Abstract
The emergence of the novel coronavirus in Wuhan, China since 2019, has put the world in an exotic state of emergency and affected millions of lives. It is five times more deadly than Influenza and causes significant morbidity and mortality. COVID-19 mainly affects the pulmonary system leading to respiratory disorders. However, earlier studies indicated that COVID-19 infection may cause cardiovascular diseases, which can be detected using an electrocardiogram (ECG). This work introduces an advanced deep learning architecture for the automatic detection of COVID-19 and heart diseases from ECG images. In particular, a hybrid combination of the EfficientNet-B0 CNN model and Vision Transformer is adopted in the proposed architecture. To our knowledge, this study is the first research endeavor to investigate the potential of the vision transformer model to identify COVID-19 in ECG data. We carry out two classification schemes, a binary classification to identify COVID-19 cases, and a multi-class classification, to differentiate COVID-19 cases from normal cases and other cardiovascular diseases. The proposed method surpasses existing state-of-the-art approaches, demonstrating an accuracy of 100% and 95.10% for binary and multiclass levels, respectively. These results prove that artificial intelligence can potentially be used to detect cardiovascular anomalies caused by COVID-19, which may help clinicians overcome the limitations of traditional diagnosis.
1. Introduction
In recent years, the world has been affected by the COVID-19 outbreak. This perilous virus has affected millions of lives []. COVID-19 is now known to affect all major systems in the body []. The most reliable diagnostic technique is the RT-PCR, but this test remains scarce and costly in developing countries and rural areas. In addition, it requires extended waiting periods of at least 6 h, the expertise of qualified personnel, and the requirement for a logistically centralized installation. More effective methods are therefore required to improve the rapidity and effectiveness of the diagnosis.
The literature has shown that medical imaging, involving both radiography X-ray and computed tomography offers information that can be useful for COVID-19 diagnosis [,,]. However, these techniques are very expensive and require the interpretation of radiographs by expert radiologists, as COVID-19 patterns present a close similarity to other viral and bacterial pneumonia on chest radiographs, making diagnosis difficult. Also, medical imaging presents certain challenges as it involves transferring the patient to the appropriate room, which requires careful cleaning of the machines every time they are used and entails an increased risk of radiation exposure. Hence, novel approaches are needed to complement the COVID-19 diagnosis while the outbreak persists. This perilous virus mainly affects either the superior respiratory system (including sinuses, nose, and throat) or the inferior respiratory system (involving the windpipe and lungs), but some clinical research has also shown a potential relation between COVID-19 and cardiac disorders. It was found that COVID-19 causes severe damage to the cardiovascular system and it may be the cause of many heart problems, including myocardial injury (MI) [], arrhythmias, acute myocarditis [], and venous thromboembolism []. These abnormalities can be identified using electrocardiogram (ECG). The purpose of electrocardiography is to measure the electric activity of the heart []. Observation and analysis of those activities are important steps in diagnosing the heart rate reflected in ECG sequences. Each sequence includes five waves: P, Q, R, S, and T representing the phases of cardiac activity. One way to detect cardiac disease is to determine the presence of abnormalities in the PQRST interval []. Given the considerable benefits of the electrocardiogram application, such as portability, accessibility, safety, and real-time monitoring, it can be useful for assessing early cardiovascular involvement in COVID-19 cases. Typically, COVID-19 patients show several forms of abnormality in their ECG, such as ST changes [,], PR interval shortening [,], and QT prolongation [,].
The major impact of cardiovascular diseases on mortality and morbidity requires a global assessment of the disease and efficient detection techniques. Accurate and rapid detection of cardiac disorders is of vital importance in healthcare practice. The rapid detection of high-risk patients makes it possible to apply preventive actions, proactive procedures, and customized treatment modalities to ensure effective control of the disease’s evolution and mitigate its adverse effects. In the last decade, advances in artificial intelligence have led to considerable progress in automatic disease detection. The application of deep learning (DL) in the field of medicine has spurred many studies focused on the diagnosis of diverse diseases, including brain tumors from MR images [], several types of brain problems from EEG [], and skin diseases []. Moreover, in some medical imaging cases, it has been shown that the deep learning model’s classification performance can reach or even surpass that of medical experts [].
In this work, we present an approach that can help to diagnose cardiac manifestations of coronavirus from ECG images and overcome the limitations of traditional diagnostic approaches. The aim of this study is to differentiate COVID-19 ECGs from others (normal or with cardiovascular disorders). We perform two classification levels, a binary classification to identify COVID-19 cases, and a multi-class classification, to differentiate between COVID-19, other cardiovascular diseases, and patients with no findings. The relevance and novelty of our work lies in the fact that it integrates a vision transformer (ViT) [] with a lightweight CNN model (EfficientNetB0) []. The choice for using the ViT architecture is that it demonstrates excellent performance in terms of computational efficiency and accuracy compared to existing CNN models for several computer vision applications. The results demonstrate that our solution surpasses state-of-the-art (SOTA) approaches without any pre-processing or post-processing.
The main contributions of this work are as follows:
- (1)
- A hybrid vision transformer-CNN for cardiac anomaly detection from ECG images.
- (2)
- Ablation study to evaluate the effect of fusion CNN with vision transformer architecture.
- (3)
- A high-performance approach on ECG dataset from subjects with both cardiovascular conditions and COVID-19.
- (4)
- Fast solution adapted for real-world applications.
The remainder of the paper is as follows. In Section 2 we briefly summarize existing studies. In Section 3 we explain our proposed solution. Section 4 presents the evaluation of the approach using an ECG dataset from patients with cardiovascular diseases and COVID-19. Then, in Section 5 we conclude and suggest potential improvements.
2. Related Work
Several works in the automatic detection of cardiac anomalies from ECG records are reported in the literature [,,,,,]. Heart anomaly detection has become more and more popular since the expansion of healthcare data and the progress of big data analytics. Two distinct approaches are used to study ECG data by AI: low-level features and deep features. The low-level features generally are used with machine learning classifiers, while DL methods are used to automatically extract useful features from ECG sequences. Most of the proposed approaches share a common configuration comprising four processing stages: data pre-processing, feature extraction (hand-crafted or deep), dimension reduction, and classification. In recent years, deep learning has outperformed hand-crafted methods and shown remarkable performance in ECG classification [,]. Zadeh et al. [] presented an approach to identify five classes from ECG data. Their method includes three main steps: feature extraction based on discrete wavelet transform, classification based on SVM, and optimization based on genetic algorithm. They obtained an accuracy of 95.89%. Li et al. [] trained a convolution network to classify ECG signals. They achieve an accuracy exceeding 97.50%, outperforming multiple ECG classification methods. Ribeiro et al. [] constructed a large dataset of labeled 12-lead ECG records for diagnostic purposes. Then, they train a deep network to identify six categories of ECG abnormalities, comprising rhythmic and morphological findings. The authors reported that their model outperformed cardiology residents in identifying these anomalies, achieving F1 scores of 80.00% and specificities of 99.00%.
Despite the success of these methods on standardized datasets, their adaptation in real medical environments remains challenging. The majority of such studies rely on ECG signals-based datasets and therefore cannot be easily used in a real medical environment, while most ECG data in real medical practice is stored as images. The most suitable solution is to convert the ECG image to a digital signal but the transformation step is complex and the generated signal is of low quality, which consequently may affect the performance of AI techniques. Also, the digital ECG signal is acquired at a high sampling frequency while the ECG image is acquired in a few hertz. The significant reduction in the sampling rate results in a significant loss of information. For these reasons, some studies are based on ECG images to classify heart abnormalities. In recent work, Du et al. [] have presented a DL framework to identify anomalies in ECG images. They used a weakly supervised fine-grained classification mechanism. Then, an RNN model was used to achieve remarkable performance, with a sensitivity of 83.59% and precision of 90.42%. Khan et al. [] presented a method to detect four main cardiac abnormalities in 12-lead ECG images, using the MobileNet v2 model. They reached an accuracy of 98.00%. Hao et al. [] presented a multi-branch fusion system designed for automated diagnosis of myocardial infarction in ECG images. The 12 leads are introduced in the network, generating 12 feature maps. Then, these feature maps are concatenated by fusion, followed by classification to determine the presence of a myocardial infarction in the ECG image. The proposed approach reaches good performance with an accuracy of 94.73%, a sensitivity of 96.41%, a specificity of 95.94%, and an F1 score of 93.79%. Li et al. [] used transfer learning with Inception-V3 to diagnose seven kinds of arrhythmia. They obtained a balanced accuracy of 98.46%, sensitivity of 95.43%, and specificity of 96.75%.
In recent years, with the release of the ECG images dataset [,], which contains subjects with both COVID-19 and cardiovascular diseases, several research efforts have been undertaken to assess the relevance of using DL techniques with ECG images to identify COVID-19. Rahman et al. [] investigate the potential application of CNN architectures for identifying COVID-19 patterns in electrocardiogram images. Employing a variety of models such as ResNet18, ResNet50, ResNet101, InceptionV3, DenseNet201, and MobileNetv2, they explore their efficacy in detecting COVID-19 signals within ECG records. They achieved the best performance using DensNet201 with 99.10% and 97.36% for binary and multiclass, respectively. Ozdemir et al. [] used a novel and efficient approach referred to as hexaxial feature mapping to depict ECG signals in 2D colored images. The resulting images were then introduced to a CNN network to identify COVID-19. They obtained an accuracy of 96.20% for binary classification. Irmak [] presented a novel CNN network for the classification task. An overall accuracy of 98.57% and 86.55% is reported for binary and multiclass classifications, respectively. Sobahi et al. [] proposed a 3D CNN model with an attention mechanism. Using 10-fold cross-validation, an average accuracy of 99.0% for the binary level and 92.00% for the multi-class level were reported. Prashant et al. [] used an ensemble technique with three pre-trained CNN models. They achieved an average accuracy of 100% for the binary level and 95.29 % for the multiclass level. Attallah [] designed a framework called ECG-BiCoNet to identify COVID-19. This approach introduces five DL architectures and extracts distinct levels of features from separate layers of every DL model. Next, a feature selection approach is used to select the relevant features. Then, an ensemble learning method with three classifiers is used to perform the classification task. This approach achieves an accuracy of 98.80% and 91.73% for binary and multiclass classifications, respectively. Sakr et al. [] presented a novel CNN model to differentiate between COVID-19 ECGs and normal ECGs. They achieved an average accuracy of 94.91%. Chorney et al. [] proposed AttentionCOVIDNet, a CNN architecture based on attention. They achieved an average accuracy of 99.29% for the binary task and 91.26% for the binary multiclass.
3. Materials and Methods
This study explores the possibility of using a hybrid CNN vision transformer to identify COVID-19 and other cardiovascular abnormalities in ECG images.
3.1. Dataset
In this paper, we used an open-source ECG dataset from subjects with both cardiovascular conditions and COVID-19 []. So far, to our knowledge, it is the main and unique open-source dataset for COVID-19 ECG recordings. It provides 1937 12-lead ECG images of unique patients grouped into 5 classes: (COVID-19, myocardial infarction, history of MI, abnormal heartbeats, and normal patients). All collected data have been reviewed and annotated by several medical professionals []. Table 1 reports the distribution of images in each class. Some sample images are shown in Figure 1.

Table 1.
ECG images distribution among classes (original distribution) [].
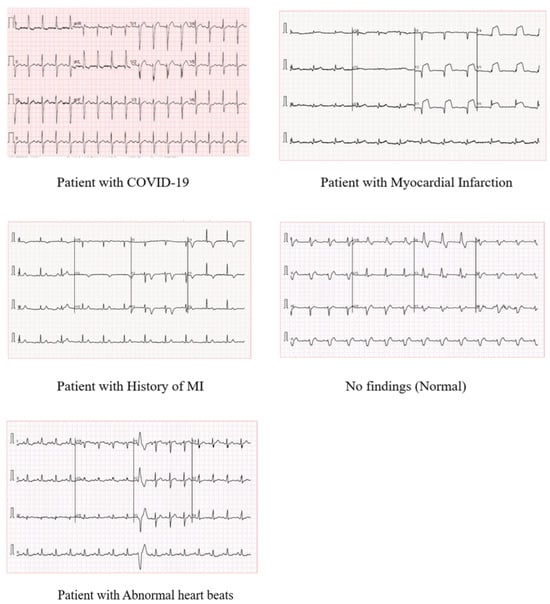
Figure 1.
Samples of ECG images from each class from [].
For a fair comparison with other methods, we evaluate our approach using the same class distribution for each classification level. For binary classification, we consider images from COVID-19 and normal patients, while for multiclass, three categories are used (COVID-19, Abnormal heartbeats, and Normal).
3.2. Pre-Processing
To improve the ECG image quality, each ECG image is passed through a minimal image pre-processing pipeline. The ECG data were pre-processed in the following steps:
Cropping: Each image is cut from top to bottom to eliminate unnecessary parts (date, patient data, etc.)
Binarization: The 12-leads ECG image is converted to a binary image by Otsu’s method [], such that each ECG lead can be represented by a contrasting black signal on a white background.
Resizing: The image is scaled to 500 × 500, to match the input shape of the proposed architecture.
3.3. Adapted EfficientNet Network-Based Vision Transformer for COVID-19 Diagnosis
In this paper, we explore the fusion of the CNN network vision transformer to identify COVID-19 and normal and abnormal heart rhythms from ECG images. By integrating CNN components with the vision transformer architecture, the model benefits from the complementary strengths of both approaches. CNNs excel at identifying spatial dependencies and local patterns, while vision transformers can identify global relationships and long-term dependencies through self-attention mechanisms. The combination of local and global information enables the model to capture fine-grained details and subtle patterns crucial for accurate classification, particularly in multi-class scenarios with distinct classes like COVID-19, normal, and abnormal. In particular, the feature maps generated by the CNN serve as informative input embeddings for the subsequent layers of the vision transformer. These feature maps encapsulate relevant spatial information extracted from the input images, providing the ViT with rich and detailed representations of the visual content.
3.4. EfficientNet-B0 CNN Model
In recent years, Tan et al. [] explored the relationship between the depth and width of CNN models, demonstrating a more effective way to design models with reduced parameters while improving classification accuracy. The authors published a paper introducing a new family of CNNs called EfficientNet (EfficientNetB0....EfficientNetB7). The key component in the EfficientNet family [] is the mobile inverted bottleneck convolution (MBConv), enhanced with SE optimization (Figure 2). This concept is derived from MobileNet architecture []. Also, EfficientNet models introduce Swish as a new activation function. It takes a comparable form to ReLU and LeakyReLU, sharing some of their performance advantages. However, swish provides a continuous curve along the loss optimization procedure with gradient descent. These architectures have shown superior accuracy to a large number of convolutional neural networks while maintaining considerably enhanced computational capabilities.
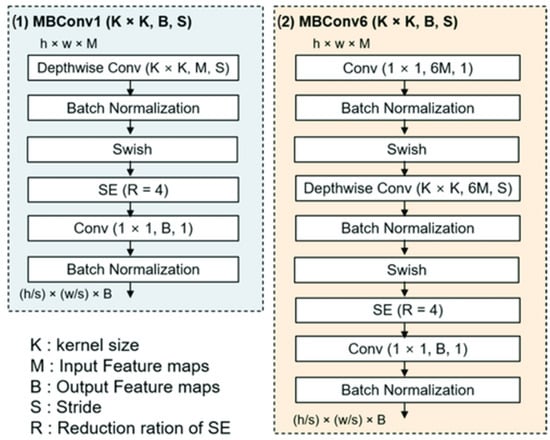
Figure 2.
The architecture of the MBConv block. MBConv1 vs. MBConv6 [].
In this study, we use EfficientNetB0 Figure 3, as a feature extractor to extract deep features from the ECG records. Then, this features map is fed into a ViT model, which uses attention mechanisms to process the feature representations and identify patterns. The proposed architecture is illustrated in Figure 4. The motivation behind the fusion approach lies in addressing the inherent limitations of individual architectures. While vision transformers excel in capturing global context and semantic relationships in images through self-attention mechanisms, they may struggle with capturing fine-grained details and local spatial information effectively. Conversely, CNNs are adept at learning hierarchical representations and extracting low-level features but may lack the ability to capture long-range dependencies efficiently.
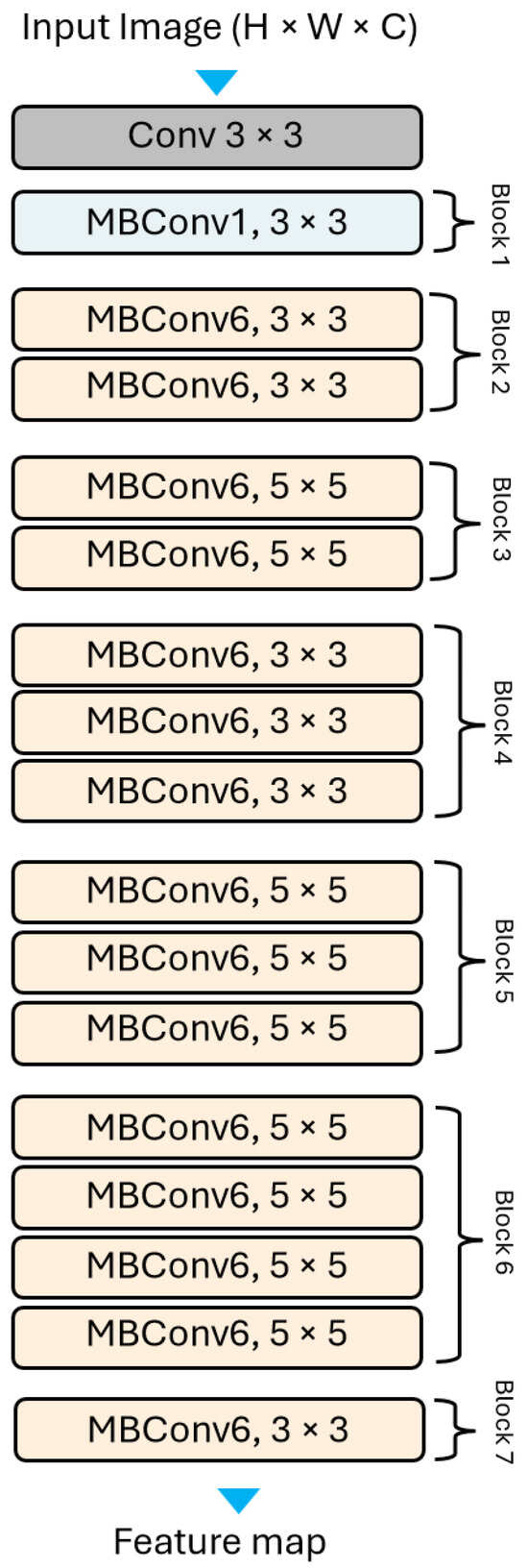
Figure 3.
The EffecientNet-B0 general architecture.
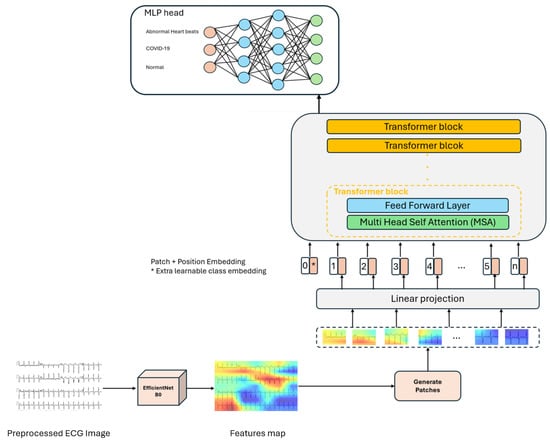
Figure 4.
The overview of the proposed approach based on the vision transformer architecture, multi-class scenario. Ecg images are passed through EfficientNetB0 to extract feature maps, then these maps are fed into the ViT model, which uses self-attention to extract patterns in the images and MLP head for the classification task.
3.5. Vision Transformer-EfficientNet-B0
The vision transformer (ViT) [] is a pioneering deep learning architecture that extends the success of natural language processing (NLP) transformers to the field of computer vision. The backbone of the ViT model [] is vanilla transformer architecture []. Unlike CNNs, which use convolution operations to extract spatial features from input data, the transformer is based only on the attention mechanism without recurrent or convolutional layers and focuses on the relationship between different parts of the input. The initial part of the network has a patch encoder layer that divides the features map generated from the EfficientNetB0 model (16 × 16 × 1280) into 2D patches of P × P (we choose P = 4); because only sequential data are compatible with the transformer bloc. Specifically, for a given features map of 16 × 16 × 1280 size, it is split into N = patches, where each patch is flattened to a vector of length . Then, the flattened patches are passed through a trainable linear transformation layer, which reduces their dimensionality while preserving important features; the patches are mapped from E to D dimensions to get patch embedding (D = 64). Each patch embedding provides an input for the transformer bloc. Since transformers do not inherently understand the spatial relationships between image patches, positional embeddings are added to provide information about the position of each patch within the image. These embeddings help the model to learn the spatial relationships between different patches in the image []. The sequence of lower-dimensional embeddings (including positional embeddings) is fed into a transformer encoder. The transformer blocks used in our architecture consist mainly of three sublayers: a multi-headed self-attention (MSA), feed forward (MLP), and normalization layer (Norm).
The MSA layers are one of the main mechanisms used in transformer architecture. They enable the model to take into account interactions between different parts of the input, generating attention maps from the input patches. Within the multi-head attention mechanism, the model computes attention scores between all pairs of input token embeddings. Each attention head (we used 4 heads), calculates a weighting for each pair of positions in the input sequence, focusing on different combinations of information. By combining the outputs of several heads, the model can capture different and complementary aspects of the relationships between the input elements. Three vectors are learned: Query, Key, and Value. These vectors are linearly projected from the input embeddings, allowing each head to focus on distinct parts of the entry simultaneously. The attention scores are computed independently for each attention head using the query and key vectors, using a dot product. Then, they are rescaled and undergo a softmax to get attention weights, and a weighted sum is then calculated from these weights. It represents the output of the attention mechanism for each attention head. Specifically, it determines how much each element in the input sequence contributes to the output of the attention bloc. In this way, the network learns to focus only on relevant information while filtering out noise [].
The outputs of the multiple attention heads are concatenated and linearly transformed to generate the final output of the MSA block. This aggregated representation is then passed through feed-forward neural network layers to project the attentional output, giving it a richer representation. The output of the second layer is fed back into the network as input, and a normalization layer is applied. The norm layer is performed to stabilize the network, considerably reducing training time and improving generalization capabilities. Residual connections are added to tackle the vanishing gradient and enable each bloc to flow directly in the network instead of passing through non-linear activations. For each layer, we added dropout and we used GELU as an activation function since it is considered a smoother version of the ReLU [].
At the end, the transformer output is then transformed to the MLP Head to perform the classification task. Unlike the original technique [], which combines the sequence of encoded patches with a learnable embedding to serve as the image representation, the final transformer block outputs are reshaped before being used as input for the MLP head. The MLP Head consists of two hidden layers of 1024 units and a classification layer with softmax activation. It gives the probabilities for each class: COVID-19, abnormal heartbeats, and normal.
4. Experiments
4.1. Environment Setup
The experiments were performed using Python 3.8.0 and TensorFlow 2.14.0 framework with the following specifications:
- RAM of 12.70 GB.
- NVIDIA T4 GPU of 16 GB.
4.2. Implementation Details and Hyper-Parameters Tuning
To deal with the problem of classification bias deriving from the unbalanced data, we selected 250 ECG images from each class, resulting in a total of 500 images for binary classification and 750 images for multi-class classification, maintaining the same proportionate distribution across classes.
When the dataset is small, as is the in case our studies, cross-validation is an important technique to have a fair evaluation of the classification system. We evaluate our approach using 10-fold cross-validation. The dataset is split into ten equal folds, and there was no patient overlap between the folds. Each time, 90% of the data are used as a training set, while the remaining 10% are kept for the test.
For binary classification, normal and COVID-19 classes are considered. Each fold consists of 50 images (25 images for each class). Nine folds are used for training (450 images) and the remaining folds for testing (50 images). For the multiclass level, normal, abnormal heartbeats, and COVID-19 classes are considered. Each fold consists of 75 images (25 images for each class). Nine folds are used for training (675 images) and the remaining folds for testing (75 images), Table 2.

Table 2.
Distribution of images between training and test sets.
As mentioned at the beginning, we are dealing with a classification problem. We use categorical cross-entropy. It is specifically designed for scenarios where the target variable has multiple classes, and it measures the dissimilarity between the predicted value and the ground truth, as described below:
where represents the predicted probability, and is the relevant class label. The dimension is the number of categories (2 for binary level and 3 for multiclass level).
We train the entire architecture using the NovoGrad [] optimizer. Unlike, Adam, NovoGrad is an SGD-based algorithm that operates at a first-order level, calculating second moments per layer rather than per weight. Also, this optimizer is known for its lower memory requirements and has demonstrated enhanced numerical stability []. Table 3 shows the initialization of the hyperparameters.

Table 3.
Hyper-parameters configuration.
4.3. Results
The binary classification results for each fold are summarized in Table 4. We achieve a performance of 100% for all classification metrics with a cross-validation strategy for both models (ViT-EfficientNetB0 and ViT) using the same folds. This result suggests that both architectures are highly effective in identifying the presence or absence of COVID-19 from medical images with no observable performance difference between them. The identical performance of both models indicates strong generalization capabilities in distinguishing between COVID-19 and normal cases across the dataset used in the study. We hypothesize that it is due to the presence of remarkable cardiac findings in the COVID-19 ECG, which are easily detectable by the vision transformer model. This suggests that the inherent capabilities of the vision transformer architecture alone are sufficient for binary classification tasks.

Table 4.
Binary classification result (COVID-19 vs. normal) of an ablation study with cross-validation using the same folds.
For the three class levels (COVID-19 vs abnormal heartbeats vs no finding), the average performance metrics are summarized in Table 5. The ViT–EfficientNetB0 model outperformed the ViT alone in multiclass classification scenarios, achieving higher precision, recall, F1-score, and accuracy across all classes (COVID, normal, and abnormal). The integration of CNN features enhances the discriminative power of the vision transformer by providing it with a more comprehensive understanding of the visual context. While ViT excels at capturing global relationships within data, EfficientNetB0 can capture local spatial information. These local patterns and anomalies are crucial for accurate classification. By incorporating EfficientNetB0, the model can effectively capture these local features, complementing the global context provided by the ViT. Also, EfficientNetB0 is proficient at extracting hierarchical features from images. It utilizes convolutional layers to detect low-level features like edges and textures, gradually building up to higher-level features such as shapes and patterns. Furthermore, it efficiently extracts relevant features from the raw images, such as waveform patterns indicative of different cardiac conditions. We achieve an average accuracy of 95.10%, precision of 95.30%, sensitivity of 95.10%, and F1-score of 95.10%. The performance metrics at each fold for abnormal heartbeats, COVID-19, and the normal class are shown in Table 6.

Table 5.
Multi class classification result (abnormal heartbeats vs. COVID-19 vs. normal) of an ablation study with cross-validation using the same folds.

Table 6.
Performance metrics of our approach (ViT–EfficientNetB0) for multiclass classification using a 10-fold cross-validation strategy.
4.4. Comparative Study
In this study, the proposed solution is compared with some recent SOTA methods on the same dataset. As given in Table 7. The majority of these methods give better results for binary classification and this is due to the presence of cardiac findings in the ECG of COVID-19 patients, which are easily detectable. However, for multi-class classification, the methods suffer from identifying COVID patients from other cardiac pathologies due to the resemblance of abnormalities in the ECG. Our solution outperforms the majority of SOTA methods in terms of binary and multi-class classification levels. We achieve an average accuracy of 100% and 95.10% for binary and multiclass, respectively.

Table 7.
Comparison of the proposed approach with SOTA methods on the same data distribution.
Despite the higher performance achieved by the different approaches, we think that the proposed model still performs very strongly, on the evidence that it is much more efficient than many models. In particular, the proposed model is lightweight and capable of effectively distinguishing the classes in question, being only slightly outperformed by much larger models. Our method was outperformed at the multiclass level by Rahman et al. [] with 2%. However, our method demonstrates superior adaptability due to its less complex architecture. In contrast to their approach, which is based on a DenseNet model with over 20 million parameters, ours is based on the simpler Vit and EfficientNetB0 models, with only 10 million parameters. This lighter approach makes our method better suited to real-life scenarios. In addition, our approach is validated by a rigorous 10-fold cross-validation process, doubling the robustness of the validation compared to their method, which uses only 5-fold. This extensive validation scheme guarantees the reliability and generalizability of our results across a wide range of data distributions.
On the other hand, our method introduces a new architecture that combines a CNN network with a vision transformer. The combination of this CNN model and the attention mechanism used enables our model to learn patterns sophisticated enough to differentiate COVID-19 from other pathologies.
5. Conclusions and Future Works
In this work, we have developed an artificial intelligence approach that complements the clinician’s diagnosis of COVID-19. Experimental results show that the integration of CNN features improves the discriminative power of the vision transformer, providing it with a more complete understanding of the visual context. Looking ahead, the proposed approach opens up new research and application possibilities in the field of medical imaging and diagnostics. As we continue to refine and optimize this technique, we can strive for more accurate and effective healthcare solutions, leading to improved patient outcomes and quality of care.
As a technical limitation of this research, it is important to highlight the problem of the restricted COVID-19 images. It can be observed that the majority of state-of-the-art methods (including our method) have used a small dataset with limited images in which only a few samples from COVID-19-positive patients are included. In addition, this study did not use optimization techniques for hyperparameter tuning. Future work could include, hyperparameter optimization, data augmentation, and, with the continued collection of data, we would like to extend the experimental work by validating the method with larger datasets. On the other hand, we are excited to apply our diagnostic system to a broader spectrum of cardiovascular conditions, including, but not limited to, myocardial infarction, arrhythmia, and other cardiac pathologies. To this end, we intend to conduct comprehensive validation studies to assess the performance of our system in various datasets and patient populations. In addition, we recognize the importance of integrating advanced technologies into our diagnostic system to enhance its capabilities. This may involve incorporating data from wearable devices or integrating with telemedicine platforms to enable remote monitoring and diagnosis. By harnessing these technologies, we aim to improve the diagnostic accuracy, accessibility, and scalability of our approach.
Author Contributions
Conceptualization, M.R.N.; Methodology, M.R.N. and Z.E.; Software, M.R.N.; Validation, M.R.N. and Z.E.; Writing—original draft, M.R.N.; Writing—review and editing, Z.E. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
The dataset generated during and/or analyzed during the current study is available to download from the following links: https://data.mendeley.com/datasets/gwbz3fsgp8/1, accessed on 15 June 2023.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Piroth, L.; Cottenet, J.; Mariet, A.S.; Bonniaud, P.; Blot, M.; Tubert-Bitter, P.; Quantin, C. Comparison of the characteristics, morbidity, and mortality of COVID-19 and seasonal influenza: A nationwide, population-based retrospective cohort study. Lancet Respir. Med. 2021, 9, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Kaliyaperumal, D.; Bhargavi, K.; Ramaraju, K.; Nair, K.S.; Ramalingam, S.; Alagesan, M. Electrocardiographic Changes in COVID-19 Patients: A Hospital-based Descriptive Study. Indian J. Crit. Care Med. 2022, 26, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Ng, M.Y.; Lee, E.; Yang, J.; Yang, F.; Li, X.; Wang, H.; Lui, M.; Lo, C.; Leung, B.S.T.; Khong, P.; et al. Imaging Profile of the COVID-19 Infection: Radiologic Findings and Literature Review. Radiol. Cardiothorac. Imaging 2020, 2, e200034. [Google Scholar] [CrossRef] [PubMed]
- Rousan, L.A.; Elobeid, E.; Karrar, M.; Khader, Y. Chest X-ray findings and temporal lung changes in patients with COVID-19 pneumonia. BMC Pulm. Med. 2020, 20, 245. [Google Scholar] [CrossRef] [PubMed]
- Raptis, C.A.; Hammer, M.M.; Short, R.G.; Shah, A.; Bhalla, S.; Bierhals, A.J.; Filev, P.D.; Hope, M.D.; Jeudy, J.; Kligerman, S.J.; et al. Chest CT and Coronavirus Disease (COVID-19): A Critical Review of the Literature to Date. Am. J. Roentgenol. 2020, 215, 839–842. [Google Scholar] [CrossRef]
- Driggin, E.; Madhavan, M.V.; Bikdeli, B.; Chuich, T.; Laracy, J.; Biondi-Zoccai, G.; Brown, T.S.; Nigoghossian, C.D.; Zidar, D.A.; Haythe, J.; et al. Cardiovascular Considerations for Patients, Health Care Workers, and Health Systems During the COVID-19 Pandemic. J. Am. Coll. Cardiol. 2020, 75, 2352–2371. [Google Scholar] [CrossRef]
- Long, B.; Brady, W.J.; Koyfman, A.; Gottlieb, M. Cardiovascular complications in COVID-19. Am. J. Emerg. Med. 2020, 38, 1504–1507. [Google Scholar] [CrossRef]
- Nishiga, M.; Wang, D.W.; Han, Y.; Lewis, D.B.; Wu, J.C. COVID-19 and cardiovascular disease: From basic mechanisms to clinical perspectives. Nat. Rev. Cardiol. 2020, 17, 543–558. [Google Scholar] [CrossRef] [PubMed]
- Luz, E.J.D.S.; Schwartz, W.R.; Cámara-Chávez, G.; Menotti, D. ECG-based heartbeat classification for arrhythmia detection: A survey. Comput. Methods Programs Biomed. 2016, 127, 144–164. [Google Scholar] [CrossRef]
- Madona, P.; Basti, R.I.; Zain, M.M. PQRST wave detection on ECG signals. Gac. Sanit. 2021, 35, S364–S369. [Google Scholar] [CrossRef]
- Vidovich, M.I. Transient Brugada-Like Electrocardiographic Pattern in a Patient with COVID-19. JACC Case Rep. 2020, 2, 1245–1249. [Google Scholar] [CrossRef] [PubMed]
- Bergamaschi, L.; D’Angelo, E.C.; Paolisso, P.; Toniolo, S.; Fabrizio, M.; Angeli, F.; Donati, F.; Magnani, I.; Rinaldi, A.; Bartoli, L.; et al. The value of ECG changes in risk stratification of COVID-19 patients. Ann. Noninvasive Electrocardiol. 2021, 26, e12815. [Google Scholar] [CrossRef] [PubMed]
- Pavri, B.B.; Kloo, J.; Farzad, D.; Riley, J.M. Behavior of the PR interval with increasing heart rate in patients with COVID-19. Heart Rhythm 2020, 17, 1434–1438. [Google Scholar] [CrossRef] [PubMed]
- Chorin, E.; Wadhwani, L.; Magnani, S.; Dai, M.; Shulman, E.; Nadeau-Routhier, C.; Knotts, R.; Bar-Cohen, R.; Kogan, E.; Barbhaiya, C.; et al. QT interval prolongation and torsade de pointes in patients with COVID-19 treated with hydroxychloroquine/azithromycin. Heart Rhythm 2020, 17, 1425–1433. [Google Scholar] [CrossRef]
- Santoro, F.; Monitillo, F.; Raimondo, P.; Lopizzo, A.; Brindicci, G.; Gilio, M.; Musaico, F.; Mazzola, M.; Vestito, D.; Benedetto, R.D.; et al. QTc Interval Prolongation and Life-Threatening Arrhythmias During Hospitalization in Patients With Coronavirus Disease 2019 (COVID-19): Results From a Multicenter Prospective Registry. Clin. Infect. Dis. 2020, 73, e4031–e4038. [Google Scholar] [CrossRef] [PubMed]
- Ghassemi, N.; Shoeibi, A.; Rouhani, M. Deep neural network with generative adversarial networks pre-training for brain tumor classification based on MR images. Biomed. Signal Process. Control 2019, 57, 101678–101688. [Google Scholar] [CrossRef]
- He, H.; Liu, X.; Hao, Y. A progressive deep wavelet cascade classification model for epilepsy detection. Artif. Intell. Med. 2021, 118, 102117. [Google Scholar] [CrossRef]
- Hameed, N.; Shabut, A.M.; Hossain, M.A. Multi-Class Skin Diseases Classification Using Deep Convolutional Neural Network and Support Vector Machine. In Proceedings of the 2018 12th International Conference on Software, Knowledge, Information Management Applications (SKIMA), Phnom Penh, Cambodia, 3–5 December 2018; pp. 1–7. [Google Scholar] [CrossRef]
- Rajpurkar, P.; Irvin, J.; Zhu, K.; Yang, B.; Mehta, H.; Duan, T.; Ding, D.Y.; Bagul, A.; Langlotz, C.P.; Shpanskaya, K.S.; et al. CheXNet: Radiologist-Level Pneumonia Detection on Chest X-Rays with Deep Learning. arXiv 2017, arXiv:1711.05225. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16 × 16 Words: Transformers for Image Recognition at Scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Tan, M.; Le, Q.V. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. In Proceedings of the 36th International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019. [Google Scholar] [CrossRef]
- Huang, J.; Chen, B.; Yao, B.; He, W. ECG Arrhythmia Classification Using STFT-Based Spectrogram and Convolutional Neural Network. IEEE Access 2019, 7, 92871–92880. [Google Scholar] [CrossRef]
- De Chazal, P.; O’Dwyer, M.; Reilly, R. Automatic classification of heartbeats using ECG morphology and heartbeat interval features. IEEE Trans. Biomed. Eng. 2004, 51, 1196–1206. [Google Scholar] [CrossRef] [PubMed]
- Kiranyaz, S.; Ince, T.; Gabbouj, M. Real-Time Patient-Specific ECG Classification by 1-D Convolutional Neural Networks. IEEE Trans. Biomed. Eng. 2016, 63, 664–675. [Google Scholar] [CrossRef]
- Rouhi, R.; Clausel, M.; Oster, J.; Lauer, F. An Interpretable Hand-Crafted Feature-Based Model for Atrial Fibrillation Detection. Front. Physiol. 2021, 12, 657304. [Google Scholar] [CrossRef]
- Eltrass, A.S.; Tayel, M.B.; Ammar, A.I. A new automated CNN deep learning approach for identification of ECG congestive heart failure and arrhythmia using constant-Q non-stationary Gabor transform. Biomed. Signal Process. Control 2021, 65, 102326. [Google Scholar] [CrossRef]
- Ertuğrul, Ö.F.; Acar, E.; Aldemir, E.; Öztekin, A. Automatic diagnosis of cardiovascular disorders by sub images of the ECG signal using multi-feature extraction methods and randomized neural network. Biomed. Signal Process. Control 2021, 64, 102260. [Google Scholar] [CrossRef]
- Acharya, U.R.; Fujita, H.; Oh, S.L.; Hagiwara, Y.; Tan, J.H.; Adam, M. Application of deep convolutional neural network for automated detection of myocardial infarction using ECG signals. Inf. Sci. 2017, 415-416, 190–198. [Google Scholar] [CrossRef]
- Baloglu, U.B.; Talo, M.; Yildirim, O.; Tan, R.S.; Acharya, U.R. Classification of myocardial infarction with multi-lead ECG signals and deep CNN. Pattern Recognit. Lett. 2019, 122, 23–30. [Google Scholar] [CrossRef]
- Zadeh, A.E.; Khazaee, A. High Efficient System for Automatic Classification of the Electrocardiogram Beats. Ann. Biomed. Eng. 2010, 39, 996–1011. [Google Scholar] [CrossRef]
- Li, D.; Zhang, J.; Zhang, Q.; Wei, X. Classification of ECG signals based on 1D convolution neural network. In Proceedings of the 2017 IEEE 19th International Conference on e-Health Networking, Applications and Services (Healthcom), Dalian, China, 12–15 October 2017; pp. 1–6. [Google Scholar] [CrossRef]
- Ribeiro, A.H.; Ribeiro, M.H.; Paixão, G.M.M.; Oliveira, D.M.; Gomes, P.R.; Canazart, J.A.; Ferreira, M.P.S.; Andersson, C.R.; Macfarlane, P.W.; Meira, W.; et al. Automatic diagnosis of the 12-lead ECG using a deep neural network. Nat. Commun. 2020, 11, 1760. [Google Scholar] [CrossRef]
- Du, N.; Cao, Q.; Yu, L.; Liu, N.; Zhong, E.; Liu, Z.; Shen, Y.; Chen, K. FM-ECG: A fine-grained multi-label framework for ECG image classification. Inf. Sci. 2021, 549, 164–177. [Google Scholar] [CrossRef]
- Khan, A.H.; Hussain, M.; Malik, M.K. Cardiac Disorder Classification by Electrocardiogram Sensing Using Deep Neural Network. Complexity 2021, 2021, 5512243. [Google Scholar] [CrossRef]
- Hao, P.; Gao, X.; Li, Z.; Zhang, J.; Wu, F.; Bai, C. Multi-branch fusion network for Myocardial infarction screening from 12-lead ECG images. Comput. Methods Programs Biomed. 2020, 184, 105286. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhao, H.; Lu, W.; Leng, X.; Wang, L.; Lin, X.; Pan, Y.; Jiang, W.; Jiang, J.; Sun, Y.; et al. DeepECG: Image-based electrocardiogram interpretation with deep convolutional neural networks. Biomed. Signal Process. Control 2021, 69, 102824. [Google Scholar] [CrossRef]
- Khan, A.H.; Hussain, M.; Malik, M.K. ECG Images dataset of Cardiac and COVID-19 Patients. Data Brief 2021, 34, 106762. [Google Scholar] [CrossRef]
- Khan, A.H. ECG Images dataset of Cardiac and COVID-19 Patients. Data Brief 2020, 34, 106762. [Google Scholar] [CrossRef] [PubMed]
- Rahman, T.; Akinbi, A.; Chowdhury, M.E.H.; Rashid, T.A.; Şengür, A.; Khandakar, A.; Islam, K.R.; Ismael, A.M. COV-ECGNET: COVID-19 detection using ECG trace images with deep convolutional neural network. Health Inf. Sci. Syst. 2022, 10, 1. [Google Scholar] [CrossRef] [PubMed]
- Ozdemir, M.A.; Ozdemir, G.D.; Guren, O. Classification of COVID-19 electrocardiograms by using hexaxial feature mapping and deep learning. BMC Med. Inform. Decis. Mak. 2021, 21, 170. [Google Scholar] [CrossRef] [PubMed]
- Irmak, E. COVID-19 disease diagnosis from paper-based ECG trace image data using a novel convolutional neural network model. Phys. Eng. Sci. Med. 2022, 45, 167–179. [Google Scholar] [CrossRef]
- Sobahi, N.; Sengur, A.; Tan, R.S.; Acharya, U.R. Attention-based 3D CNN with residual connections for efficient ECG-based COVID-19 detection. Comput. Biol. Med. 2022, 143, 105335. [Google Scholar] [CrossRef]
- Prashant, K.; Choudhary, P.; Agrawal, T.; Kaushik, E. OWAE-Net: COVID-19 detection from ECG images using deep learning and optimized weighted average ensemble technique. Intell. Syst. Appl. 2022, 16, 200154. [Google Scholar] [CrossRef]
- Attallah, O. ECG-BiCoNet: An ECG-based pipeline for COVID-19 diagnosis using Bi-Layers of deep features integration. Comput. Biol. Med. 2022, 142, 105210. [Google Scholar] [CrossRef] [PubMed]
- Sakr, A.S.; Pławiak, P.; Tadeusiewicz, R.; Pławiak, J.; Sakr, M.; Hammad, M. ECG-COVID: An end-to-end deep model based on electrocardiogram for COVID-19 detection. Inf. Sci. 2023, 619, 324–339. [Google Scholar] [CrossRef] [PubMed]
- Chorney, W.; Wang, H.; Fan, L.W. AttentionCovidNet: Efficient ECG-based diagnosis of COVID-19. Comput. Biol. Med. 2024, 168, 107743. [Google Scholar] [CrossRef]
- Otsu, N. A threshold selection method from gray level histograms. IEEE Trans. Syst. Man Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. MobileNetV2: Inverted Residuals and Linear Bottlenecks. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 4510–4520. [Google Scholar] [CrossRef]
- Gang, S.; Fabrice, N.; Chung, D.; Lee, J. Character Recognition of Components Mounted on Printed Circuit Board Using Deep Learning. Sensors 2021, 21, 2921. [Google Scholar] [CrossRef] [PubMed]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention is all you need. In Proceedings of the Advances in Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- Alamri, F.; Dutta, A. Multi-Head Self-Attention via Vision Transformer for Zero-Shot Learning. arXiv 2021, arXiv:2108.00045. [Google Scholar]
- Ginsburg, B.; Castonguay, P.; Hrinchuk, O.; Kuchaiev, O.; Lavrukhin, V.; Leary, R.; Li, J.; Nguyen, H.; Cohen, J.M. Stochastic Gradient Methods with Layer-wise Adaptive Moments for Training of Deep Networks. arXiv 2019, arXiv:1905.11286. [Google Scholar]
- Attallah, O. An Intelligent ECG-Based Tool for Diagnosing COVID-19 via Ensemble Deep Learning Techniques. Biosensors 2022, 12, 299. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).