Advancing Skin Cancer Prediction Using Ensemble Models
Abstract
1. Introduction
- This study’s primary goal is to examine the unique application of the Max Voting ensemble approach in order to enhance the accuracy and reliability of skin cancer lesion classifications.
- The proposed approach shows improved accuracy and robustness by combining the strengths of many pre-trained ML models. These models include Random Forest, Gradient Boosting, AdaBoost, CatBoost, and Extra Trees.
- A genetic algorithm (GA) generated the best feature vectors from a set of images. More complex ensemble learning classification techniques utilize these vectors. We assess these models using several measures, including accuracy, F1-score, recall, and precision.
2. Literature Review
- Diversity and Larger Datasets: To ensure robustness and generalizability, we will train and test the proposed model on a variety of datasets, including real-world clinical data.
- Multi-class Classification: The model will identify skin lesions other than melanoma and non-melanoma.
- Hybrid Model Architecture: The proposed model combines the strengths of different ML techniques, such as GA for feature extraction and ensemble learning for accuracy, to increase performance.
- Standardized Evaluation Metrics: The suggested methodology would use standardized procedures and metrics to make the study comparisons more uniform and meaningful.
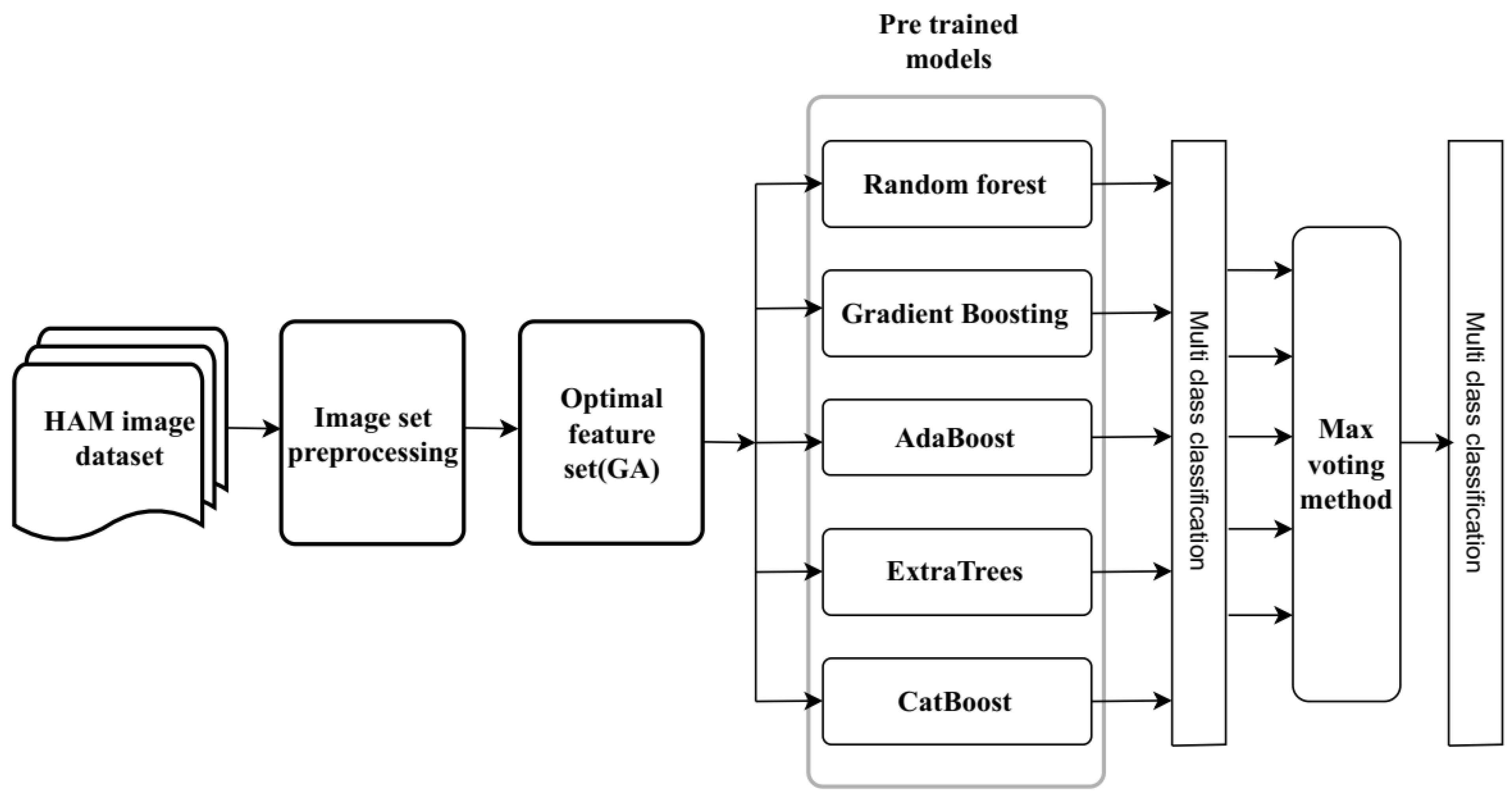
3. Proposed Method
3.1. Abstract Model View of the Proposed Method
- 1.
- Selecting a Proper Dataset: Choose an extensive dataset of skin lesion images, which includes seven different types of skin cancer lesions. For this investigation, we used the HAM10000 Skin Cancer Dataset [19].
- 2.
- Image Preprocessing: The following stage, picture preprocessing, involves enhancing the quality and analytical applicability of raw images through various transformations.
- 3.
- Optimal Feature Set Selection: Using a genetic algorithm (GA), the optimal feature set selection of an image set is produced.
- 4.
- Pre-trained Ensemble Models: Select a variety of pre-trained ensemble learning models for multi-class skin cancer classification, including Random Forest (RF), Gradient Boosting (GB), AdaBoost (AB), CatBoost (CB), and Extra Trees (ET).
- 5.
- Max Voting Method: Use the Maximum Voting technique to aggregate the predictions from each model separately. According to the majority vote taken by the predictions produced by the different models, this method produces predictions for each image.
- 6.
- Performance Evaluation: Evaluate the performance of the ensemble models on the test set by utilizing metrics such as confusion matrices, F1-score, accuracy, precision, and recall.
3.2. Comprehensive View of the Suggested Model
- Cross-validation is a crucial strategy for reducing overfitting. K-fold cross-validation allows us to guarantee that the model’s performance does not change when it is used on different data subsets.
- Using regularization methods like L1 and L2 can help to reduce the risk of overfitting and minimize a model’s complexity. By including a regularization term in the loss function, these techniques encourage the model to have lower weights and reduce the impact of unimportant features.
- One other method to reduce overfitting is early stopping. Using this method, the model’s performance on a validation set is tracked during training and the training process is stopped when the performance stops improving. This helps keep the model from overly optimizing the training data.
- Tree-based models like Random Forest and Gradient Boosting can be pruned to remove irrelevant components that do not really affect the broad generalization of the data. This lessens the model’s complexity and enhances its capacity for predicting new data.
- Ensemble techniques, which combine the predictions of several models, can help to reduce the variance related to individual models, hence reducing overfitting. Because ensemble methods combine the findings of several models, they improve the accuracy and resilience of predictions.
| Algorithm 1 Proposed Method for Skin Cancer Lesions Classification |
Input: Select pre-trained ensemble models (selectedensembleModels) Select skin lesion images (input Images) Equivalent Target labels (TargetLabels) Output: Each individual input image is predicted by an ensemble model (ensemble predictions) |
Step 1: Ensemble Model selection selected models ⟸ [Random Forest, Gradient Boosting, AdaBoost, CatBoost, Extra Trees] |
Step 2: Image preprocessing while InputImages ≠ eachImage do Preprocessed Image ⟸ [Resizing and Cropping, Rescale, Normalization, Color Correction, Noise Reduction] |
Step 3: Optimal feature extraction of Images Optimal feature extraction of an Image ⟸ [Genetic algorithm] |
Step 4: Load Ensemble Pre-trained Models Loaded Models ⟸ load Models (selected Models) |
Step 5: Generate Predictions while eachloadedmodel ≠ LoadedModels do ModelPrediction ⟸ PredictImage (LoadedModel, PreprocessedImage) |
Step 6: Max Voting method while eachImagePrediction ≠ setof predictions do MajorityVote ⟸ calculateMajorityVote (imagePrediction) |
Step 7: Evaluate Models’ Performance Accuracy ⟸ evaluateAccuracy(ensemblePredictions, TargetLabels) |
3.3. DATASET
3.4. Image Preprocessing
- Resizing: Resizing refers to altering the size of an image by modifying its dimensions. Within this particular context, the initial images of skin cancer are resized to a consistent dimension of 256 × 256 pixels. Ensuring uniformity in the image dimensions is essential for maintaining consistency in the analysis and processing procedures. It guarantees uniform dimensions for all images, hence simplifying computational tasks like feature extraction and model training.
- Color Space Conversion: Converting images to grayscale improves the study by eliminating color information and concentrating exclusively on the light’s intensity. Since this conversion enhances the visibility of features linked to texture and shape in skin lesions, it is particularly useful in medical image analysis. Images in grayscale only exist in levels of gray, where the values of the individual pixels represent the intensity of the light, from black to white.
- Contrast Enhancement: Contrast enhancement refers to the application of techniques that increase the intensity difference between different regions in an image in order to improve the visibility of its characteristics. One often-used technique for increasing contrast is histogram equalization. It changes how pixels’ intensity values are arranged in an image, extending the range of intensities to include the whole grayscale range. The contrast in the image is much increased by this technique, which makes the intricacies inside the skin lesions more visible than the background.
- Denoising: Unwanted noise, which can originate from a variety of causes, including ambient conditions or sensor errors, is removed from an image by a technique called denoising. Preserving the integrity of lesion boundaries and raising the accuracy of later analysis depend on noise reduction in skin cancer images. One often-used method for reducing noise in images is the median filter. The procedure is replacing the median value of the surrounding pixels for every pixel in the image. This filter works well to improve contrast-enhanced images’ clarity by reducing noise while maintaining edge details.
- Edge Detection: A key stage in image processing, edge detection is used especially to find object boundaries. The diagnosis of skin cancer depends on precise definition of the boundaries of lesions. One often used technique for detecting major changes in intensity in an image to identify edges is the Canny edge detector. The way this system works is by image smoothing, edge magnitude and direction computation using gradients, and edge detection and connection using hysteresis thresholding.
- Data Augmentation: A method known as data augmentation modifies current images to deliberately expand and diversify a collection. Among the many possibilities for these adjustments are rotation, scaling, flipping, and cropping. The analysis of skin cancer images requires data augmentation since it improves the flexibility and applicability of machine learning models to a variety of situations. The model enhances its ability to identify a wide range of patterns and features by producing more variants of skin lesion images, which enhances its performance on unknown data.
3.5. Feature Extraction
- Color histograms: They show the lesion’s color distribution by calculating the proportion of pixels that belong to each color bin. Histograms can be generated for each channel in RGB, HSV, or other color spaces to capture the overall color distribution.
- Color Moments:
- −
- Mean (First Moment): Displays the dominant color and the average color intensity for each channel during the lesion.
- −
- Standard Deviation (Second Moment): Calculates color variability and displays the degree of uniformity in the color distribution.
- −
- Skewness (Third Moment): Determines how symmetrically the color distribution is, which may point to the existence of uneven pigmentation.
- 1.
- Initialization: Start with a zero matrix P of size .
- 2.
- Pixel Pair Counting: For each pixel in the image, consider another pixel at distance b in direction c. If the intensity of the first pixel is u and the second pixel is v, increment by 1.
- 3.
- Normalization: After counting all relevant pixel pairs, normalize the matrix so that the sum of its elements equals 1. This converts frequency counts into probabilities.
- 1.
- Contrast measures the local variations in the GLCM.
- 2.
- Energy (or Angular Second Moment) reflects the uniformity of the GLCM.
- 3.
- Correlation indicates how correlated a pixel is to its neighbor over the entire image.Here, and are the means and and are the standard deviations of the row and column sums of the GLCM, respectively.
- 4.
- Homogeneity (or Inverse Difference Moment) shows the closeness of the distribution of elements in the GLCM to the GLCM diagonal.
- 5.
- Entropy measures the randomness in the GLCM.
3.6. Feature Selection Optimization
- Step 1: Define the GA Components:
- −
- Chromosome Representation: In feature selection tasks, chromosomes are usually shown using a binary string, where each bit in the string represents a different feature from the set of features, where the value of each position indicates whether a particular feature is selected (1) or not selected (0).
- −
- Fitness Function: By utilizing this fitness function, the GA is directed to find the best subset of features that both simplify the model and achieve high classification accuracy.
- Step 2: Initialize Population: Start with a randomly generated population of chromosomes.
- Step 3: GA Operations:
- −
- Selection: To choose parent chromosomes for breeding, use tournament selection.
- −
- Crossover: To produce offspring, use a single-point crossover. For instance, the first 10 features of one parent are combined with the remaining features of the other parent if the crossover point is at position 10.
- −
- Mutation: Introduce variety into the offspring by applying mutation to them with a given probability (mutation rate).
- −
- Replacement: Incorporate the new offspring into the population to replace the least-perfect chromosomes.
- Step 4: Iterate: Continue selecting, crossover, mutating, and replacing until the fitness score no longer increases noticeably, or for a predetermined number of generations.
- Step 5: Evaluate the Best Solution: Select the best-performing chromosome based on the optimization function result. This chromosome stands for the ideal selection of characteristics.
- quantifies the predictive accuracy or effectiveness of the classifier when using the subset of features selected by X. This could be measured through accuracy, F1-score, or area under the ROC curve (AUC).
- reflects the complexity of the model, often represented by the number of features selected (i.e., the number of 1s in X). The rationale is to penalize solutions that use more features than necessary to prevent overfitting and ensure model simplicity.
- and are weighting parameters that balance model accuracy and complexity.
- is the optimal subset of features that we aim to find.
- X represents a candidate solution, which is a subset of the available features in the dataset.
- is the fitness function that quantifies the quality of the solution X.
3.7. Ensemble Models
- Bagging (Bootstrap Aggregating): Bootstrapping [29] is the process of training a model many times using different replacement-sampled portions of the training data. The ultimate forecast is acquired by classifying the forecasts of individual models through voting. A well-liked technique called Random Forest employs bagging, with decision trees serving as the foundational models.
- Boosting: The purpose of boosting [30] is to make underperforming models more accurate by assigning greater importance to instances that were incorrectly classified. To fix the misclassifications, a new weak model is trained for each iteration. The sum of the weighted predictions made by each separate model is the final prediction. Gradient boosting, AdaBoost, and CatBoost are a few examples.
3.7.1. Random Forest (RF)
3.7.2. Gradient Boosting
3.7.3. AdaBoost
3.7.4. CatBoost
3.7.5. Extra Trees
3.8. Max Voting Mechanism
- D: The final decision or prediction made by the Max Voting algorithm.
- argmax: A function that finds the argument j that maximizes the given expression.
- j: An index representing the class labels .
- k: Number of unique classes.
- ∑: The symbol that sums all N models’ votes.
- N: The total number of models in the ensemble.
- : The indicator function, which returns 1 if the condition inside is true (i.e., if model i predicts class ) and 0 otherwise.
- : The prediction made by model i for the input sample.
- : The jth class label among the possible class labels.
4. Simulation Results and Discussion
4.1. Experimental Setup
4.2. Features analysis
4.3. Evaluation Parameters
- Confusion Matrix: The table provides a concise overview of the classification model’s performance. It aids in comprehending the model’s performance across several classes by providing a thorough breakdown of the FN, FP, TP, and TN.
- Accuracy: This performance metric, which is the most logical one, is just the proportion of correctly predicted observations to the total number of observations. When the target classes are evenly distributed, it is helpful.
- Precision: Precision is the proportion of TP predictions out of all the positive predictions generated by the method. It aids in analyzing the model’s ability to detect FPs.
- Recall (Sensitivity): A dataset’s true positive cases corresponding to true positive predictions are determined by the recall function. It facilitates the comprehension of the model’s capacity to identify every positive case.
- F1-score: The harmonic mean of precision and recall is known as the F1-score. Because it attains high values for both precision and recall, a high F1-score is an excellent indicator of your model’s performance. For classes that are unbalanced, this metric is preferable to accuracy.
4.4. Error Analysis and Confusion Matrix (CM)
- Diagonal Dominance: In every confusion matrix, the diagonal elements have greater values, indicating a majority number of accurate classifications.
- Misclassification Patterns: The off-diagonal elements of the matrix indicate distinct tendencies of misclassification that differ between the models. For example, certain models may more commonly misidentify specific types of lesions.
- Performance Improvement: The Max Voting method exhibits superior performance by minimizing misclassifications, hence highlighting the efficacy of ensemble techniques in enhancing classification accuracy.
4.5. Performance of Multiple Models on the HAM10000 Dataset
4.6. Performance of Multiple Models on the ISIC 2018 Dataset
4.7. Comparison of State-of-the-Art Models vs. Proposed Max Voting Model
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Apalla, Z.; Nashan, D.; Weller, R.B.; Castellsagué, X. Skin cancer: Epidemiology, disease burden, pathophysiology, diagnosis, and therapeutic approaches. Dermatol. Ther. 2017, 7, 5–19. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Fang, L.; Ni, R.; Zhang, H.; Pan, G. Changing trends in the disease burden of non-melanoma skin cancer globally from 1990 to 2019 and its predicted level in 25 years. BMC Cancer 2022, 22, 836. [Google Scholar] [CrossRef] [PubMed]
- Zelin, E.; Zalaudek, I.; Agozzino, M.; Dianzani, C.; Dri, A.; Di Meo, N.; Giuffrida, R.; Marangi, G.F.; Neagu, N.; Persichetti, P.; et al. Neoadjuvant therapy for non-melanoma skin cancer: Updated therapeutic approaches for basal, squamous, and merkel cell carcinoma. Curr. Treat. Options Oncol. 2021, 22, 35. [Google Scholar] [CrossRef] [PubMed]
- Magnus, K. The Nordic profile of skin cancer incidence. A comparative epidemiological study of the three main types of skin cancer. Int. J. Cancer 1991, 47, 12–19. [Google Scholar] [CrossRef]
- Bhatt, H.; Shah, V.; Shah, K.; Shah, R.; Shah, M. State-of-the-art machine learning techniques for melanoma skin cancer detection and classification: A comprehensive review. Intell. Med. 2023, 3, 180–190. [Google Scholar] [CrossRef]
- Goyal, M.; Knackstedt, T.; Yan, S.; Hassanpour, S. Artificial intelligence-based image classification methods for diagnosis of skin cancer: Challenges and opportunities. Comput. Biol. Med. 2020, 127, 104065. [Google Scholar] [CrossRef] [PubMed]
- Raval, D.; Undavia, J.N. A Comprehensive assessment of Convolutional Neural Networks for skin and oral cancer detection using medical images. Healthc. Anal. 2023, 3, 100199. [Google Scholar] [CrossRef]
- Iqbal, S.N.; Qureshi, A.; Li, J.; Mahmood, T. On the analyses of medical images using traditional machine learning techniques and convolutional neural networks. Arch. Comput. Methods Eng. 2023, 30, 3173–3233. [Google Scholar] [CrossRef] [PubMed]
- Elgamal, M. Automatic skin cancer images classification. Int. J. Adv. Comput. Sci. Appl. 2013, 4, 287–294. [Google Scholar] [CrossRef]
- Kanca, E.; Ayas, S. Learning Hand-Crafted Features for K-NN based Skin Disease Classification. In Proceedings of the International Congress on Human-Computer Interaction, Optimization and Robotic Applications (HORA), Ankara, Turkey, 9–11 June 2022; pp. 1–4. [Google Scholar]
- Bansal, P.; Vanjani, A.; Mehta, A.; Kavitha, J.C.; Kumar, S. Improving the classification accuracy of melanoma detection by performing feature selection using binary Harris hawks optimization algorithm. Soft Comput. 2022, 26, 8163–8181. [Google Scholar] [CrossRef]
- Moradi, N.; Mahdavi-Amiri, N. Kernel sparse representation based model for skin lesions segmentation and classification. Comput. Methods Programs Bio Med. 2019, 182, 105038. [Google Scholar] [CrossRef] [PubMed]
- Balaji, V.R.; Suganthi, S.T.; Rajadevi, R.; Krishna Kumar, V.; Saravana Balaji, B.; Pandiyan, S. Skin disease detection and segmentation using dynamic graph cut algorithm and classification through Naive Bayes classifier. Measurement 2020, 163, 107922. [Google Scholar] [CrossRef]
- Brinker, T.J.; Hekler, A.; Utikal, J.S.; Grabe, N.; Schadendorf, D.; Klode, J.; Berking, C.; Steeb, T.; Enk, A.H.; Von Kalle, C. Skin cancer classification using convolutional neural networks: Systematic review. J. Med. Internet Res. 2018, 20, e11936. [Google Scholar] [CrossRef] [PubMed]
- Shokouhifar, A.; Shokouhifar, M.; Sabbaghian, M.; Soltanian-Zadeh, H. Swarm intelligence empowered three-stage ensemble deep learning for arm volume measurement in patients with lymphedema. Biomed. Signal Process. Control 2023, 85, 105027. [Google Scholar] [CrossRef]
- Bao, J.; Hou, Y.; Qin, L.; Zhi, R.; Wang, X.-M.; Shi, H.-B.; Sun, H.-Z.; Hu, C.-H.; Zhang, Y.-D. High-throughput precision MRI assessment with integrated stack-ensemble deep learning can enhance the preoperative prediction of prostate cancer Gleason grade. Br. J. Cancer 2023, 128, 1267–1277. [Google Scholar] [CrossRef] [PubMed]
- Guergueb, T.; Akhloufi, M.A. Skin Cancer Detection using Ensemble Learning and Grouping of Deep Models. In Proceedings of the 19th International Conference on Content-Based Multimedia Indexing, Graz, Austria, 14–16 September 2022; pp. 121–125. [Google Scholar]
- Avanija, J.; Reddy, C.C.; Reddy, C.S.; Reddy, D.H.; Narasimhulu, T.; Hardhik, N.V. Skin Cancer Detection using Ensemble Learning. In Proceedings of the International Conference on Sustainable Computing and Smart Systems (ICSCSS), Coimbatore, India, 14–16 June 2023; IEEE: Piscataway, NJ, USA; pp. 184–189. [Google Scholar]
- Tschandl, P.; Rosendahl, C.; Kittler, H. The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci. Data 2018, 5, 180161. [Google Scholar] [CrossRef] [PubMed]
- Jain, S.; Singhania, U.; Tripathy, B.; Nasr, E.A.; Aboudaif, M.K.; Kamrani, A.K. Deep learning-based transfer learning for classification of skin cancer. Sensors 2021, 21, 8142. [Google Scholar] [CrossRef]
- Shorten, C.; Khoshgoftaar, T.M. A survey on image data augmentation for deep learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Ibraheem, N.A.; Hasan, M.M.; Khan, R.Z.; Mishra, P.K. Understanding color models: A review. ARPN J. Sci. Technol. 2012, 2, 265–275. [Google Scholar]
- Sharma, B.; Rupali Nayyer, R. Use and analysis of color models in image processing. J. Food Process. Technol. 2016, 7, 533. [Google Scholar] [CrossRef]
- Nguyen, T.P.; Vu, N.-S.; Manzanera, A. Statistical binary patterns for rotational invariant texture classification. Neurocomputing 2016, 173, 1565–1577. [Google Scholar] [CrossRef]
- Song, T.; Feng, J.; Wang, S.; Xie, Y. Spatially weighted order binary pattern for color texture classification. Expert Syst. Appl. 2020, 147, 113167. [Google Scholar] [CrossRef]
- Liu, L.; Chen, J.; Fieguth, P.; Zhao, G.; Chellappa, R.; Pietikäinen, M. From BoW to CNN: Two decades of texture representation for texture classification. Int. J. Comput. Vis. 2019, 127, 74–109. [Google Scholar] [CrossRef]
- Hong, H.; Zheng, L.; Pan, S. Computation of Gray Level Co-Occurrence Matrix Based on CUDA and Optimization for Medical Computer Vision Application. IEEE Access 2018, 6, 67762–67770. [Google Scholar] [CrossRef]
- Shukla, A.K. Simultaneously feature selection and parameters optimization by teaching–learning and genetic algorithms for diagnosis of breast cancer. Int. J. Data Sci. Anal. 2024, 1–22. [Google Scholar] [CrossRef]
- Ngo, G.; Beard, R.; Chandra, R. Evolutionary bagging for ensemble learning. Neurocomputing 2022, 510, 1–14. [Google Scholar] [CrossRef]
- Chen, W.; Lei, X.; Chakrabortty, R.; Chandra Pal, S.; Sahana, M.; Janizadeh, S. Evaluation of different boosting ensemble machine learning models and novel deep learning and boosting framework for head-cut gully erosion susceptibility. J. Environ. Manag. 2021, 284, 112015. [Google Scholar] [CrossRef]
- Lin, W.; Wu, Z.; Lin, L.; Wen, A.; Li, J. An ensemble random forest algorithm for insurance big data analysis. IEEE Access 2017, 5, 16568–16575. [Google Scholar] [CrossRef]
- Bentéjac, C.; Csörgő, A.; Martínez-Muñoz, G. A comparative analysis of gradient boosting algorithms. Artif. Intell. Rev. 2021, 54, 1937–1967. [Google Scholar] [CrossRef]
- Mienye, I.D.; Yanxia Sun, Y. A survey of ensemble learning: Concepts, algorithms, applications, and prospects. IEEE Access 2022, 10, 99129–99149. [Google Scholar] [CrossRef]
- Hancock, J.T.; Khoshgoftaar, T.M. CatBoost for big data: An interdisciplinary review. J. Big Data 2020, 7, 94. [Google Scholar] [CrossRef]
- Sharma, D.; Kumar, R.; Jain, A. Breast cancer prediction based on neural networks and extra tree classifier using feature ensemble learning. Meas. Sens. 2022, 24, 100560. [Google Scholar] [CrossRef]
- Dogan, A.; Birant, D. A weighted majority voting ensemble approach for classification. In Proceedings of the 2019 4th International Conference on Computer Science and Engineering (UBMK), Samsun, Turkey, 11–15 September 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 1–6. [Google Scholar]
- Backes, A.R.; Casanova, D.; Bruno, O.M. Color texture analysis based on fractal descriptors. Pattern Recognit. 2012, 45, 1984–1992. [Google Scholar] [CrossRef]
- Bianconi, F.; Harvey, R.; Southam, P.; Fernández, A. Theoretical and experimental comparison of different approaches for color texture classification. J. Electron. Imaging 2011, 20, 043006. [Google Scholar] [CrossRef]
- Palm, C. Color texture classification by integrative co-occurrence matrices. Pattern Recognit. 2004, 37, 965–976. [Google Scholar] [CrossRef]
- Humeau-Heurtier, A. Texture feature extraction methods: A survey. IEEE Access 2019, 7, 8975–9000. [Google Scholar] [CrossRef]
- Liu, L.; Fieguth, P.; Guo, Y.; Wang, X.; Pietikäinen, M. Local binary features for texture classification: Taxonomy and experimental study. Pattern Recognit. 2017, 62, 135–160. [Google Scholar] [CrossRef]
- Qi, X.; Zhao, G.; Shen, L.; Li, Q.; Pietikäinen, M. LOAD: Local orientation adaptive descriptor for texture and material classification. Neurocomputing 2016, 184, 28–35. [Google Scholar] [CrossRef]
- Tohka, J.; Van Gils, M. Evaluation of machine learning algorithms for health and wellness applications: A tutorial. Comput. Biol. Med. 2021, 132, 104324. [Google Scholar] [CrossRef]
- Heydarian, M.; Doyle, T.E.; Samavi, R. MLCM: Multi-label confusion matrix. IEEE Access 2022, 10, 19083–19095. [Google Scholar] [CrossRef]
- Gouda, W.; Sama, N.U.; Al-waakid, G.; Humayun, M.; Jhanjhi, N.Z. Detection of Skin-Cancer Based on Skin Lesion Images Using Deep Learning. Healthcare 2022, 10, 1183. [Google Scholar] [CrossRef]
- Khan, M.A.; Muhammad, K.; Sharif, M.; Akram, T.; Kadry, S. Intelligent fusion-assisted skin lesion localization and classification for smart healthcare. Neural Comput. Appl. 2024, 36, 37–52. [Google Scholar] [CrossRef]
- Hossain, M.M.; Hossain, M.M.; Arefin, M.B.; Akhtar, F.; Blake, J. Combining State-of-the-Art Pre-Trained Deep Learning Models: A Noble Approach for Skin Cancer Detection Using Max Voting Ensemble. Diagnostics 2023, 14, 89. [Google Scholar] [CrossRef] [PubMed]




| Study | ML Technique | Classification Task | Accuracy (%) | Demerits |
|---|---|---|---|---|
| [10] | KNN | Melanoma vs. Seborrhoeic Nevi-keratoses | 85 | Sensitive to distance metric and k value. |
| [11] | SVM | Melanoma vs. Non-Melanoma | 90 | Requires careful tuning, computationally expensive. |
| [12] | SKR | Multi-class, Binary (Melanoma/Normal). | 87 (Multi-class), 92 (Binary) | Complex to implement |
| [13] | Naive Bayes | Skin Disease Classification | 80 | Assumes feature independence, may limit performance. |
| [14] | CNNs | Systematic Review | Up to 95 | Requires large labeled datasets, computationally intensive. |
| [15,16] | Ensemble Learning | Biological Imaging | Not specified | Complexity in implementation and interpretation. |
| [17] | Ensemble Learning | Melanoma Prediction | 97 | Single dataset reliance, generalizability concerns. |
| [18] | VGG16, ResUNet, CapsNet | ISIC Skin Cancer Dataset | 86 | Complex ensemble approach, interpretability challenges. |
| Name of Lesion | Total Images (10,015) | Distribution after Data Augmentation |
|---|---|---|
| Actinic Keratosis | 327 | 3235 |
| Basal cell Carcinoma | 514 | 3283 |
| Benign keratosis | 1099 | 3170 |
| Dermatofibroma | 155 | 3252 |
| Melanocytic nevi | 6705 | 3179 |
| Melanoma | 1113 | 3195 |
| Vascular Lesion | 142 | 3189 |
| Key Parameters of GA | Values |
|---|---|
| Population | 30 chromosomes |
| Fitness Function | Support Vector Machine (SVM) |
| Tournament Selection | size of 3 |
| Crossover | crossover rate of 0.7 |
| Mutation | Bit flip mutation rate of 0.01 |
| Termination | stops early if the accuracy exceeds 95%. |
| Key Parameters of RF | Values |
|---|---|
| Number of Trees | 60 |
| Criterion | gini |
| Maximum Depth | 30 |
| Minimum Samples Split | 2 |
| Minimum Samples Leaf | 5 |
| Bootstrap | True |
| Key Parameters of GB | Values |
|---|---|
| Loss Function | Multinomial logistic loss |
| Number of Estimators | 60 trees |
| Learning Rate | 0.1 |
| Maximum depth | 15 |
| Minimum Samples Split | 2 |
| Minimum Samples Leaf | 5 |
| Sub-sample | 0.8 |
| Key Parameters of AB | Values |
|---|---|
| Base Estimator | decision tree with depth = 1 |
| Number of Estimators | 50 weak learners |
| Learning Rate | 0.1 |
| Algorithm | SAMME.R |
| Random State | random state = 42 |
| Key Parameters of CB | Values |
|---|---|
| Number of Trees | 70 |
| Number of Estimators | 50 weak learners |
| Learning Rate | 0.1 |
| Loss Function | cross-entropy loss |
| Evaluation Metric | F1-score |
| Boosting _ type | Ordered |
| auto_class_weights | Balanced |
| l2_leaf_reg to a value | 3 |
| Key Parameters of ET | Values |
|---|---|
| Number of Estimators | 50 trees |
| Criterion | gini |
| Minimum Samples Split | 4 |
| Max Features | auto |
| Min Samples Leaf | 2 |
| Bootstrap | False |
| number of jobs | n_jobs = −1 |
| Key Parameters of MV | Values |
|---|---|
| Individual Classifiers | RF, GB, AB, CB, ET |
| Voting | Majority-Weighted votes |
| random_state | 42 |
| Key Components of Hardware | Specifications |
|---|---|
| NVIDIA Windows Driver | 546.12 |
| NVIDIA GPU | GeForce Titan Xp |
| CPU Chip | Intel core i7 |
| CPU Cores | 4 |
| Chip Speed | 3.2 GHz |
| CPU RAM | 12 GB |
| Disk Space | 320 GB |
| Iteration | Accuracy (%) | Precision (%) | Recall (%) | F1-Score (%) |
|---|---|---|---|---|
| Iteration1 | 92.03 | 91.37 | 91.70 | 98.55 |
| Iteration2 | 97.48 | 90.63 | 93.93 | 91.37 |
| Iteration3 | 93.10 | 97.83 | 95.41 | 94.12 |
| Iteration4 | 96.73 | 98.01 | 97.37 | 93.54 |
| Iteration5 | 97.79 | 96.73 | 97.26 | 96.24 |
| Iteration6 | 93.50 | 91.78 | 93.39 | 92.45 |
| Iteration7 | 92.60 | 90.08 | 92.59 | 93.66 |
| Iteration8 | 94.90 | 92.46 | 91.69 | 95.87 |
| Iteration9 | 94.90 | 95.48 | 97.39 | 91.22 |
| Iteration10 | 96.08 | 90.74 | 93.33 | 92.65 |
| Average | 95.80 | 95.04 | 95.44 | 95.20 |
| Model | Accuracy (%) | Recall (%) | Precision (%) | F1-Score (%) | Key Features |
|---|---|---|---|---|---|
| Random Forest | 82.41 | 79.53 | 78.35 | 78.77 | Trees are trained independently. |
| Gradient Boosting | 86.20 | 73.74 | 84.06 | 84.21 | Trees are trained sequentially. |
| AdaBoost | 90.64 | 88.88 | 89.64 | 89.14 | Automatically handles missing values |
| Extra Trees | 91.12 | 90.23 | 90.82 | 90.43 | Uses the whole learning sample to construct trees |
| CatBoost | 93.06 | 92.46 | 93.71 | 92.93 | Specialized in dealing with categorical data |
| Max Voting | 95.80 | 95.04 | 95.44 | 95.20 | Combine the predictions from multiple models |
| Class | Avg. Precision (%) | Avg. Recall (%) | Avg. F1-Score (%) |
|---|---|---|---|
| Class 0 | 96.08 | 90.74 | 93.33 |
| Class 1 | 92.03 | 91.37 | 91.70 |
| Class 2 | 97.48 | 90.63 | 93.93 |
| Class 3 | 93.10 | 97.83 | 95.41 |
| Class 4 | 96.73 | 98.01 | 97.37 |
| Class 5 | 97.79 | 96.73 | 97.26 |
| Class 6 | 94.90 | 99.98 | 97.39 |
| Model | Accuracy (%) | Recall (%) | Precision (%) | F1-Score (%) |
|---|---|---|---|---|
| Random Forest | 84.50 | 82.30 | 81.20 | 81.74 |
| Gradient Boosting | 88.70 | 76.50 | 87.90 | 87.20 |
| AdaBoost | 92.10 | 90.50 | 91.30 | 90.90 |
| Extra Trees | 92.80 | 91.70 | 92.50 | 92.10 |
| CatBoost | 94.30 | 93.80 | 94.90 | 94.20 |
| Max Voting | 96.20 | 95.50 | 96.30 | 95.63 |
| Class | Avg. Precision (%) | Avg. Recall (%) | Avg. F1-Score (%) |
|---|---|---|---|
| Class 0 | 97.10 | 91.20 | 94.10 |
| Class 1 | 93.20 | 92.50 | 92.80 |
| Class 2 | 96.30 | 91.70 | 94.80 |
| Class 3 | 94.00 | 98.50 | 96.20 |
| Class 4 | 97.50 | 98.70 | 98.10 |
| Class 5 | 98.20 | 97.10 | 97.65 |
| Class 6 | 95.80 | 96.00 | 97.36 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Natha, P.; RajaRajeswari, P. Advancing Skin Cancer Prediction Using Ensemble Models. Computers 2024, 13, 157. https://doi.org/10.3390/computers13070157
Natha P, RajaRajeswari P. Advancing Skin Cancer Prediction Using Ensemble Models. Computers. 2024; 13(7):157. https://doi.org/10.3390/computers13070157
Chicago/Turabian StyleNatha, Priya, and Pothuraju RajaRajeswari. 2024. "Advancing Skin Cancer Prediction Using Ensemble Models" Computers 13, no. 7: 157. https://doi.org/10.3390/computers13070157
APA StyleNatha, P., & RajaRajeswari, P. (2024). Advancing Skin Cancer Prediction Using Ensemble Models. Computers, 13(7), 157. https://doi.org/10.3390/computers13070157





