol. 2004, 78, 2994–3002. [Google Scholar] [CrossRef]
- Kato, Y.; Aizawa, T.; Hoshino, H.; Kawano, K.; Nitta, K.; Zhang, H. Abf-1 and Abf-2, ASABF-Type Antimicrobial Peptide Genes in Caenorhabditis Elegans. Biochem. J. 2002, 361, 221–230. [Google Scholar] [CrossRef]
- Li, X.; Wang, L.; Zhao, D.; Zhang, G.; Luo, J.; Deng, R.; Yang, Y. Identification of Host Cell Binding Peptide from an Overlapping Peptide Library for Inhibition of Classical Swine Fever Virus Infection. Virus Genes 2011, 43, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Loose, C.; Jensen, K.; Rigoutsos, I.; Stephanopoulos, G. A Linguistic Model for the Rational Design of Antimicrobial Peptides. Nature 2006, 443, 867–869. [Google Scholar] [CrossRef]
- Hall, P.R.; Hjelle, B.; Njus, H.; Ye, C.; Bondu-Hawkins, V.; Brown, D.C.; Kilpatrick, K.A.; Larson, R.S. Phage Display Selection of Cyclic Peptides That Inhibit Andes Virus Infection. J. Virol. 2009, 83, 8965–8969. [Google Scholar] [CrossRef]
- Lombardi, S.; Massi, C.; Indino, E.; Rosa, C.L.; Mazzetti, P.; Falcone, M.L.; Rovero, P.; Fissi, A.; Pieroni, O.; Bandecchi, P.; et al. Inhibition of Feline Immunodeficiency Virus Infectionin Vitroby Envelope Glycoprotein Synthetic Peptides. Virology 1996, 220, 274–284. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Treffers, C.; Chen, L.; Anderson, R.C.; Yu, P.-L. Isolation and Characterisation of Antimicrobial Peptides from Deer Neutrophils. Int. J. Antimicrob. Agents 2005, 26, 165–169. [Google Scholar] [CrossRef] [PubMed]
- Owens, B.J.; Anantharamaiah, G.M.; Kahlon, J.B.; Srinivas, R.V.; Compans, R.W.; Segrest, J.P. Apolipoprotein A-I and Its Amphipathic Helix Peptide Analogues Inhibit Human Immunodeficiency Virus-Induced Syncytium Formation. J. Clin. Investig. 1990, 86, 1142–1150. [Google Scholar] [CrossRef]
- Shiba, K. Natural and Artificial Peptide Motifs: Their Origins and the Application of Motif-Programming. Chem. Soc. Rev. 2010, 39, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Tompa, P.; Davey, N.E.; Gibson, T.J.; Babu, M.M. A Million Peptide Motifs for the Molecular Biologist. Mol. Cell 2014, 55, 161–169. [Google Scholar] [CrossRef]
- Neduva, V.; Linding, R.; Su-Angrand, I.; Stark, A.; Masi, F.D.; Gibson, T.J.; Lewis, J.; Serrano, L.; Russell, R.B. Systematic Discovery of New Recognition Peptides Mediating Protein Interaction Networks. PLoS Biol. 2005, 3, e405. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-J.; Cheng, C.-S.; Lai, S.-M.; Hsu, M.-P.; Chen, C.-S.; Lyu, P.-C. Solution Structure of the Plant Defensin VrD1 from Mung Bean and Its Possible Role in Insecticidal Activity against Bruchids. Proteins Struct. Funct. Bioinforma. 2006, 63, 777–786. [Google Scholar] [CrossRef]
- Lay, F.T.; Schirra, H.J.; Scanlon, M.J.; Anderson, M.A.; Craik, D.J. The Three-Dimensional Solution Structure of NaD1, a New Floral Defensin from Nicotiana Alata and Its Application to a Homology Model of the Crop Defense Protein alfAFP. J. Mol. Biol. 2003, 325, 175–188. [Google Scholar] [CrossRef]
- Ovchinnikova, T.V.; Balandin, S.V.; Aleshina, G.M.; Tagaev, A.A.; Leonova, Y.F.; Krasnodembsky, E.D.; Men’shenin, A.V.; Kokryakov, V.N. Aurelin, a Novel Antimicrobial Peptide from Jellyfish Aurelia Aurita with Structural Features of Defensins and Channel-Blocking Toxins. Biochem. Biophys. Res. Commun. 2006, 348, 514–523. [Google Scholar] [CrossRef]
- Craik, D.J.; Daly, N.L.; Bond, T.; Waine, C. Plant Cyclotides: A Unique Family of Cyclic and Knotted Proteins That Defines the Cyclic Cystine Knot Structural Motif. J. Mol. Biol. 1999, 294, 1327–1336. [Google Scholar] [CrossRef]
- Göransson, U.; Luijendijk, T.; Johansson, S.; Bohlin, L.; Claeson, P. Seven Novel Macrocyclic Polypeptides from Viola arvensis. J. Nat. Prod. 1999, 62, 283–286. [Google Scholar] [CrossRef]
- Yang, X.; Lee, W.-H.; Zhang, Y. Extremely Abundant Antimicrobial Peptides Existed in the Skins of Nine Kinds of Chinese Odorous Frogs. J. Proteome Res. 2012, 11, 306–319. [Google Scholar] [CrossRef]
- Basir, Y.J.; Knoop, F.C.; Dulka, J.; Conlon, J.M. Multiple Antimicrobial Peptides and Peptides Related to Bradykinin and Neuromedin N Isolated from Skin Secretions of the Pickerel Frog, Rana Palustris. Biochim. Biophys. Acta BBA—Protein Struct. Mol. Enzymol. 2000, 1543, 95–105. [Google Scholar] [CrossRef]
- Agopian, A.; Gros, E.; Aldrian-Herrada, G.; Bosquet, N.; Clayette, P.; Divita, G. A New Generation of Peptide-Based Inhibitors Targeting HIV-1 Reverse Transcriptase Conformational Flexibility. J. Biol. Chem. 2009, 284, 254–264. [Google Scholar] [CrossRef] [PubMed]
- Münch, J.; Ständker, L.; Adermann, K.; Schulz, A.; Schindler, M.; Chinnadurai, R.; Pöhlmann, S.; Chaipan, C.; Biet, T.; Peters, T.; et al. Discovery and Optimization of a Natural HIV-1 Entry Inhibitor Targeting the Gp41 Fusion Peptide. Cell 2007, 129, 263–275. [Google Scholar] [CrossRef]
- Umetsu, Y.; Tenno, T.; Goda, N.; Shirakawa, M.; Ikegami, T.; Hiroaki, H. Structural Difference of Vasoactive Intestinal Peptide in Two Distinct Membrane-Mimicking Environments. Biochim. Biophys. Acta BBA—Proteins Proteom. 2011, 1814, 724–730. [Google Scholar] [CrossRef]
- Krebs, D.; Maroun, R.G.; Sourgen, F.; Troalen, F.; Davoust, D.; Fermandjian, S. Helical and Coiled-Coil-Forming Properties of Peptides Derived from and Inhibiting Human Immunodeficiency Virus Type 1 Integrase Assessed by 1H-NMR. Use of NH Temperature Coefficients to Probe Coiled-Coil Structures. Eur. J. Biochem. 1998, 253, 236–244. [Google Scholar] [CrossRef]
- Spriggs, M.K.; Olmsted, R.A.; Venkatesan, S.; Coligan, J.E.; Collins, P.L. Fusion Glycoprotein of Human Parainfluenza Virus Type 3: Nucleotide Sequence of the Gene, Direct Identification of the Cleavage-Activation Site, and Comparison with Other Paramyxoviruses. Virology 1986, 152, 241–251. [Google Scholar] [CrossRef]
- Conlon, J.M.; Demandt, A.; Nielsen, P.F.; Leprince, J.; Vaudry, H.; Woodhams, D.C. The Alyteserins: Two Families of Antimicrobial Peptides from the Skin Secretions of the Midwife Toad Alytes Obstetricans (Alytidae). Peptides 2009, 30, 1069–1073. [Google Scholar] [CrossRef] [PubMed]
- Conlon, J.M.; Mechkarska, M.; Arafat, K.; Attoub, S.; Sonnevend, A. Analogues of the Frog Skin Peptide Alyteserin-2a with Enhanced Antimicrobial Activities against Gram-Negative Bacteria: ALYTESERIN-2: STRUCTURE-ACTIVITY. J. Pept. Sci. 2012, 18, 270–275. [Google Scholar] [CrossRef]
- Muñoz-Camargo, C.; Salazar, V.; Barrero-Guevara, L.; Camargo, S.; Mosquera, A.; Groot, H.; Boix, E. Unveiling the Multifaceted Mechanisms of Antibacterial Activity of Buforin II and Frenatin 2.3S Peptides from Skin Micro-Organs of the Orinoco Lime Treefrog (Sphaenorhynchus Lacteus). Int. J. Mol. Sci. 2018, 19, 2170. [Google Scholar] [CrossRef]
- Verly, R.M.; Moraes, C.M.D.; Resende, J.M.; Aisenbrey, C.; Bemquerer, M.P.; Piló-Veloso, D.; Valente, A.P.; Almeida, F.C.L.; Bechinger, B. Structure and Membrane Interactions of the Antibiotic Peptide Dermadistinctin K by Multidimensional Solution and Oriented 15N and 31P Solid-State NMR Spectroscopy. Biophys. J. 2009, 96, 2194–2203. [Google Scholar] [CrossRef]
- Lequin, O.; Ladram, A.; Chabbert, L.; Bruston, F.; Convert, O.; Vanhoye, D.; Chassaing, G.; Nicolas, P.; Amiche, M. Dermaseptin S9, an α-Helical Antimicrobial Peptide with a Hydrophobic Core and Cationic Termini. Biochemistry 2006, 45, 468–480. [Google Scholar] [CrossRef]
- Jabeen, M.; Biswas, P.; Islam, M.T.; Paul, R. Antiviral Peptides in Antimicrobial Surface Coatings—From Current Techniques to Potential Applications. Viruses 2023, 15, 640. [Google Scholar] [CrossRef]
- Manavalan, B.; Basith, S.; Lee, G. Comparative Analysis of Machine Learning-Based Approaches for Identifying Therapeutic Peptides Targeting SARS-CoV-2. Brief. Bioinform. 2022, 23, bbab412. [Google Scholar] [CrossRef]
- Pang, Y.; Wang, Z.; Jhong, J.-H.; Lee, T.-Y. Identifying Anti-Coronavirus Peptides by Incorporating Different Negative Datasets and Imbalanced Learning Strategies. Brief. Bioinform. 2021, 22, 1085–1095. [Google Scholar] [CrossRef] [PubMed]
- Timmons, P.B.; Hewage, C.M. ENNAVIA Is a Novel Method Which Employs Neural Networks for Antiviral and Anti-Coronavirus Activity Prediction for Therapeutic Peptides. Brief. Bioinform. 2021, 22, bbab258. [Google Scholar] [CrossRef]
- Singh, S.; Chauhan, P.; Sharma, V.; Rao, A.; Kumbhar, B.V.; Prajapati, V.K. Identification of Multi-Targeting Natural Antiviral Peptides to Impede SARS-CoV-2 Infection. Struct. Chem. 2022; online ahead of print. [Google Scholar] [CrossRef]
- Moretta, A.; Salvia, R.; Scieuzo, C.; Di Somma, A.; Vogel, H.; Pucci, P.; Sgambato, A.; Wolff, M.; Falabella, P. A Bioinformatic Study of Antimicrobial Peptides Identified in the Black Soldier Fly (BSF) Hermetia Illucens (Diptera: Stratiomyidae). Sci. Rep. 2020, 10, 16875. [Google Scholar] [CrossRef]
- Tucs, A.; Tran, D.P.; Yumoto, A.; Ito, Y.; Uzawa, T.; Tsuda, K. Generating Ampicillin-Level Antimicrobial Peptides with Activity-Aware Generative Adversarial Networks. ACS Omega 2020, 5, 22847–22851. [Google Scholar] [CrossRef]
- Kanehisa, M. The KEGG Databases at GenomeNet. Nucleic Acids Res. 2002, 30, 42–46. [Google Scholar] [CrossRef] [PubMed]
- Grant, C.E.; Bailey, T.L.; Noble, W.S. FIMO: Scanning for Occurrences of a given Motif. Bioinformatics 2011, 27, 1017–1018. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).