Stabilization of Nucleosomes by Histone Tails and by FACT Revealed by spFRET Microscopy
Abstract
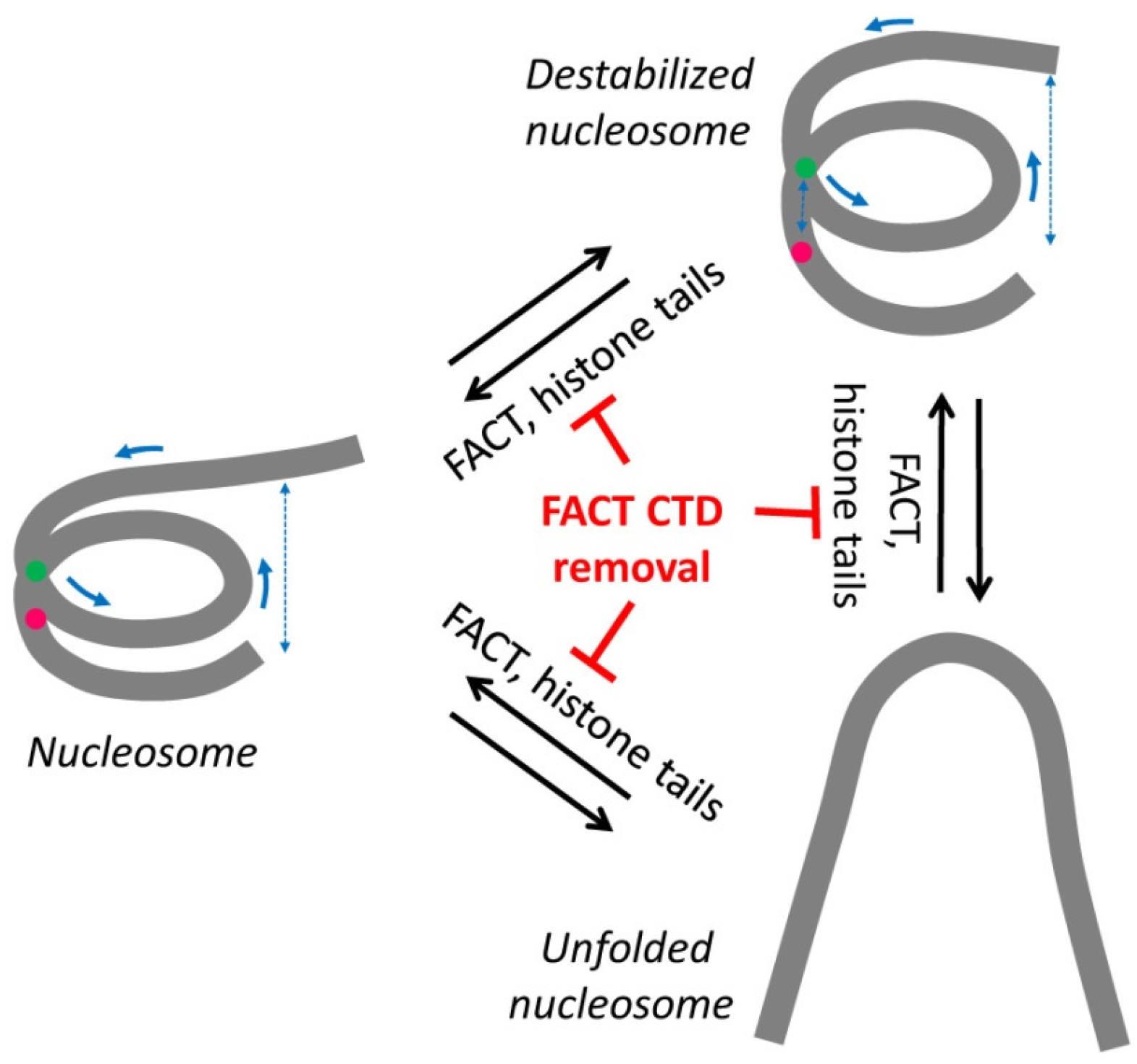
:1. Introduction
2. Results
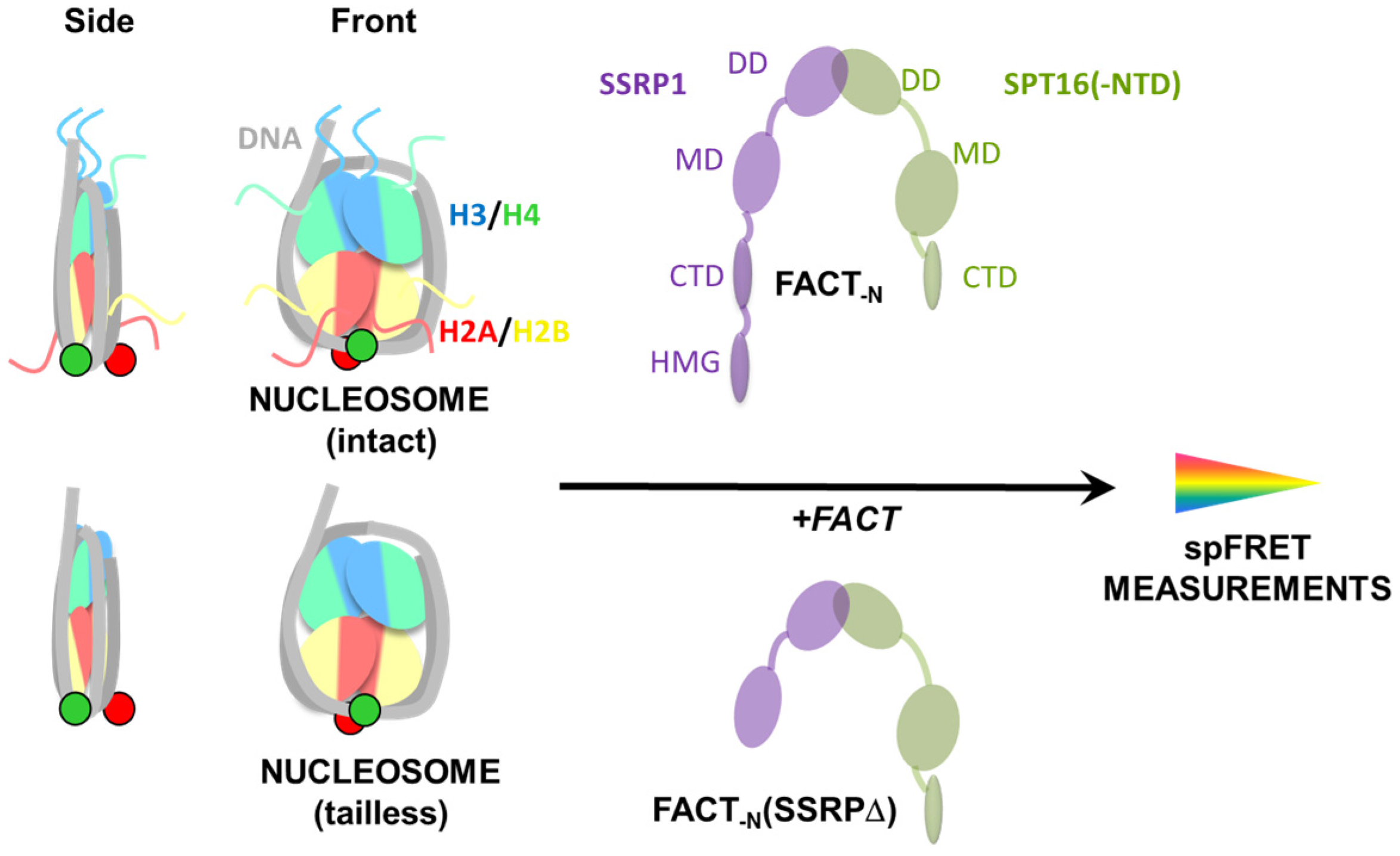
2.1. Experimental System
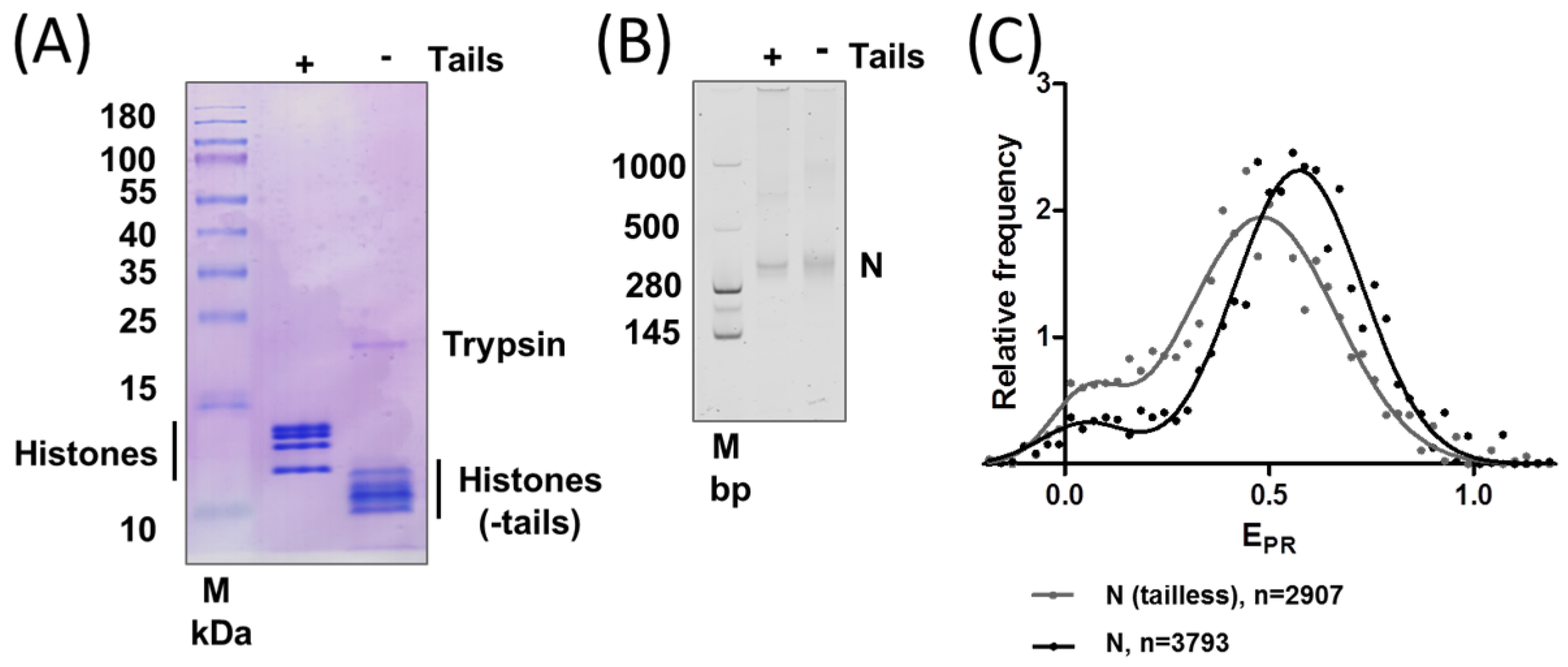
2.2. Removal of Histone Tails Affects the Structure of Nucleosomes
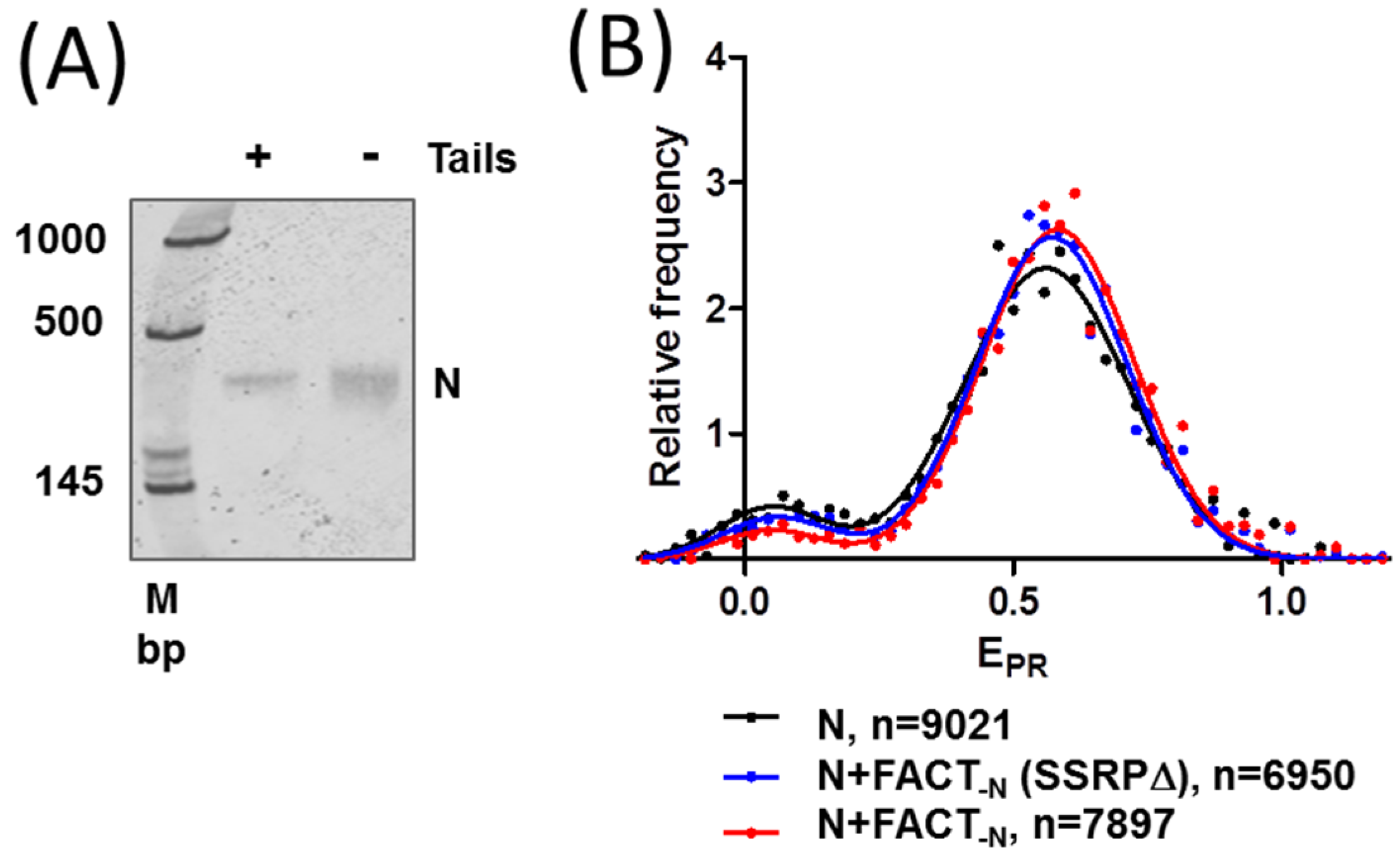
2.3. FACT Minimally Affects the Structure of Intact Nucleosomes
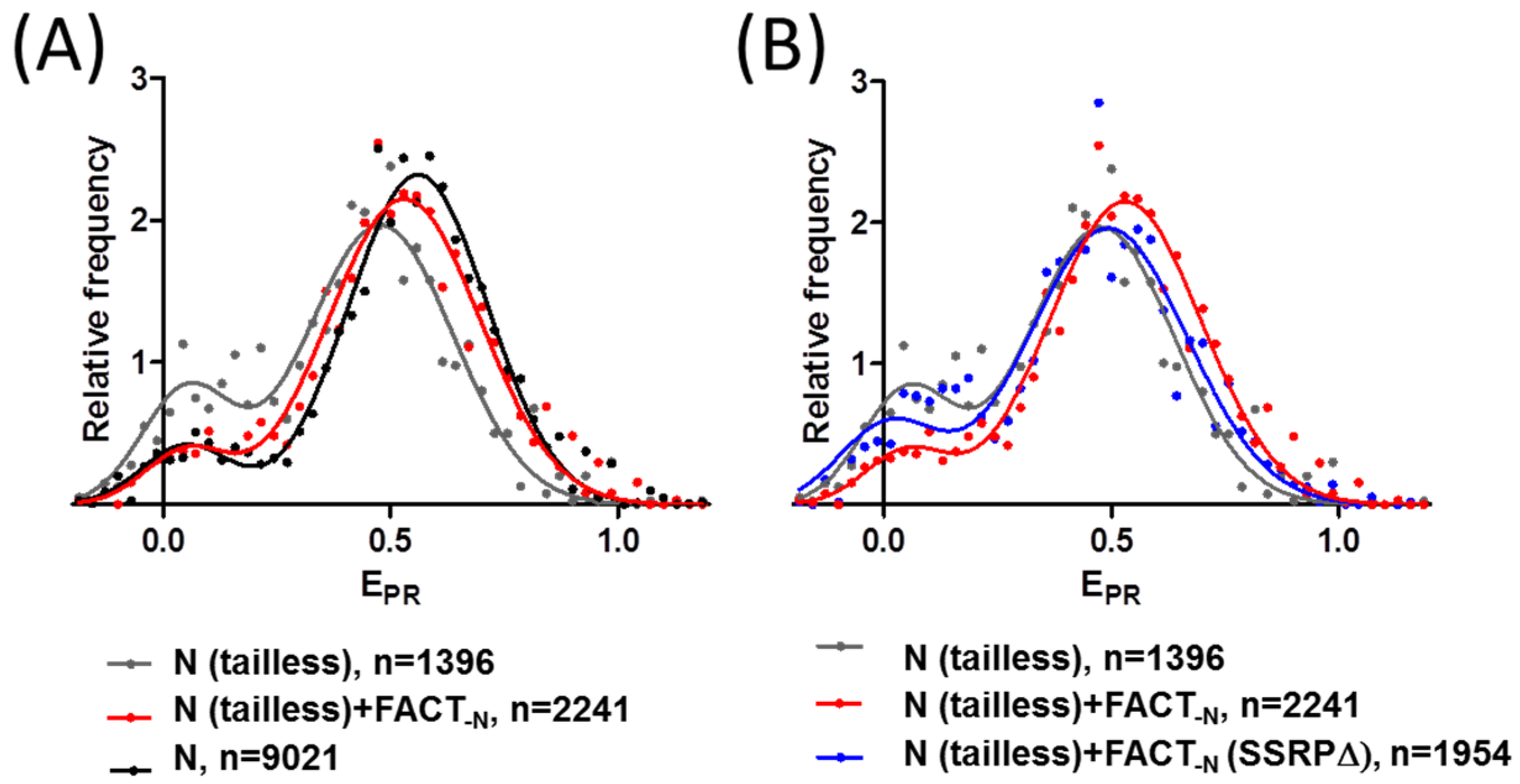
2.4. FACT Stabilizes Tailless Nucleosomes
3. Discussion
4. Materials and Methods
4.1. Purification of human FACT
4.2. DNA Templates
4.3. Purification and Tryptic Cleavage of the Donor Chromatin
4.4. Nucleosome Assembly and Purification
4.5. Incubation of the Nucleosomes with FACT
4.6. spFRET Measurements
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Maluchenko, N.V.; Chang, H.W.; Kozinova, M.T.; Valieva, M.E.; Gerasimova, N.S.; Kitashov, A.V.; Kirpichnikov, M.P.; Georgiev, P.G.; Studitsky, V.M. Inhibiting the pro-tumor and transcription factor FACT: Mechanisms. Mol. Biol. (Mosk.) 2016, 50, 599–610. [Google Scholar] [CrossRef] [PubMed]
- Gasparian, A.V.; Burkhart, C.A.; Purmal, A.A.; Brodsky, L.; Pal, M.; Saranadasa, M.; Bosykh, D.A.; Commane, M.; Guryanova, O.A.; Pal, S.; et al. Curaxins: Anticancer compounds that simultaneously suppress NF-κB and activate p53 by targeting FACT. Sci. Transl. Med. 2011, 3, 95ra74. [Google Scholar] [CrossRef] [PubMed]
- Garcia, H.; Miecznikowski, J.C.; Safina, A.; Commane, M.; Ruusulehto, A.; Kilpinen, S.; Leach, R.W.; Attwood, K.; Li, Y.; Degan, S.; et al. Facilitates chromatin transcription complex is an “accelerator” of tumor transformation and potential marker and target of aggressive cancers. Cell Rep. 2013, 4, 159–173. [Google Scholar] [CrossRef] [PubMed]
- Burkhart, C.; Fleyshman, D.; Kohrn, R.; Commane, M.; Garrigan, J.; Kurbatov, V.; Toshkov, I.; Ramachandran, R.; Martello, L.; Gurova, K.V. Curaxin CBL0137 eradicates drug resistant cancer stem cells and potentiates efficacy of gemcitabine in preclinical models of pancreatic cancer. Oncotarget 2014, 5, 11038–11053. [Google Scholar] [CrossRef] [PubMed]
- Formosa, T.; Ruone, S.; Adams, M.D.; Olsen, A.E.; Eriksson, P.; Yu, Y.; Rhoades, A.R.; Kaufman, P.D.; Stillman, D.J. Defects in SPT16 or POB3 (yFACT) in Saccharomyces cerevisiae cause dependence on the Hir/Hpc pathway: Polymerase passage may degrade chromatin structure. Genetics 2002, 162, 1557–1571. [Google Scholar] [PubMed]
- Jamai, A.; Puglisi, A.; Strubin, M. Histone chaperone spt16 promotes redeposition of the original h3-h4 histones evicted by elongating RNA polymerase. Mol. Cell 2009, 35, 377–383. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, C.D.; Laprade, L.; Winston, F. Transcription elongation factors repress transcription initiation from cryptic sites. Science 2003, 301, 1096–1099. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, F.-K.; Kulaeva, O.I.; Patel, S.S.; Dyer, P.N.; Luger, K.; Reinberg, D.; Studitsky, V.M. Histone chaperone FACT action during transcription through chromatin by RNA polymerase II. Proc. Natl. Acad. Sci. USA 2013, 110, 7654–7659. [Google Scholar] [CrossRef] [PubMed]
- Winkler, D.D.; Luger, K. The histone chaperone FACT: Structural insights and mechanisms for nucleosome reorganization. J. Biol. Chem. 2011, 286, 18369–18374. [Google Scholar] [CrossRef] [PubMed]
- Stuwe, T.; Hothorn, M.; Lejeune, E.; Rybin, V.; Bortfeld, M.; Scheffzek, K.; Ladurner, A.G. The FACT Spt16 “peptidase” domain is a histone H3-H4 binding module. Proc. Natl. Acad. Sci. USA 2008, 105, 8884–8889. [Google Scholar] [CrossRef] [PubMed]
- Hondele, M.; Stuwe, T.; Hassler, M.; Halbach, F.; Bowman, A.; Zhang, E.T.; Nijmeijer, B.; Kotthoff, C.; Rybin, V.; Amlacher, S.; et al. Structural basis of histone H2A-H2B recognition by the essential chaperone FACT. Nature 2013, 499, 111–114. [Google Scholar] [CrossRef] [PubMed]
- Kemble, D.J.; Whitby, F.G.; Robinson, H.; McCullough, L.L.; Formosa, T.; Hill, C.P. Structure of the Spt16 middle domain reveals functional features of the histone chaperone FACT. J. Biol. Chem. 2013, 288, 10188–10194. [Google Scholar] [CrossRef] [PubMed]
- Kemble, D.J.; McCullough, L.L.; Whitby, F.G.; Formosa, T.; Hill, C.P. FACT disrupts nucleosome structure by binding H2A-H2B with conserved peptide motifs. Mol. Cell 2015, 60, 294–306. [Google Scholar] [CrossRef] [PubMed]
- Mao, Z.; Pan, L.; Wang, W.; Sun, J.; Shan, S.; Dong, Q.; Liang, X.; Dai, L.; Ding, X.; Chen, S.; et al. Anp32e, a higher eukaryotic histone chaperone directs preferential recognition for H2A.Z. Cell Res. 2014, 24, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Obri, A.; Ouararhni, K.; Papin, C.; Diebold, M.-L.; Padmanabhan, K.; Marek, M.; Stoll, I.; Roy, L.; Reilly, P.T.; Mak, T.W.; et al. ANP32E is a histone chaperone that removes H2A.Z from chromatin. Nature 2014, 505, 648–653. [Google Scholar] [CrossRef] [PubMed]
- Winkler, D.D.; Muthurajan, U.M.; Hieb, A.R.; Luger, K. Histone chaperone FACT coordinates nucleosome interaction through multiple synergistic binding events. J. Biol. Chem. 2011, 286, 41883–41892. [Google Scholar] [CrossRef] [PubMed]
- Tsunaka, Y.; Fujiwara, Y.; Oyama, T.; Hirose, S.; Morikawa, K. Integrated molecular mechanism directing nucleosome reorganization by human FACT. Genes Dev. 2016, 30, 673–686. [Google Scholar] [CrossRef] [PubMed]
- Valieva, M.E.; Armeev, G.A.; Kudryashova, K.S.; Gerasimova, N.S.; Shaytan, A.K.; Kulaeva, O.I.; McCullough, L.L.; Formosa, T.; Georgiev, P.G.; Kirpichnikov, M.P.; et al. Large-scale ATP-independent nucleosome unfolding by a histone chaperone. Nat. Struct. Mol. Biol. 2016, 23, 1111–1116. [Google Scholar] [CrossRef] [PubMed]
- Gaykalova, D.A.; Kulaeva, O.I.; Volokh, O.; Shaytan, A.K.; Hsieh, F.-K.; Kirpichnikov, M.P.; Sokolova, O.S.; Studitsky, V.M. Structural analysis of nucleosomal barrier to transcription. Proc. Natl. Acad. Sci. USA 2015, 112, E5787–E5795. [Google Scholar] [CrossRef] [PubMed]
- Kulaeva, O.I.; Gaykalova, D.A.; Pestov, N.A.; Golovastov, V.V.; Vassylyev, D.G.; Artsimovitch, I.; Studitsky, V.M. Mechanism of chromatin remodeling and recovery during passage of RNA polymerase II. Nat. Struct. Mol. Biol. 2009, 16, 1272–1278. [Google Scholar] [CrossRef] [PubMed]
- Morozov, A.V.; Fortney, K.; Gaykalova, D.A.; Studitsky, V.M.; Widom, J.; Siggia, E.D. Using DNA mechanics to predict in vitro nucleosome positions and formation energies. Nucleic Acids Res. 2009, 37, 4707–4722. [Google Scholar] [CrossRef] [PubMed]
- Kudryashova, K.S.; Chertkov, O.V.; Nikitin, D.V.; Pestov, N.A.; Kulaeva, O.I.; Efremenko, A.V.; Solonin, A.S.; Kirpichnikov, M.P.; Studitsky, V.M.; Feofanov, A.V. Preparation of mononucleosomal templates for analysis of transcription with RNA polymerase using spFRET. Methods Mol. Biol. 2015, 1288, 395–412. [Google Scholar] [PubMed]
- Yang, Z.; Zheng, C.; Thiriet, C.; Hayes, J.J. The core histone N-terminal tail domains negatively regulate binding of transcription factor IIIA to a nucleosome containing a 5S RNA gene via a novel mechanism. Mol. Cell. Biol. 2005, 25, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Hizume, K.; Nakai, T.; Araki, S.; Prieto, E.; Yoshikawa, K.; Takeyasu, K. Removal of histone tails from nucleosome dissects the physical mechanisms of salt-induced aggregation, linker histone H1-induced compaction, and 30-nm fiber formation of the nucleosome array. Ultramicroscopy 2009, 109, 868–873. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Hayes, J.J. Structures and interactions of the core histone tail domains. Biopolymers 2003, 68, 539–546. [Google Scholar] [CrossRef] [PubMed]
- Bondarenko, M.T.; Maluchenko, N.V.; Valieva, M.E.; Gerasimova, N.S.; Kulaeva, O.I.; Georgiev, P.G.; Studitsky, V.M. Structure and function of histone chaperone FACT. Mol. Biol. (Mosk.) 2015, 49, 891–904. [Google Scholar] [CrossRef] [PubMed]
- Formosa, T. FACT and the reorganized nucleosome. Mol. Biosyst. 2008, 4, 1085–1093. [Google Scholar] [CrossRef] [PubMed]
- Xin, H.; Takahata, S.; Blanksma, M.; McCullough, L.; Stillman, D.J.; Formosa, T. yFACT induces global accessibility of nucleosomal DNA without H2A-H2B displacement. Mol. Cell 2009, 35, 365–376. [Google Scholar] [CrossRef] [PubMed]
- Kireeva, M.L.; Walter, W.; Tchernajenko, V.; Bondarenko, V.; Kashlev, M.; Studitsky, V.M. Nucleosome remodeling induced by RNA polymerase II: Loss of the H2A/H2B dimer during transcription. Mol. Cell 2002, 9, 541–552. [Google Scholar] [CrossRef]
- Gaykalova, D.A.; Kulaeva, O.I.; Bondarenko, V.A.; Studitsky, V.M. Preparation and analysis of uniquely positioned mononucleosomes. Methods Mol. Biol. 2009, 523, 109–123. [Google Scholar] [PubMed]
| N | N+FACT-N | FACT-N(SSRPΔ) | N(tailless) | N(tailless)+FACT-N | N(tailless)+FACT-N(SSRPΔ) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gaussians | Peak 1 | Peak 2 | Peak 1 | Peak 2 | Peak 1 | Peak 2 | Peak 1 | Peak 2 | Peak 1 | Peak 2 | Peak 1 | Peak 2 |
| EPR (max) | 0.07 ± 0.05 | 0.57 ± 0.01 | 0.06 ± 0.05 | 0.58 ± 0.01 | 0.08 ± 0.05 | 0.57 ± 0.01 | 0.08 ± 0.03 | 0.49 ± 0.01 | 0.06 ± 0.03 | 0.54 ± 0.01 | 0.08 ± 0.03 | 0.51 ± 0.01 |
| Subpopulation (%) | 9.3 ± 1.4 | 91 ± 2 | 7 ± 6 | 93 ± 6 | 9 ± 3 | 92 ± 3 | 20 ± 3 | 80 ± 3 | 8 ± 6 | 93 ± 9 | 17 ± 2 | 83 ± 2 |
© 2017 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valieva, M.E.; Gerasimova, N.S.; Kudryashova, K.S.; Kozlova, A.L.; Kirpichnikov, M.P.; Hu, Q.; Botuyan, M.V.; Mer, G.; Feofanov, A.V.; Studitsky, V.M. Stabilization of Nucleosomes by Histone Tails and by FACT Revealed by spFRET Microscopy. Cancers 2017, 9, 3. https://doi.org/10.3390/cancers9010003
Valieva ME, Gerasimova NS, Kudryashova KS, Kozlova AL, Kirpichnikov MP, Hu Q, Botuyan MV, Mer G, Feofanov AV, Studitsky VM. Stabilization of Nucleosomes by Histone Tails and by FACT Revealed by spFRET Microscopy. Cancers. 2017; 9(1):3. https://doi.org/10.3390/cancers9010003
Chicago/Turabian StyleValieva, Maria E., Nadezhda S. Gerasimova, Kseniya S. Kudryashova, Anastasia L. Kozlova, Mikhail P. Kirpichnikov, Qi Hu, Maria Victoria Botuyan, Georges Mer, Alexey V. Feofanov, and Vasily M. Studitsky. 2017. "Stabilization of Nucleosomes by Histone Tails and by FACT Revealed by spFRET Microscopy" Cancers 9, no. 1: 3. https://doi.org/10.3390/cancers9010003
APA StyleValieva, M. E., Gerasimova, N. S., Kudryashova, K. S., Kozlova, A. L., Kirpichnikov, M. P., Hu, Q., Botuyan, M. V., Mer, G., Feofanov, A. V., & Studitsky, V. M. (2017). Stabilization of Nucleosomes by Histone Tails and by FACT Revealed by spFRET Microscopy. Cancers, 9(1), 3. https://doi.org/10.3390/cancers9010003







