Abstract
Deep learning models yield remarkable results in skin lesions analysis. However, these models require considerable amounts of data, while accessibility to the images with annotated skin lesions is often limited, and the classes are often imbalanced. Data augmentation is one way to alleviate the lack of labeled data and class imbalance. This paper proposes a new data augmentation method based on image fusion technique to construct large dataset on all existing tones. The fusion method consists of a pulse-coupled neural network fusion strategy in a non-subsampled shearlet transform domain and consists of three steps: decomposition, fusion, and reconstruction. The dermoscopic dataset is obtained by combining ISIC2019 and ISIC2020 Challenge datasets. A comparative study with current algorithms was performed to access the effectiveness of the proposed one. The first experiment results indicate that the proposed algorithm best preserves the lesion dermoscopic structure and skin tones features. The second experiment, which consisted of training a convolutional neural network model with the augmented dataset, indicates a more significant increase in accuracy by 15.69%, and 15.38% respectively for tanned, and brown skin categories. The model precision, recall, and F1-score have also been increased. The obtained results indicate that the proposed augmentation method is suitable for dermoscopic images and can be used as a solution to the lack of dark skin images in the dataset.
1. Introduction
Advances in neural network architecture, computation power, and access to big data have favored the application of computer vision to many tasks. Esteva et al. in [] have demonstrated the effectiveness of convolutional neural networks (CNN) in computer vision tasks such as skin lesion classification. CNNs identify and extract useful and best features to classify images. Research has shown that training deep models with millions of parameters requires relatively large-scale datasets to reach high accuracy. It is then according to [], a generally accepted notion that larger dataset improves classification performance.
However, assembling huge datasets can quickly become tricky due to the manual work required to collect and label the data. Building big medical image datasets is especially tricky due to the rarity of diseases, patient privacy [], requirement of medical experts for labeling, and the high cost of medical imaging acquisition systems. These obstacles have led to the creation of several data augmentation methods such as color space transformations, geometric transformations, kernel filters, mixing images, and random erasing []. More complex augmentation methods based on generative models and image fusion strategy have recently been developed for medical image classification, but these methods are not all adapted to all task [].
This paper proposes a novel image augmentation algorithm that combines the structure of a dermoscopic image with the color appearance of another to construct augmented images considering all the existing tones. The algorithm is based on a pulse-coupled neural network fusion strategy in a nonsubsampled shearlet transform domain.
The remainder of the paper is structured as follows. Section 2 is a comprehensive review of deep-learning-based data augmentation techniques and image fusion methods. Section 3 presents the proposed approach, followed by experimental results and discussion in Section 4. The article ends with the conclusion and future perspectives.
2. Literature Review
2.1. Data Augmentation Methods
In the literature, there are two categories of data augmentation. The first category is based on basic image manipulations, and the other category is based on deep learning.
Data augmentations from basic image manipulations commonly consist of image rotation, reflection, scaling (zoom in/out), shearing, histogram equalization, enhancing contrast or brightness, white balancing, sharpening, and blurring []. Those easy-to-understand methods have been proven to be fast, reproducible, and reliable and their implementation code is relatively easy and available to download for the most known deep learning frameworks, and thus more popular [].
The literature distinguishes two forms of deep-learning-based data augmentation: convolutional-layer-based methods and generative adversarial networks (GANs) based methods. Hui et al. in [] proposed a deep-learning-based method named Densefuse. The method combines convolutional layers and a dense block as encoder to extract deep features and convolutional layers as decoder to reconstruct the final fused image. Addition strategy and l1-norm strategy are used to combine features. The results indicate the effectiveness of the proposed architecture for infrared and visible image fusion tasks. Zhang et al. in [] proposed a method named IFCNN. This method framework consists of feature selection with convolutional layers, fusion rule, and features reconstruction with convolutional layers. Th results demonstrate good generalization potential. Subbiah Parvathy et al. in [] proposed a deep learning concepts based method that optimizes the threshold of fusion rules in shearlet transform. The proposed method has high efficiency for different input images.
GAN-based data augmentation originally proposed by Goodfellow et al. brought a breakthrough in the synthetic data generation research field. A GAN framework consists of two separate networks called the discriminator and generator, training competitively. According to Bowles et al. in [], GANs generate additional information from a dataset. The intended task of the discriminator is to distinguish synthesized samples from original ones, whereas the generator is tasked with generating realistic images that can fool the discriminator. Since then, GANs were introduced in 2014 [] and various works on GAN extensions such as DCGANs, CycleGANs, and progressively growing GANs [] were published in 2015, 2017, and 2018, respectively. In medical image analysis, GANs are widely used for image reconstruction [,,], segmentation [], classification [,], detection [], registration, and image synthesis such as brain MRI image [,], liver lesion [], and skin lesion synthesis [,].
Zhiwei Qin et al. [] proposed a style-based GANs model. The model modifies the structure of style control and noise input in the original generator, adjusts both the generator and discriminator to efficiently synthesize 256 × 256 skin lesion images. By adding the synthesized 800 melanoma images to the training set, the accuracy, sensitivity, specificity, average precision, and balanced multiclass accuracy of the classifier were improved by 1.6%, 24.4%, 3.6%, 23.2%, and 5.6% respectively. Alceu Bissoto and al. in [] proposed an image-to-image translation model named pix2pixHD. Instead of generating the image from noise (usual procedure with GANs), the model synthesizes new images from a semantic label map (segmentation mask) and an instance map (an image where each pixel belongs to a class). Synthetic images generated contain features that characterize a lesion as malignant or benign. Even more, synthetic images contain relevant features that improve the classification network by an average of 1.3 percentage points and keep the network more stable. Kora Venu et al. in [] generated X-ray images for the underrepresented class using a deep convolutional generative adversarial network (DCGAN). Experiments results show an improvement of a CNN classifier trained with the augmented data.
In conclusion, Table 1 presents the strengths and the limitations of deep-learning-based methods. As presented, several works applied deep-learning-based data augmentation to correct the class imbalance, by generating realistic images. However, although the generated images by GANs are realistic, there is a problem of partial collapse mode [,]. Mode collapse refers to scenarios in which the generator produces multiple images containing the same color or texture themes, which favors duplicates in generated images.

Table 1.
Summary of medical fusion methods and deep-learning-based data-augmentation methods.
2.2. Images Fusion Methods
Image fusion generates an informative image via the integration of images obtained from multiple source images in the same scene. The input source images in an image fusion system can be acquired either from various kinds of imaging sensors or from one sensor with different optical parameter settings. An efficient image fusion can preserve relevant features by extracting all important information from the images without producing any inconsistencies in the output image []. Image fusion techniques have been widely used in computer vision, surveillance, medical imaging, and remote sensing. According to the literature, there are two main branches of image fusion, namely the spatial domain method and the transform domain method []. Spatial domain methods consist in merging the source images without transformation by choosing the pixels regions or blocks. Transform domain fusion techniques consist of three steps: source images domain transformation, a fusion of corresponding transform coefficients, and finally inverse transformation to produce the fused image.
There are a variety of transforms that have been used for image fusion, such as those based on sparse representation [], discrete wavelet transform [,], curvelet transform, contourlet transform, dual-tree complex wavelet transform, non-subsampled contourlet transform [], and shearlet transform []. There are also a variety of fusion strategies that have been used for images fusion, such as sparse representation (SR) [,], enhanced sparse representation [], modified sum-modified Laplacian (SML) [], coupled neural P (CNP) systems [], pulse coupled neural network (PCNN) [], and PCNN variant [].
The literature on medical image fusion is growing and methods combining decomposition methods and fusion strategies have been proposed. Sarmad Maqsood et al. in [] proposed a method for computed tomography and magnetic resonance imaging images fusion. In this method, they have used spatial gradient-based edge detection technique to transform into detail layer and base layer each source image, an enhanced sparse representation approach as fusion strategy and have formed the fused image by linear integration of final fused detail layer and fused base layer. Five metrics, entropy, spatial structural similarity, mutual information, feature mutual information, and visual information fidelity, were used to confirm the superiority of the proposed method on other methods. Yuanyuan Li et al. in [] proposed a fusion technique based on non-subsampled contour transformation (NSCT) and sparse representation (SR). NSCT is applied for the source images decomposition to obtain the corresponding low pass and high pass coefficients. The low pass and high pass coefficients are fused using SR and the sum-modified Laplacian (SML), respectively. The final fused image is obtained by applying the inverse transform on the fused coefficients. Experiments show that the proposed solutions achieve better performance on structural similarity and detail preservation in fused images. Similarly, Li Liangliang et al. in [] applied NSCT for images decomposition and refine the fused image based on energy of the gradient (EOG). Visual results and evaluation fusion metrics results show a significant performance of the proposed technique. Xiaosong et al. in [] proposed an image fusion and denoising method that decomposes images into high-frequency layer, low-frequency structure and low-frequency texture. They applied sparse representation, absolute maximum, and neighborhood spatial frequency as fusion rules on the different layers respectively to generate the fused layers. The fusion result is finally obtained by reconstructing the three fused layers. The results show that the method responds well to noisy image fusion problems. Bo Li et al. in [] proposed a method based on coupled neural P (CNP) systems in the NSST domain. They first compared the method to others fusion methods and then compare the method to deep-learning-based fusion methods. Experimental results have demonstrated the advantages of the proposed fusion method for multimodality medical images fusion. Shehanaz, S. et al. in [] proposed a multimodalities fusion method based on discrete wavelet transform (DWT) for image decomposition and using particle swarm optimization for optimal fusion of coefficients. Wang et al. [] proposed an image fusion method based on wavelet transformation. The method also uses the discrete wavelet transform (DWT) to decompose the source images, then fuse the coefficients with dual-channel pulse coupled neural network (PCNN) and applied inverse DWT for fused image reconstruction. The effectiveness of the proposed method was demonstrated by experimental comparisons of different fusion methods. Li et al. in [] combined PCNN and weighted sum of eight neighborhood-based modified Laplacian (WSEML) integrating guided image filtering (GIF) fusion rules in non-subsampled contourlet transform (NSCT) domain. The proposed method fused multimodal medical images well.
In conclusion, according to the literature, there are several fusion methods, and their efficiency depends on the decomposition method and the fusion strategy. Table 1 summarizes the strengths and limitations of the presented image fusion methods. The main advantages of fusion methods are that they preserve features, are easy to implement and fast.
3. Proposed Method
This paper proposes a solution to correct skin tones imbalance observe in all skin lesion datasets. The proposed method illustrated in Figure 1 is composed of two main parts. The first part consists of source images decomposition and fused coefficients reconstruction using non-subsampled shearlet transform (NSST) because NSST-based algorithms are shift invariant and can eliminate edge effects efficiently. The second one performs coefficients fusion using an updated pulse-coupled neural network (PCNN) for its simplicity, speed, and efficiency.
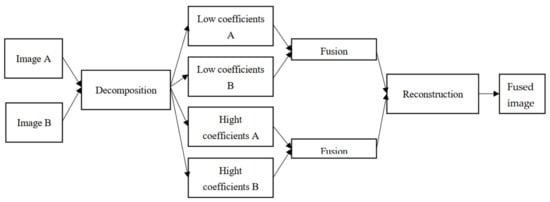
Figure 1.
Schematic diagram of proposed method.
3.1. Non-Subsampled Shearlet Transform (NSST)
NSST, as shown in Figure 2, is a multiscale decomposition used to efficiently represent high and low-frequency information of source image []. Firstly, the source image is decomposed into low-pass and high-pass bands using the non-subsampled Laplacian pyramid (NLSP) transform. For each level of decomposition, the high-pass bands are submitted to translation invariance shearlet filters and the low-pass bands are further decomposed into low-pass and high-pass bands for the following level. Then inverse NSST transform is applied by taking the sum of all shift-invariant shearlet filter responses at the respective levels of decomposition, and inverse non-subsampled Laplacian pyramid transform is finally applied to get the reconstructed image.
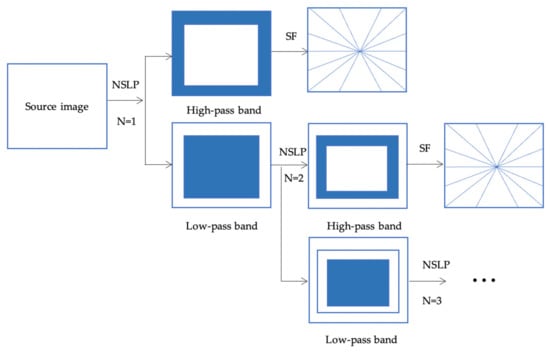
Figure 2.
NSST decomposition process.
3.2. Pulse Coupled Neural Network (PCNN)
The PCNN introduced by Johnson, J.L. et al. [] is a neuron based on the visual cortex of small mammals as cats and is composed of three modules: the receptive field, the modulation field, and the pulse generator [].
PCNN is a two-dimensional M × N network, in which each neuron corresponds to a specific pixel of the image. Figure 3 presents an original PCNN neuron. By an iterative calculation combining these different modules, the following equations are used to activate the neuron. The index
refers to a pixel location in image, refers to neighborhood pixels around a pixel, and denotes the current iteration.
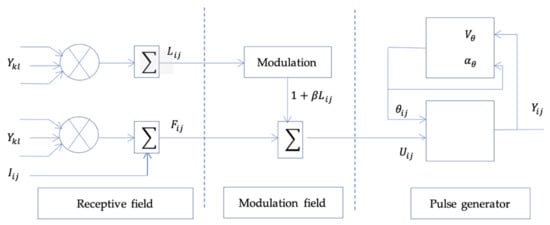
Figure 3.
Original PCNN neuron.
The receptive field described by Equations (1) and (3), consists of and channel. , the linking parameter receives local stimulus from surrounding neurons. On simplified PCNN neuron the feeding neuron receives external stimulus from the input signal. On original PCNN is described by (2).
The modulation field consists of as presented in Equation (4). the internal activation modulates the information of the above module with the linking strength.
Pulse generator is described in Equations (5) and (6). The output module compares with the dynamic threshold. If is larger than , then the neuron is activated and generate a pulse, which is characterized by , otherwise . The excitation time of each neuron is denoting represented in Equation (7).
The weight matrices and are local interconnections and is a large impulse, and are the magnitudes scaling terms. and are the time decayed constants associated with , and respectively.
According to Equation (4), the parameter β highly influence the neuron internal activation. The PCNN neuron has been improved by changing β as adaptive local value instead of global value. As demonstrated in Equation (8), it consists in using a sigmoid function to normalize between 0 and 1 the gradient magnitude of 3 × 3 local region of source images.
The linking strength is therefore dynamically adjusted according to the magnitude gradient. This modification allows the PCNN model to better preserve the image details in the final image.
3.3. Detailed Algorithm
The schematic diagram of the proposed data augmentation method is shown in Figure 1. The proposed algorithm can be summarized as the following steps in Algorithm 1.
| Algorithm 1. Proposed fusion algorithm |
| Input: Dermoscopic images in dataset A and unaffected darker tones images in dataset B Output: Fused image Step 1: Image decomposition with NSST |
| Randomly select source images and decompose each source image into five levels with NSST, to obtain (,) and (,). and are the low-frequency coefficients of A and B. and represent the l-th high-frequency sub-band coefficients in the kth decomposition layer of A and B. |
| Step 2: Fusion strategy |
| The low-frequency sub-band contains texture structure and background of source images. The fused low-frequency coefficients are obtained as follows: |
| High-frequency sub-bands contain information about details in images. The fused high-frequency sub-bands are obtained by computing the following operations on each pixel of each high-frequency sub-bands. |
| 1. Normalize between 0 and 1 high-frequency coefficients; |
| 2. Initialize , , and to 0 and to 1 to accelerate neurons activation; |
| 3. Stimulate the PCNN respectively with the normalized coefficients; |
| 4. Compute , , , and until the neuron is activated (equal to 1); |
| 5. Fuse high-frequency sub-bands coefficients as follows: |
| Step 3: Image reconstruction with inverse NSST |
4. Experimental Results and Discussion
4.1. Dataset
The dataset contained two source images: source A and source B. Source A images were dermoscopic images of melanomas and nevus obtained by combining the ISIC2019 and ISIC2020 Challenge datasets [,,]. Source B images were RGB images of darker skin tones. Some of the source images used in the experiments are shown in Figure 4.
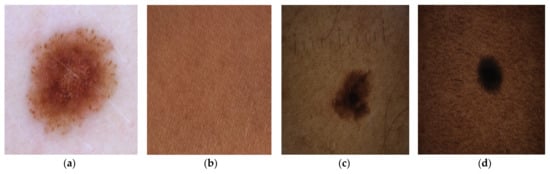
Figure 4.
(a) Source A dermoscopic features of melanoma in very light skin [,,]. (b) Source B healthy brown skin image. (c) Dermoscopic features of melanocytic nevus in tanned skin [,,]. (d) Dermoscopic features of melanocytic nevus in brown skin [,,].
To quantify the skin tone categories present in source A dataset, skin images were segmented to extract non diseased regions and the individual typological angle (ITA) metric [] was used to characterize the skin tone of that region. ITA is an objective classification tool computed from images. The pixels from the nonaffected part are converted to CIELab-space to obtain the luminance L of each pixel and b the amount of yellow in each pixel. The mean ITA value is in degrees and is calculated using Equation (11) []. As presented in Table 2, ITA values classify skin tones into six categories: very light, light, intermediate, tanned, brown, and dark.

Table 2.
Skin color categories based on the individual typology angle (ITA°) [].
In Table 1, source A images are categorized into six groups: very light (1901 images), light (8956 images), intermediate (471 images), tanned (51 images), brown (13 images), and undefined (46 images). The results show that darker tones are underrepresented with 94.92% of lighter tone images (very light and light categories) compared to 4.68% darker tone images (intermediate, tanned, brown, and dark categories). Matthew Groh et al. in [] also reveal the imbalance of skin types in the Fitzpatrick 17k dataset and in datasets in general.
4.2. Experimental Setup
The proposed method was compared to other fusion approaches to certify its efficiency and superiority. The comparative study was performed with color transfer (CT) [] method, wavelet and sparse representation-based method (DWT_SR) [], wavelet and color transfer-based method (DWT_CT), sparse representation, and sum-modified Laplacian in NSCT domain-based method (NSCT_SR_SML) [] and the proposed method (NSST_PCNN).
Six objective image evaluation metrics were adopted for quantitative evaluation: gradient-based fusion performance QG [] to evaluate the amount of edge information that is transferred from sources images to the fused image; QS [], QC [], and QY [] to evaluate similarities between saliency maps and structural information of the fused image and sources images; and the Chen–Blum metric QCB [], which is a human perception inspired fusion metric to evaluate the human visualization performance of fused images.
Additionally, the augmented dataset and the real dataset were used separately to train two Gabor-based convolutional neural network [] inspired models. A comparative study was performed on the accuracy, precision, recall, and F1 score of each model for the different skin tones to assess the effectiveness of the proposed data augmentation method on skin lesion classification for underrepresenting skin tones.
Experiments were conducted by MATLAB R2020b with an Apple M1 chip, eight cores and 16GB memory. A five-level NSST decomposition was performed in source images. For PCNN, the number of iterations was set to 100 and the parameters were set as .
4.3. Results and Discussion
4.3.1. Visual and Qualitative Evaluation
A visual quality comparison of three fused images using six different methods is displayed in Figure 5. Figure 5(a1)–(a3) indicates dermoscopic features in very light and light skin tones. Images obtained by CT, DWT_SR, DWT_CT, NSCT_SR_SML, and proposed method NSST_PCNN respectively are displayed in Figure 5(b1)–(f3).
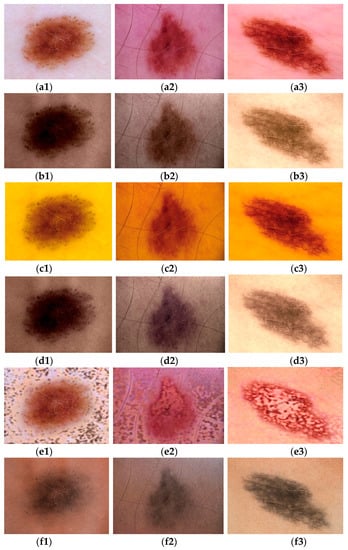
Figure 5.
(a1–a3) Dermoscopic source A images [,,]. (b1–f3) Results obtained using respectively CT, DWT_SR, DWT_CT, NSCT_SR_SML, and the proposed method light skin tones. Images obtained by CT, DWT_SR, DWT_CT, NSCT_SR_SML, and proposed method NSST_PCNN respectively are displayed in Figure 5(b1)–(f3).
The result obtained by NSCT_SR_SML was unnatural. Compared with DWT_SR, the proposed method combined input images effectively and preserved distinctly dermoscopic structures such as pigment networks, amorphous structureless areas (blotches), and dots and globules between the dermoscopic images, as seen in Figure 5(a1)–(a3) and the generated images Figure 5(f1)–(f3). Compared to CT and DWT_CT, the proposed method achieved better performance on skin tone preservation. Visually, the pigmentation of Figure 5(a1)–(a3) lesions was different for the fused images in Figure 5(f1)–(f3) but, according to [], the result is real as skin lesions on dark skin are characterized by central hyperpigmentation and a dark brown peripheral network.
Although visual evaluation results show that the dermoscopic structures were preserved, it should be noted that visual evaluation is a subjective method. Table 3 lists the results of five objective evaluation metrics applied on different methods. QG, QS, QC, QY metrics assess the amount of transferred edge information and similarities between fused and source images, the highest values of the metrics are 1. The proposed method, compared to other methods, exceeded on average in QG, QS, and QCB metrics. This performance was followed by those of the DWT_CT and DWT_SR methods, which showed better results for the QC and QY metrics respectively. It can then be indicated that result images obtained by the proposed method better preserved details and similarity with source images. Furthermore, human visualization performance metric QCB values support the pigmentation difference observed on visual evaluation. The fused images were not just duplicates of skin lesions images.

Table 3.
Evaluation metrics results of different methods.
Table 4 shows the average time taken by each algorithm to generate an image. The results indicate that the proposed method had a longer execution time than most methods. This high value can be explained by the fact that the linking strength is adaptive, which increases the computation time.

Table 4.
Execution time.
4.3.2. Impact of Data Augmentation in Skin Lesion Classification on All Existing Tones
First, a convolutional neural network inspired by the model proposed in [] was trained with 80% of the dataset and tested with 20% of the dataset. The accuracy, precision, recall, and F1-score of Model 1 are reported in Table 5.

Table 5.
Evaluation metrics results of model 1.
Second, the training model was reinforced with the augmented images and then tested with the same 20% of the dataset. To verify the neural network’s generalization, the models were tested with only real images. The accuracy, precision, recall, and F1-score of model 2 are reported in Table 6.

Table 6.
Evaluation metrics results of model 2.
Figure 6 shows the accuracy, precision, recall, and F1-score of models based on these experiments. Compared to model 1, model 2’s accuracy increased by 2.22%, 0.73%, 1.67%, 15.69%, and 15.38% respectively for the very light, light, intermediate, tanned and brown categories. The model precision also increased by 1.36%, 0.68%, 4.00%, 13.04%, and 16.67% respectively on different categories. Similarly, for true positive recall and F1_score, the results are more significant with a larger increase observed in medium-brown and dark-brown tones. As for GAN-based models proposed by Zhiwei Qin et al. in [] and Alceu Bissoto and al. in [], data augmentation improved the model, with the particularity that the proposed data augmentation also corrected skin tone imbalance. Data augmentation therefore favored the reinforcement and generalization of the classifier. Introducing new images inspired by real images but which also contain other features, made it possible to promote the generalization of the classifier.
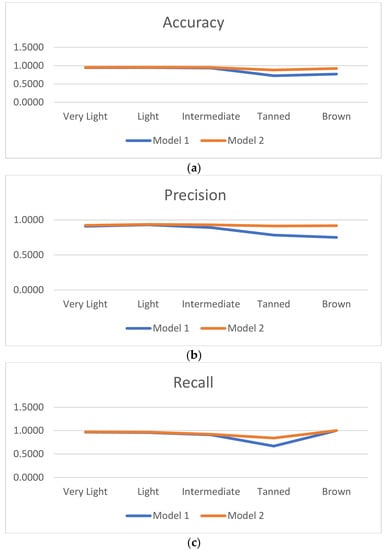
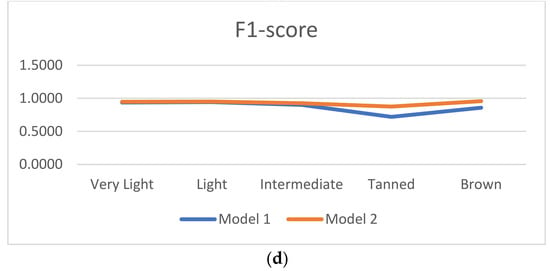
Figure 6.
Comparison of evaluation metrics results. (a) Accuracy, (b) precision, (c) recall and (d) F1-score.
5. Conclusions and Future Works
In this paper, a data augmentation method based on multiscale image decomposition and PCNN fusion strategy is proposed as a solution to alleviate the lack of labeled dermoscopic data in all existing skin tones. Compared to the existing methods, the proposed method presented more informative dermoscopic structure and detail. Particularly, the proposed method has the advantage of being suitable for dermoscopic images. Experiment results also prove this method improved the performance and accuracy of a convolutional neural network-based skin lesion classifier, even for under-represented skin tones. To conclude, this work is innovative because the proposed method is simple, does not require training, and effectively augments dermoscopic images.
A limitation of this method is the computation time. The high value can be explained by the fact that the linking strength is adaptive, which increases the computation time. Therefore, reducing the processing time of the algorithm is an improvement that can be made in the future. Future work will also focus on applying the proposed algorithm in other areas of medical imaging to test and improve the efficiency and generalization of the algorithm. Finally, the development of these results should also focus on increasing the skin lesion dataset and strengthening skin lesion classifiers on the darker tone categories.
Author Contributions
Conceptualization, E.C.A. and A.T.S.M.; methodology, E.C.A. and A.T.S.M.; software, E.C.A.; validation, E.C.A.; A.T.S.M. and P.G.; formal analysis, E.C.A.; A.T.S.M. and P.G.; resources, E.C.A.; data curation, E.C.A.; writing—original draft preparation, E.C.A.; writing—review and editing, E.C.A.; A.T.S.M. and P.G.; visualization, E.C.A. and A.T.S.M.; supervision, E.C.A.; A.T.S.M.; P.G. and J.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: https://challenge2020.isic-archive.com (accessed on 9 February 2022), https://challenge2019.isic-archive.com/data.htm (accessed on 9 February 2022).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.; Li, X.; Wang, L.; He, J.; Li, D.; Zhou, J. How Does the Data set Affect CNN-based Image Classification Performance? In Proceedings of the 2018 5th International Conference on Systems and Informatics (ICSAI), Nanjing, China, 10–12 November 2018; pp. 361–366. [Google Scholar] [CrossRef]
- Cunniff, C.; Byrne, J.L.; Hudgins, L.M.; Moeschler, J.B.; Olney, A.H.; Pauli, R.M.; Seaver, L.H.; Stevens, C.A.; Figone, C. Informed consent for medical photographs. Genet. Med. 2000, 2, 353–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simard, P.Y.; Steinkraus, D.; Platt, J.C. Best practices for convolutional neural networks applied to visual document analysis. In Proceedings of the 7th International Conference on Document Analysis and Recognition, Edinburgh, UK, 3–6 August 2003; pp. 958–963. [Google Scholar] [CrossRef]
- Li, S.; Kang, X.; Fang, L.; Hu, J.; Yin, H. Pixel-level image fusion: A survey of the state of the art. Inf. Fusion 2017, 33, 100–112. [Google Scholar] [CrossRef]
- Mikołajczyk, A.; Grochowski, M. Data augmentation for improving deep learning in image classification problem. In Proceedings of the International Interdisciplinary PhD Workshop (IIPhDW), Swinoujscie, Poland, 9–12 May 2018; pp. 117–122. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, B.; Wang, L.; Zu, C.; Lalush, D.S.; Lin, W.; Wu, X.; Zhou, J.; Shen, D.; Zhou, L. 3D conditional generative adversarial networks for high-quality PET image estimation at low dose. NeuroImage 2018, 174, 550–562. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wu, X.-J. DenseFuse: A Fusion Approach to Infrared and Visible Images. IEEE Trans. Image Process. 2019, 28, 2614–2623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Liu, Y.; Sun, P.; Yan, H.; Zhao, X.; Zhang, L. IFCNN: A general image fusion framework based on convolutional neural network. Inf. Fusion 2020, 54, 99–118. [Google Scholar] [CrossRef]
- Parvathy, V.S.; Pothiraj, S.; Sampson, J. A novel approach in multimodality medical image fusion using optimal shearlet and deep learning. Int. J. Imaging Syst. Technol. 2020, 30, 847–859. [Google Scholar] [CrossRef]
- Bowles, C.; Chen, L.; Guerrero, R.; Bentley, P.; Gunn, R.; Hammers, A.; Dickie, D.A.; Hernández, M.V.; Wardlaw, J.; Rueckert, D. GAN Augmentation: Augmenting Training Data using Generative Adversarial Networks. arXiv 2018, arXiv:1810.10863. [Google Scholar]
- Goodfellow, I.J.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative Adversarial Networks. arXiv 2014, arXiv:1406.2661. [Google Scholar] [CrossRef]
- Karras, T.; Aila, T.; Laine, S.; Lehtinen, J. Progressive Growing of Gans for Improved Quality, Stability, and Variation. arXiv 2018, arXiv:1710.10196. [Google Scholar]
- Wolterink, J.M.; Leiner, T.; Viergever, M.A.; Isgum, I. Generative Adversarial Networks for Noise Reduction in Low-Dose CT. IEEE Trans. Med. Imaging 2017, 36, 2536–2545. [Google Scholar] [CrossRef] [PubMed]
- Shitrit, O.; Raviv, T.R. Accelerated Magnetic Resonance Imaging by Adversarial Neural Network. In Lecture Notes in Computer Science; Metzler, J.B., Ed.; Springer: Cham, Switzerland, 2017; pp. 30–38. [Google Scholar]
- Mahapatra, D.; Bozorgtabar, B. Retinal Vasculature Segmentation Using Local Saliency Maps and Generative Adversarial Networks For Image Super Resolution. arXiv 2018, arXiv:1710.04783. [Google Scholar]
- Madani, A.; Moradi, M.; Karargyris, A.; Syeda-Mahmood, T. Chest x-ray generation and data augmentation for cardiovascular abnormality classification. In Medical Imaging 2018: Image Processing; SPIE Medical Imaging: Houston, TX, US, 2018; p. 105741M. [Google Scholar] [CrossRef]
- Lu, C.-Y.; Rustia, D.J.A.; Lin, T.-T. Generative Adversarial Network Based Image Augmentation for Insect Pest Classification Enhancement. IFAC-PapersOnLine 2019, 52, 1–5. [Google Scholar] [CrossRef]
- Chuquicusma, M.J.M.; Hussein, S.; Burt, J.; Bagci, U. How to fool radiologists with generative adversarial networks? A visual turing test for lung cancer diagnosis. arXiv 2018, arXiv:1710.09762. [Google Scholar]
- Calimeri, F.; Marzullo, A.; Stamile, C.; Terracina, G. Biomedical Data Augmentation Using Generative Adversarial Neural Networks. In Artificial Neural Networks and Machine Learning—ICANN 2017, Proceedings of the 26th International Conference on Artificial Neural Networks, Alghero, Italy, 11–14 September 2017; Springer: Cham, Switzerland, 2017; pp. 626–634. [Google Scholar] [CrossRef]
- Plassard, A.J.; Davis, L.T.; Newton, A.T.; Resnick, S.M.; Landman, B.A.; Bermudez, C. Learning implicit brain MRI manifolds with deep learning. In Medical Imaging 2018: Image Processing; International Society for Optics and Photonics: Bellingham, WA, USA, 2018; Volume 10574, p. 105741L. [Google Scholar] [CrossRef]
- Frid-Adar, M.; Diamant, I.; Klang, E.; Amitai, M.; Goldberger, J.; Greenspan, H. GAN-based synthetic medical image augmentation for increased CNN performance in liver lesion classification. Neurocomputing 2018, 321, 321–331. [Google Scholar] [CrossRef] [Green Version]
- Ding, S.; Zheng, J.; Liu, Z.; Zheng, Y.; Chen, Y.; Xu, X.; Lu, J.; Xie, J. High-resolution dermoscopy image synthesis with conditional generative adversarial networks. Biomed. Signal Process. Control 2021, 64, 102224. [Google Scholar] [CrossRef]
- Bissoto, A.; Perez, F.; Valle, E.; Avila, S. Skin lesion synthesis with Generative Adversarial Networks. In OR 2.0 Context-Aware Operating Theaters, Computer Assisted Robotic Endoscopy, Clinical Image-Based Procedures, and Skin Image Analysis; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2018; Volume 11041, pp. 294–302. [Google Scholar] [CrossRef] [Green Version]
- Qin, Z.; Liu, Z.; Zhu, P.; Xue, Y. A GAN-based image synthesis method for skin lesion classification. Comput. Methods Programs Biomed. 2020, 195, 105568. [Google Scholar] [CrossRef]
- Venu, S.K.; Ravula, S. Evaluation of Deep Convolutional Generative Adversarial Networks for Data Augmentation of Chest X-ray Images. Future Internet 2020, 13, 8. [Google Scholar] [CrossRef]
- Goodfellow, I. NIPS 2016 Tutorial: Generative Adversarial Networks. arXiv 2016, arXiv:1701.00160. [Google Scholar]
- Wang, K.; Gou, C.; Duan, Y.; Lin, Y.; Zheng, X.; Wang, F.-Y. Generative adversarial networks: Introduction and outlook. IEEE/CAA J. Autom. Sin. 2017, 4, 588–598. [Google Scholar] [CrossRef]
- Wang, N.; Wang, W. An image fusion method based on wavelet and dual-channel pulse coupled neural network. In Proceedings of the 2015 IEEE International Conference on Progress in Informatics and Computing (PIC), Nanjing, China, 18–20 December 2015; pp. 270–274. [Google Scholar]
- Li, Y.; Sun, Y.; Huang, X.; Qi, G.; Zheng, M.; Zhu, Z. An Image Fusion Method Based on Sparse Representation and Sum Modified-Laplacian in NSCT Domain. Entropy 2018, 20, 522. [Google Scholar] [CrossRef] [Green Version]
- Biswas, B.; Sen, B.K. Color PET-MRI Medical Image Fusion Combining Matching Regional Spectrum in Shearlet Domain. Int. J. Image Graph. 2019, 19, 1950004. [Google Scholar] [CrossRef]
- Li, B.; Peng, H.; Luo, X.; Wang, J.; Song, X.; Pérez-Jiménez, M.J.; Riscos-Núñez, A. Medical Image Fusion Method Based on Coupled Neural P Systems in Nonsubsampled Shearlet Transform Domain. Int. J. Neural Syst. 2021, 31, 2050050. [Google Scholar] [CrossRef]
- Li, L.; Ma, H. Pulse Coupled Neural Network-Based Multimodal Medical Image Fusion via Guided Filtering and WSEML in NSCT Domain. Entropy 2021, 23, 591. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Ma, H.; Jia, Z.; Si, Y. A novel multiscale transform decomposition based multi-focus image fusion framework. Multimed. Tools Appl. 2021, 80, 12389–12409. [Google Scholar] [CrossRef]
- Li, X.; Zhou, F.; Tan, H. Joint image fusion and denoising via three-layer decomposition and sparse representation. Knowl.-Based Syst. 2021, 224, 107087. [Google Scholar] [CrossRef]
- Shehanaz, S.; Daniel, E.; Guntur, S.R.; Satrasupalli, S. Optimum weighted multimodal medical image fusion using particle swarm optimization. Optik 2021, 231, 166413. [Google Scholar] [CrossRef]
- Maqsood, S.; Javed, U. Multi-modal Medical Image Fusion based on Two-scale Image Decomposition and Sparse Representation. Biomed. Signal Process. Control 2020, 57, 101810. [Google Scholar] [CrossRef]
- Qi, G.; Hu, G.; Mazur, N.; Liang, H.; Haner, M. A Novel Multi-Modality Image Simultaneous Denoising and Fusion Method Based on Sparse Representation. Computers 2021, 10, 129. [Google Scholar] [CrossRef]
- Wang, M.; Tian, Z.; Gui, W.; Zhang, X.; Wang, W. Low-Light Image Enhancement Based on Nonsubsampled Shearlet Transform. IEEE Access 2020, 8, 63162–63174. [Google Scholar] [CrossRef]
- Johnson, J.L.; Ritter, D. Observation of periodic waves in a pulse-coupled neural network. Opt. Lett. 1993, 18, 1253–1255. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.; Zhang, L.; Lei, Y. Novel fusion method for visible light and infrared images based on NSST–SF–PCNN. Infrared Phys. Technol. 2014, 65, 103–112. [Google Scholar] [CrossRef]
- Tschandl, P.; Rosendahl, C.; Kittler, H. The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci. Data 2018, 5, 180161. [Google Scholar] [CrossRef] [PubMed]
- Codella, N.C.F.; Gutman, D.; Emre Celebi, M.; Helba, B.; Marchetti, M.A.; Dusza, S.W.; Kalloo, A.; Liopyris, K.; Mishra, N.; Kittler, H. Skin Lesion Analysis Toward Melanoma Detection: A Challenge at the 2017 International Symposium on Biomedical Imaging (ISBI), Hosted by the International Skin Imaging Collaboration (ISIC). arXiv 2017, arXiv:1710.05006. [Google Scholar]
- Combalia, M.; Codella, N.C.F.; Rotemberg, V.; Helba, B.; Vilaplana, V.; Reiter, O.; Carrera, C.; Barreiro, A.; Halpern, A.C.; Puig, S.; et al. BCN20000: Dermoscopic Lesions in the Wild. arXiv 2019, arXiv:1908.02288. [Google Scholar]
- Kinyanjui, N.M.; Odonga, T.; Cintas, C.; Codella, N.C.F.; Panda, R.; Sattigeri, P.; Varshney, K.R. Fairness of Classifiers Across Skin Tones in Dermatology. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2017; Springer: Cham, Switzerland, 2020; pp. 320–329. [Google Scholar]
- Groh, M.; Harris, C.; Soenksen, L.; Lau, F.; Han, R.; Kim, A.; Koochek, A.; Badri, O. Evaluating Deep Neural Networks Trained on Clinical Images in Dermatology with the Fitzpatrick 17k Dataset. arXiv 2021, arXiv:2104.09957. [Google Scholar]
- Xiao, Y.; Decenciere, E.; Velasco-Forero, S.; Burdin, H.; Bornschlogl, T.; Bernerd, F.; Warrick, E.; Baldeweck, T. A New Color Augmentation Method for Deep Learning Segmentation of Histological Images. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 886–890. [Google Scholar]
- Liu, Z.; Blasch, E.; Xue, Z.; Zhao, J.; Laganiere, R.; Wu, W. Objective Assessment of Multiresolution Image Fusion Algorithms for Context Enhancement in Night Vision: A Comparative Study. IEEE Trans. Pattern Anal. Mach. Intell. 2012, 34, 94–109. [Google Scholar] [CrossRef]
- Adjobo, E.C.; Mahama, A.T.S.; Gouton, P.; Tossa, J. Proposition of Convolutional Neural Network Based System for Skin Cancer Detection. In Proceedings of the 2019 15th International Conference on Signal-Image Technology & Internet-Based Systems (SITIS), Sorrento, Italy, 26–29 November 2019; pp. 35–39. [Google Scholar]
- Lallas, A.; Reggiani, C.; Argenziano, G.; Kyrgidis, A.; Bakos, R.; Masiero, N.; Scheibe, A.; Cabo, H.; Ozdemir, F.; Sortino-Rachou, A.; et al. Dermoscopic nevus patterns in skin of colour: A prospective, cross-sectional, morphological study in individuals with skin type V and VI. J. Eur. Acad. Dermatol. Venereol. 2014, 28, 1469–1474. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).