Deep Learning Based Breast Cancer Detection Using Decision Fusion
Abstract
1. Introduction
- Dedicated CNN architecture: A custom CNN architecture, trained on the subset of the DDSM dataset, is used for direct classification.
- SVM with ResNet50 features: SVM is applied to features extracted from a pre-trained ResNet50 CNN model.
- SVM with LBP features: SVM is used with local binary pattern (LBP) features for classification.
2. Materials and Methods
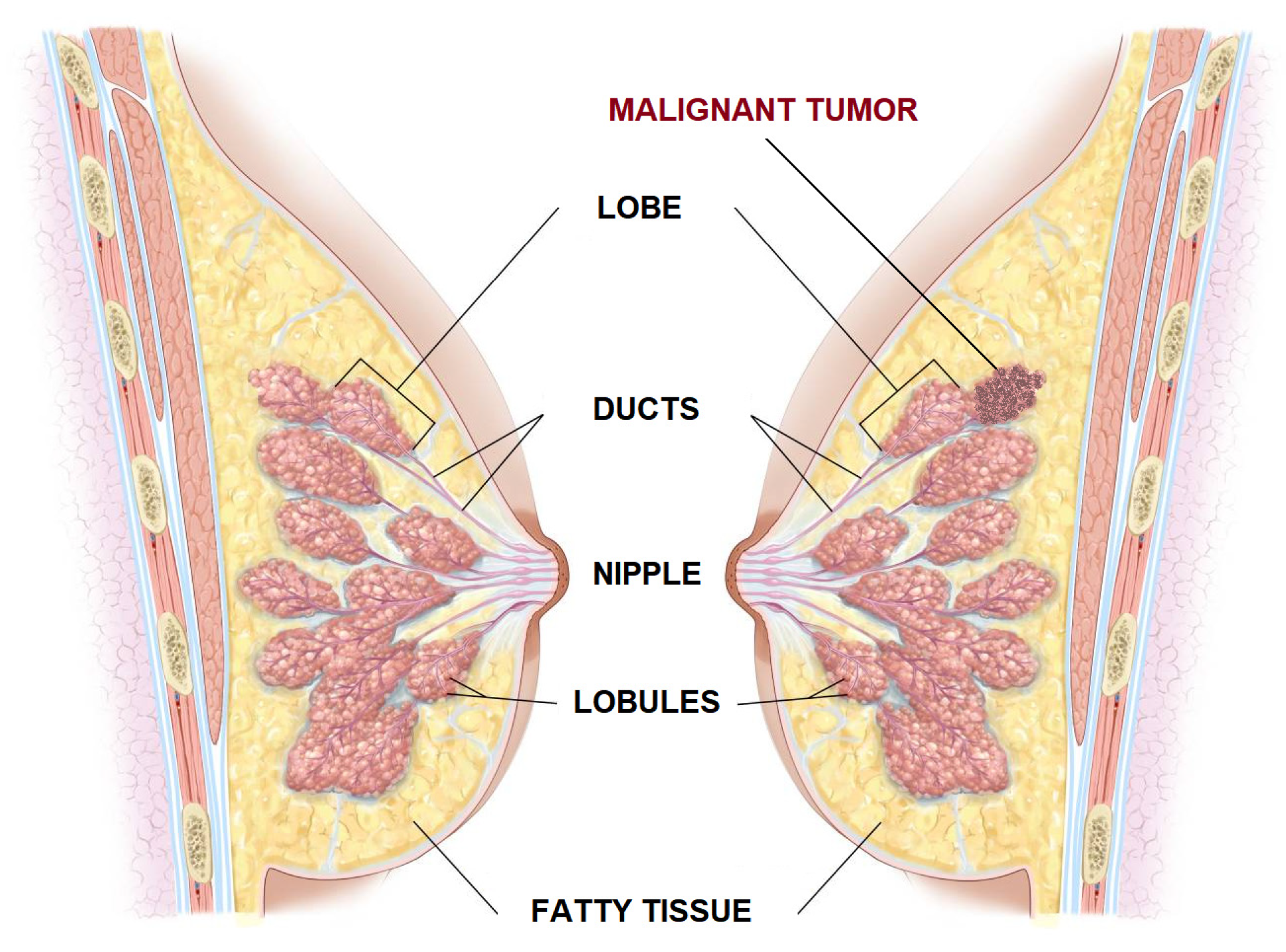
2.1. Dataset
2.2. Software Platform
2.3. Decision Fusion
- Sum rule: The probabilities from each model are simply added together.
- Product rule: The probabilities are multiplied together.
- Majority voting: The class with the highest number of votes from the individual models is selected.
- : Combined probability for each class;
- : The number of classifiers;
- : The probability assigned by the ith classifier for a particular class;
- : The predicted class by the ith classifier;
- : Each of the possible classes;
- : The indicator function;
- : Returns the class index j that maximizes the sum.
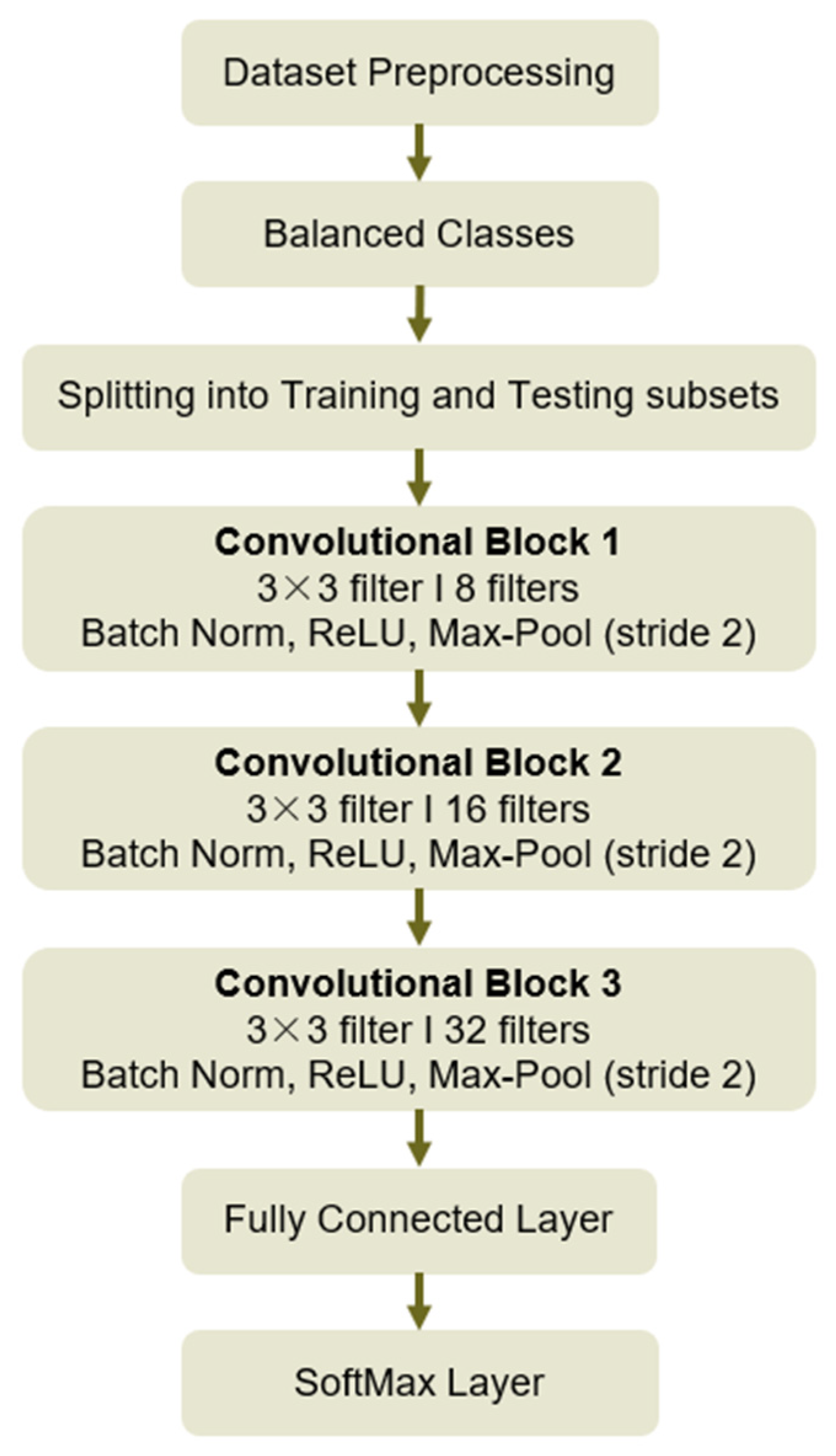
2.4. Proposed Breast Cancer Detection System
- Pre-trained ResNet50: The final classification layer of ResNet50 was removed, and the extracted feature vector after flattening was used by an SVM classifier.
- SVM with LBP features: LBP feature vectors were used by an SVM classifier.
- Dedicated CNN architecture: A custom CNN architecture with three convolutional layers was used.
2.4.1. CNN Architecture
- Convolution Operation
- Output Size Calculation
- Max-Pooling Operation
- Batch Normalization
- Fully Connected Layer
- SoftMax Function
- Cross-Entropy Loss
2.4.2. Ablation Study
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- American College of Obstetricians and Gynecologists. Breast cancer risk assessment and screening in average-risk women. Pract. Bull. 2017, 130, 1–16. [Google Scholar] [CrossRef]
- Lakhtakia, R. A Brief History of Breast Cancer: Part I: Surgical domination reinvented. Sultan Qaboos Univ. Med. J. 2014, 14, e166–e169. [Google Scholar] [PubMed]
- Breast Cancer. Available online: https://www.who.int/news-room/fact-sheets/detail/breast-cancer (accessed on 2 October 2024).
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Hossain, A.; Islam, M.T.; Islam, M.T.; Chowdhury, M.E.H.; Rmili, H.; Samsuzzaman, M. A Planar Ultrawideband Patch Antenna Array for Microwave Breast Tumor Detection. Materials 2020, 13, 4918. [Google Scholar] [CrossRef]
- Ara, S.; Das, A.; Dey, A. Malignant and Benign Breast Cancer Classification using Machine Learning Algorithms. In Proceedings of the 2021 International Conference on Artificial Intelligence (ICAI), Islamabad, Pakistan, 5–7 April 2021; pp. 97–101. [Google Scholar] [CrossRef]
- Naseem, U.; Rashid, J.; Ali, L.; Kim, J.; Ul Haq, Q.E.; Awan, M.J.; And Imran, M. An Automatic Detection of Breast Cancer Diagnosis and Prognosis Based on Machine Learning Using Ensemble of Classifiers. IEEE Access 2022, 10, 78242–78252. [Google Scholar] [CrossRef]
- Eleyan, A. Breast cancer classification using moments. In Proceedings of the 2012 20th Signal Processing and Communications Applications Conference (SIU), Mugla, Turkey, 18–20 April 2012; pp. 1–4. [Google Scholar] [CrossRef]
- Amin, S.A.; Al Shanabari, H.; Iqbal, R.; Karyotis, C. An Intelligent Framework for Automatic Breast Cancer Classification Using Novel Feature Extraction and Machine Learning Techniques. J. Signal Process. Syst. 2023, 95, 293–303. [Google Scholar] [CrossRef]
- Alzubaidi, L.; Zhang, J.; Humaidi, A.J.; Al-Dujaili, A.; Duan, Y.; Al-Shamma, O.; Santamaría, J.; Fadhel, M.A.; Al-Amidie, M.; Farhan, L. Review of deep learning: Concepts, CNN architectures, challenges, applications, future directions. J. Big Data 2021, 8, 53. [Google Scholar] [CrossRef]
- Papageorgiou, V. Brain Tumor Detection Based on Features Extracted and Classified Using a Low-Complexity Neural Network. Trait. Du Signal 2021, 38, 547–554. [Google Scholar] [CrossRef]
- Koti, M.S.; Nagashree, B.A.; Geetha, V.; Shreyas, K.P.; Mathivanan, S.K.; Dalu, G.T. Lung cancer diagnosis based on weighted convolutional neural network using gene data expression. Sci. Rep. 2024, 14, 3656. [Google Scholar]
- Eleyan, A.; Bayram, F.; Eleyan, G. Spectrogram-Based Arrhythmia Classification Using Three-Channel Deep Learning Model with Feature Fusion. Appl. Sci. 2024, 14, 9936. [Google Scholar] [CrossRef]
- Hasan, M.Z.; Ahamed, M.S.; Rakshit, A.; Hasan, K.Z. Recognition of jute diseases by leaf image classification using convolutional neural network. In Proceedings of the 2019 10th International Conference on Computing, Communication and Networking Technologies (ICCCNT), Kanpur, India, 6–8 July 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 1–5. [Google Scholar]
- Papageorgiou, V.E.; Dogoulis, P.; Papageorgiou, D.P. A convolutional neural network of low complexity for tumor anomaly detection. In Proceedings of the International Congress on Information and Communication Technology, London, UK, 20–23 February 2023; Springer Nature: Singapore, 2023; pp. 973–983. [Google Scholar]
- Shrestha, A.; Mahmood, A. Review of deep learning algorithms and architectures. IEEE Access 2019, 7, 53040–53065. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Carriero, A.; Groenhoff, L.; Vologina, E.; Basile, P.; Albera, M. Deep Learning in Breast Cancer Imaging: State of the Art and Recent Advancements in Early 2024. Diagnostics 2024, 14, 848. [Google Scholar] [CrossRef] [PubMed]
- Nasser, M.; Yusof, U.K. Deep Learning Based Methods for Breast Cancer Diagnosis: A Systematic Review and Future Direction. Diagnostics 2023, 13, 161. [Google Scholar] [CrossRef] [PubMed]
- Khourdifi, Y.; Bahaj, M. Applying best machine learning algorithms for breast cancer prediction and classification. In Proceedings of the 2018 International Conference on Electronics, Control, Optimization and Computer Science (ICECOCS), Kenitra, Morocco, 5–6 December 2018; IEEE: Piscataway, NJ, USA, 2018; pp. 1–5. [Google Scholar]
- Bah, A.; Davud, M. Analysis of Breast Cancer Classification with Machine Learning based Algorithms. In Proceedings of the 2022 2nd International Conference on Computing and Machine Intelligence (ICMI), Istanbul, Turkey, 15–16 July 2022; IEEE: Piscataway, NJ, USA, 2022; pp. 1–4. [Google Scholar]
- Mahmud, M.I.; Mamun, M.; Abdelgawad, A. A Deep Analysis of Transfer Learning Based Breast Cancer Detection Using Histopathology Images. In Proceedings of the 2023 10th International Conference on Signal Processing and Integrated Networks (SPIN), Delhi, India, 23–24 March 2023; IEEE: Piscataway, NJ, USA, 2023; pp. 198–204. [Google Scholar]
- Amgad, N.; Ahmed, M.; Haitham, H.; Zaher, M.; Mohammed, A. A Robust Ensemble Deep Learning Approach for Breast Cancer Diagnosis. In Proceedings of the 2023 Intelligent Methods, Systems, and Applications (IMSA), Giza, Egypt, 15–16 July 2023; IEEE: Piscataway, NJ, USA, 2023; pp. 452–457. [Google Scholar]
- Jaffar, M.A. Deep learning-based computer aided diagnosis system for breast mammograms. Int. J. Adv. Comput. Sci. Appl. 2017, 8, 286–290. [Google Scholar]
- Liu, M.Z.; Swintelski, C.; Sun, S.; Siddique, M.; Desperito, E.; Jambawalikar, S.; Ha, R. Weakly supervised deep learning approach to breast MRI assessment. Acad. Radiol. 2022, 29 (Suppl. S1), S166–S172. [Google Scholar] [CrossRef]
- Logan, J.; Kennedy, P.J.; Catchpoole, D. A review of the machine learning datasets in mammography, their adherence to the FaIR principles and the outlook for the future. Sci. Data 2023, 10, 595. [Google Scholar] [CrossRef]
- Lee, R.S.; Gimenez, F.; Hoogi, A.; Miyake, K.K.; Gorovoy, M.; Rubin, D.L. **A curated mammography data set for use in computer-aided detection and diagnosis research. Sci. Data 2017, 4, 170177. [Google Scholar] [CrossRef]
- Jenis, J.; Ondriga, J.; Hrcek, S.; Brumercik, F.; Cuchor, M.; Sadovsky, E. Engineering applications of artificial intelligence in mechanical design and optimization. Machines 2023, 11, 577. [Google Scholar] [CrossRef]
- Hossain, M.B.; Iqbal SH, S.; Islam, M.M.; Akhtar, M.N.; Sarker, I.H. Transfer learning with fine-tuned deep CNN ResNet50 model for classifying COVID-19 from chest X-ray images. Inform. Med. Unlocked 2022, 30, 100916. [Google Scholar] [CrossRef]
- Eleyan, A.; Alboghbaish, E. Multi-Classifier Deep Learning based System for ECG Classification Using Fourier Transform. In Proceedings of the 5th International Conference on Bio-engineering for Smart Technologies (BioSMART), Paris, France, 7–9 June 2023; pp. 1–4. [Google Scholar] [CrossRef]
- Papageorgiou, V.E.; Zegkos, T.; Efthimiadis, G.; Tsaklidis, G. Analysis of digitalized ECG signals based on artificial intelligence and spectral analysis methods specialized in ARVC. Int. J. Numer. Methods Biomed. Eng. 2022, 38, e3644. [Google Scholar] [CrossRef]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Scherer, D.; Müller, A.; Behnke, S. Evaluation of pooling operations in convolutional architectures for object recognition. In Proceedings of the International Conference on Artificial Neural Networks, Thessaloniki, Greece, 15–18 September 2010; Springer: Berlin/Heidelberg, Germany, 2010; pp. 92–101. [Google Scholar]
- Pessoa, D.; Petmezas, G.; Papageorgiou, V.E.; Rocha, B.M.; Stefanopoulos, L.; Kilintzis, V.; Maglaveras, N.; Frerichs, I.; de Carvalho, P.; Paiva, R.P. Pediatric Respiratory Sound Classification Using a Dual Input Deep Learning Architecture. In Proceedings of the 2023 IEEE Biomedical Circuits and Systems Conference (BioCAS), Toronto, ON, Canada, 19–21 October 2023; IEEE: Piscataway, NJ, USA, 2023; pp. 1–5. [Google Scholar]
- Bayram, F.; Eleyan, A. COVID-19 detection on chest radiographs using feature fusion-based deep learning. Signal Image Video Process. 2022, 16, 1455–1462. [Google Scholar] [CrossRef] [PubMed]
- Eleyan, A.; Alboghbaish, E. Electrocardiogram signals classification using deep-learning-based incorporated convolutional neural network and long short-term memory framework. Computers 2024, 13, 55. [Google Scholar] [CrossRef]
- Jiao, Z.; Gao, X.; Wang, Y.; Li, J. A deep feature-based framework for breast masses classification. Neurocomputing 2016, 197, 221–231. [Google Scholar] [CrossRef]
- Sajid, U.; Khan, R.A.; Shah, S.M.; Arif, S. Breast cancer classification using deep learned features boosted with handcrafted features. Biomed. Signal Process. Control 2023, 86, 105353. [Google Scholar] [CrossRef]
- Wang, Z.; Li, M.; Wang, H.; Jiang, H.; Yao, Y.; Zhang, H.; Xin, J. Breast cancer detection using extreme learning machine based on feature fusion with CNN deep features. IEEE Access 2019, 7, 105146–105158. [Google Scholar] [CrossRef]
- Ahmed, M.; Bibi, T.; Khan, R.A.; Nasir, S. Enhancing Breast Cancer Diagnosis in Mammography: Evaluation and Integration of Convolutional Neural Networks and Explainable AI. arXiv 2024, arXiv:2404.03892. [Google Scholar]



| Parameters | Accuracy | Specificity | Precision | Sensitivity | F1 Score |
|---|---|---|---|---|---|
| ReLU + SGD | 97.7 | 97.0 | 96.9 | 98.5 | 97.7 |
| LeakyReLU + SGD | 94.6 | 96.7 | 96.9 | 92.6 | 94.7 |
| ReLU + Adem | 93.8 | 95.2 | 95.3 | 92.5 | 93.9 |
| LeakyReLU + Adem | 96.1 | 98.3 | 98.4 | 94.1 | 96.2 |
| Hyper-Parameter | Value/Metric |
|---|---|
| Epochs | 6 |
| Optimizer | SGD with momentum |
| Batch size | 10 |
| Activation function | ReLU |
| Stride | 2 |
| Shuffle | Every epoch |
| Validation frequency | 3 |
| Initial learning rate | 10−4 |
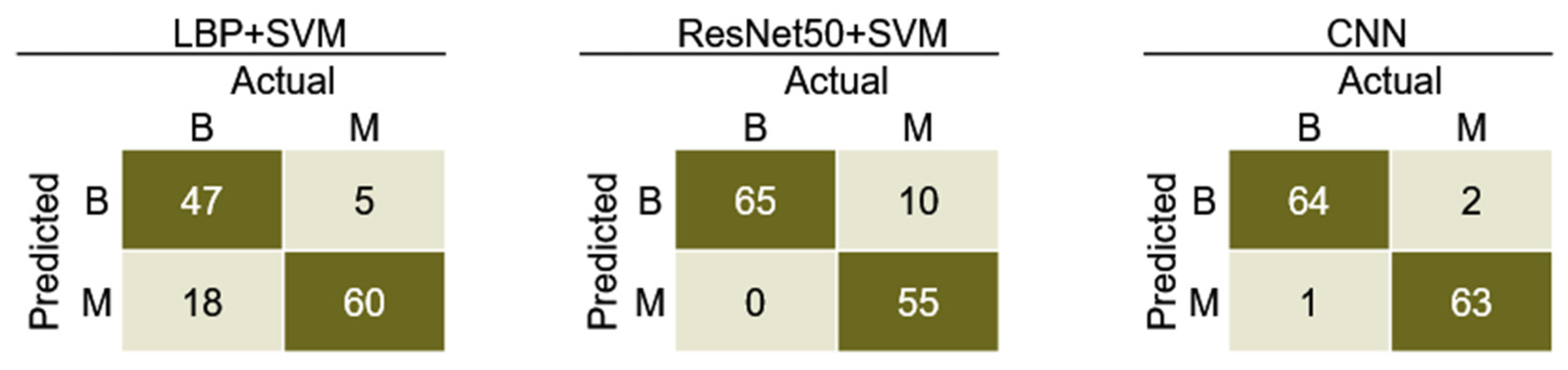
| Model | Sensitivity | Specificity | Precision | F1-Score | Accuracy |
|---|---|---|---|---|---|
| LBP + SVM | 71.7 | 90.1 | 87.9 | 79.0 | 80.9 |
| ResNet50 + SVM | 94.7 | 77.7 | 80.9 | 87.3 | 86.2 |
| CNN | 90.8 | 90.8 | 90.8 | 90.8 | 90.8 |
| Model | Sensitivity | Specificity | Precision | F1-Score | Accuracy |
|---|---|---|---|---|---|
| LBP + SVM | 71.3 | 90.7 | 88.5 | 79.0 | 81.0 |
| ResNet50 + SVM | 96.3 | 86.1 | 87.4 | 91.6 | 91.2 |
| CNN | 94.7 | 97.0 | 94.4 | 96.6 | 96.8 |
| Model | Sensitivity | Specificity | Precision | F1-Score | Accuracy |
|---|---|---|---|---|---|
| LBP + SVM | 72.3 | 92.3 | 90.4 | 80.3 | 82.3 |
| ResNet50 + SVM | 100 | 84.6 | 86.7 | 92.9 | 92.3 |
| CNN | 98.5 | 97.0 | 96.9 | 97.7 | 97.7 |
| Decision Fusion Rule | Sensitivity | Specificity | Precision | F1-Score | Accuracy |
|---|---|---|---|---|---|
| Sum rule | 98.3 | 98.9 | 98.5 | 98.7 | 98.5 |
| Product rule | 98.9 | 99.4 | 99.7 | 99.4 | 99.1 |
| Majority voting | 98.1 | 98.4 | 98.2 | 99.1 | 98.9 |
| Reference | Classes | Training/Test Ratio | Dataset | Model | Accuracy |
|---|---|---|---|---|---|
| [8] | Benign, Malignant | 40/60 | WBCD | Moments + SVM | 96.6 |
| [20] | Benign, Malignant | - | WBCD | SVM | 97.9 |
| [21] | Benign, Malignant | 80/20 | Breast Cancer Data | SVM | 87.0 |
| CNN | 89.0 | ||||
| [22] | Benign, Malignant | 80/20 | Breast Histopathology Images | ResNet50 | 90.2 |
| [23] | Benign, Malignant | 80/20 | BCI | Average Weighted Ensemble | 85.0 |
| [24] | Benign, Malignant | - | DDSM | CNN + SVM | 93.0 |
| [25] | Benign, Malignant | - | ISPY-1 Data | CNN | 94.0 |
| [37] | Benign, Malignant | - | DDSM | DCNN + SVM | 96.7 |
| [38] | Benign, Malignant | - | DDSM | LBP + HOG + CNN | 91.5 |
| [39] | Benign, Malignant | - | Hospital Images | CNN + SVM | 74.5 |
| [40] | Benign, Malignant | 90/10 | DDSM | ResNet50 | 72.0 |
| VGG16 | 56.0 | ||||
| Inception V3 | 56.0 | ||||
| Ours Individual | Benign, Malignant | 70/30 | DDSM | LBP + SVM | 82.3 |
| ResNet50 + SVM | 92.3 | ||||
| Developed CNN | 97.7 | ||||
| Ours Decision Fusion | Benign, Malignant | 70/30 | DDSM | Sum rule fusion | 98.7 |
| Majority voting fusion | 98.9 | ||||
| Product rule fusion | 99.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manalı, D.; Demirel, H.; Eleyan, A. Deep Learning Based Breast Cancer Detection Using Decision Fusion. Computers 2024, 13, 294. https://doi.org/10.3390/computers13110294
Manalı D, Demirel H, Eleyan A. Deep Learning Based Breast Cancer Detection Using Decision Fusion. Computers. 2024; 13(11):294. https://doi.org/10.3390/computers13110294
Chicago/Turabian StyleManalı, Doğu, Hasan Demirel, and Alaa Eleyan. 2024. "Deep Learning Based Breast Cancer Detection Using Decision Fusion" Computers 13, no. 11: 294. https://doi.org/10.3390/computers13110294
APA StyleManalı, D., Demirel, H., & Eleyan, A. (2024). Deep Learning Based Breast Cancer Detection Using Decision Fusion. Computers, 13(11), 294. https://doi.org/10.3390/computers13110294






