Epstein-Barr Virus-Encoded RNAs: Key Molecules in Viral Pathogenesis
Abstract
:1. Introduction
2. Structure of EBERs

3. Synthesis and Expression of EBERs
4. Localization of EBERs
5. Interactions with Cellular Proteins
5.1. La Protein
5.2. PKR
5.3. L22
5.4. Other Interacting Proteins
6. Oncogenic Roles of EBERs
7. EBERs-Mediated Pathogenesis via Modulation of Innate Immune Signals
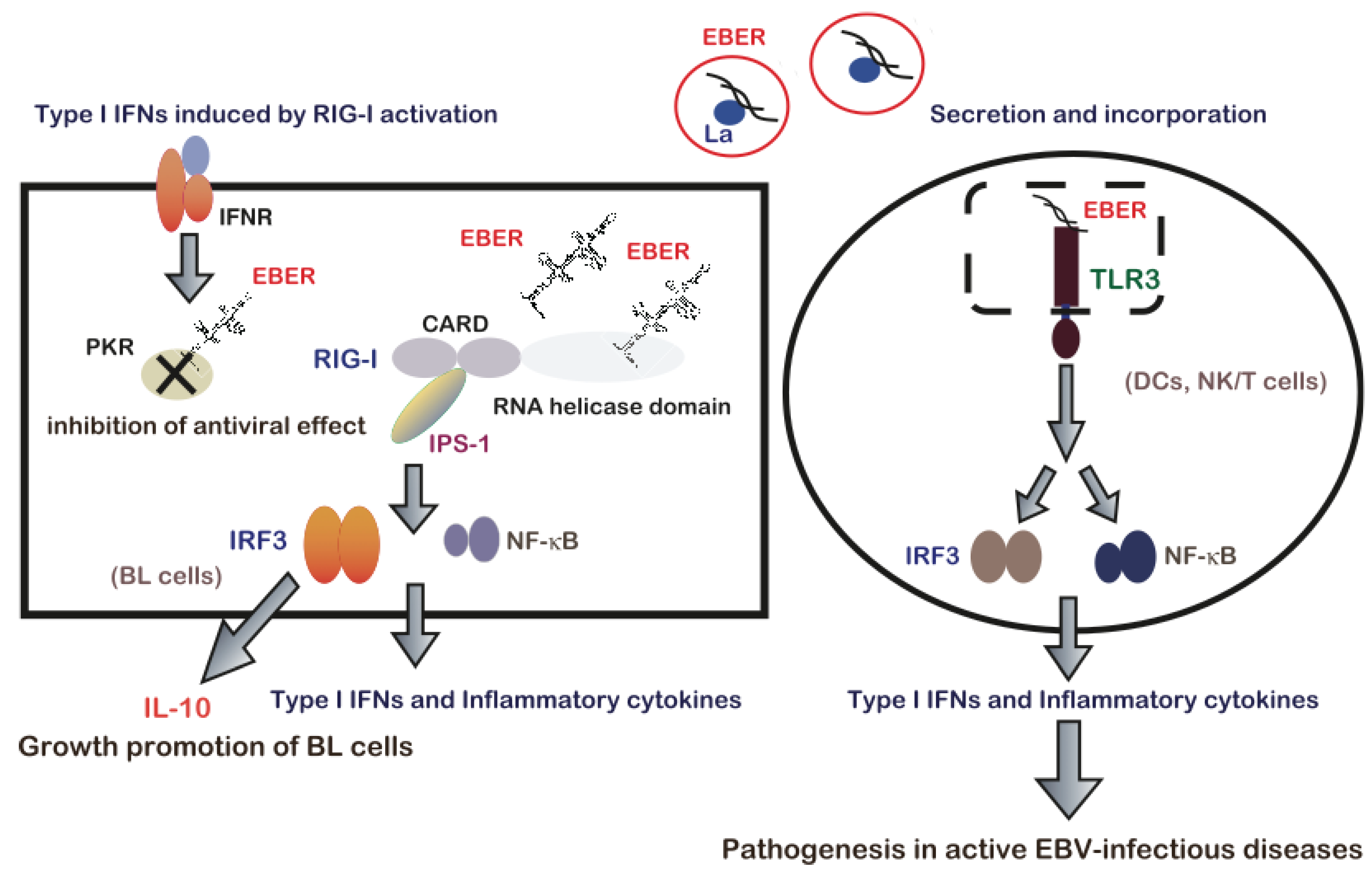
8. Conclusions and Future Direction
Acknowledgements
Conflicts of Interest
References
- Rickinson, A.B.; Kieff, E. Epstein-Barr virus. In Fields Virol; Knipe, D.M., Howley, P.M., Eds.; Walters Kluwer/Lippincott: Philadelphia, PA, USA, 2007; pp. 2655–2700. [Google Scholar]
- Lerner, M.R.; Andrews, N.C.; Miller, G.; Steitz, J.A. Two small RNAs encoded by Epstein-Barr virus and complexed with protein are precipitated by antibodies from patients with systemic lupus erythematosus. Proc. Natl. Acad. Sci. USA 1981, 78, 805–809. [Google Scholar] [CrossRef] [PubMed]
- Rymo, L. Identification of transcribed regions of Epstein-Barr virus DNA in Burkitt lymphoma-derived cells. J. Virol. 1979, 32, 8–18. [Google Scholar] [PubMed]
- Rosa, M.D.; Gottlieb, E.; Lerner, M.R.; Steitz, J.A. Striking similarities are exhibited by two small Epstein-Barr virus-encoded ribonucleic acids and the adenovirus-associated ribonucleic acids VAI and VAII. Mol. Cell. Biol. 1981, 9, 785–796. [Google Scholar]
- Chang, K.L.; Chen, Y.Y.; Shibata, D.; Weiss, L.M. Description of an in situ hybridization methodology for detection of Epstein-Barr virus RNA in paraffin-embedded tissues, with a survey of normal and neoplastic tissues. Diagn. Mol. Pahol. 1992, 1, 246–255. [Google Scholar] [CrossRef]
- Komano, J.; Maruo, S.; Kurozumi, K.; Oda, T.; Takada, K. Oncogenic role of Epstein-Barr virus-encoded RNAs in Burkitt’s lymphoma cell line Akata. J. Virol. 1999, 73, 9827–9831. [Google Scholar] [PubMed]
- Ruf, I.K.; Rhyne, P.W.; Yang, C.; Cleveland, J.L.; Sample, J.T. Epstein-Barr virus small RNAs potentiate tumorigenicity of Burkitt lymphoma cells independently of an effect on apoptosis. J. Virol. 2000, 74, 10223–10228. [Google Scholar] [CrossRef] [PubMed]
- Nanbo, A.; Inoue, K.; Adachi-Takasawa, K.; Takada, K. Epstein-Barr virus RNA confers resistance to interferon-alpha-induced apoptosis in Burkitt’s lymphoma. EMBO J. 2002, 21, 954–965. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, N.; Goto, M.; Kurozumi, K.; Maruo, S.; Fukayama, M.; Naoe, T.; Yasukaswa, M.; Hino, K.; Suzuki, T.; Todo, S.; et al. Epstein-Barr virus-encoded poly(A)(-) RNA supports Burkitt’s lymphoma growth through interleukin-10 induction. EMBO J. 2000, 19, 6742–6750. [Google Scholar] [CrossRef] [PubMed]
- Iwakiri, D.; Eizuru, Y.; Tokunaga, M.; Takada, K. Autocrine growth of Epstein-Barr virus-positive gastric carcinoma cells mediated by an Epstein-Barr virus-encoded small RNA. Cancer Res. 2003, 63, 7062–7067. [Google Scholar] [PubMed]
- Iwakiri, D.; Sheen, T.S.; Chen, J.Y.; Huang, D.P.; Takada, K. Epstein-Barr virus-encoded small RNA induces insulin-like growth factor 1 and supports growth of nasopharyngeal carcinoma-derived cell lines. Oncogene 2005, 24, 1767–1773. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Aozasa, K.; Oshimi, K.; Takada, K. Epstein-Barr virus (EBV)-encoded RNA promotes growth of EBV-infected T cells through interleukin-9 induction. Cancer Res. 2004, 64, 5332–5337. [Google Scholar] [CrossRef] [PubMed]
- Houmani, J.L.; Davis, C.I.; Ruf, I.K. Growth-promoting properties of Epstein-Barr virus EBER-1 RNA correlate with ribosomal protein L22 binding. J. Virol. 2009, 83, 9844–9853. [Google Scholar] [CrossRef]
- Repellin, C.E.; Tsimbouri, P.M.; Philbey, A.W.; Wilson, J.B. Lymphoid hyperplasia and lymphoma in transgenic mice expressing the small non-coding RNA, EBER1 of Epstein-Barr virus. PLoS One 2010, 5, e9092. [Google Scholar]
- Samanta, M.; Iwakiri, D.; Kanda, T.; Imaizumi, T.; Takada, K. EB virus-encoded RNAs are recognized by RIG-I and activate signaling to induce type I IFN. EMBO J. 2006, 25, 4207–4214. [Google Scholar] [CrossRef] [PubMed]
- Samanta, M.; Iwakiri, D.; Takada, K. Epstein-Barr virus-encoded small RNA induces IL-10 through RIG-I-mediated IRF-3 signaling. Oncogene 2008, 27, 4150–4160. [Google Scholar] [CrossRef]
- Iwakiri, D.; Zhou, L.; Samanta, M.; Matsumoto, M.; Ebihara, T.; Seya, T.; Imai, S.; Fujieda, M.; Kawa, K.; Takada, K. Epstein-Barr virus (EBV)-encoded small RNA is released from EBV-infected cells and activates signaling from Toll-like receptor 3. J. Exp. Med. 2009, 206, 2091–2099. [Google Scholar] [CrossRef] [PubMed]
- Howe, J.G.; Shu, M.D. Isolation and characterization of the genes for two small RNAs of herpesvirus papio and their comparison with Epstein-Barr virus-encoded EBER RNAs. J. Virol. 1988, 62, 2790–2798. [Google Scholar] [PubMed]
- Katze, M.G.; Wambach, M.; Wong, M.L.; Garfinkel, M.; Meurs, E.; Chong, K.; Williams, B.R.; Hovanessian, A.G.; Barber, G.N. Functional expression and RNA binding analysis of the interferon-induced, double-stranded RNA-activated, 68,000-Mr protein kinase in a cell-free system. Mol. Cell. Biol. 1991, 11, 5497–5505. [Google Scholar] [PubMed]
- Arrand, J.R.; Young, L.S.; Tugwood, J.D. Two families of sequences in the small RNA-encoding region of Epstein-Barr virus (EBV) correlate with EBV types A and B. J. Virol. 1989, 63, 983–986. [Google Scholar] [PubMed]
- Yao, Q.Y.; Tierney, R.J.; Croom-Carter, D.; Cooper, G.M.; Ellis, C.J.; Rowe, M.; Rickinson, A.B. Isolation of intertypic recombinants of Epstein-Barr virus from T-cell-immunocompromised individuals. J. Virol. 1996, 70, 4895–4903. [Google Scholar] [PubMed]
- Howe, J.G.; Shu, M.D. Epstein-Barr virus small RNA (EBER) genes: Unique transcription units that combine RNA polymerase II and III promoter elements. Cell 1989, 57, 825–834. [Google Scholar] [CrossRef] [PubMed]
- Arraud, J.R.; Ryomo, L. Characterization of the major Epstein-Barr virus-specific RNA in Burkitt lymphoma-derived cells. J. Virol. 1982, 41, 376–389. [Google Scholar] [PubMed]
- Rooney, C.; Howe, J.G.; Speck, S.H.; Miller, G. Influence of Burkitt’s lymphoma and primary B cells on latent gene expression by the nonimmortalizing P3J-HR-1 strain of Epstein-Barr virus. J. Virol. 1989, 63, 1531–1539. [Google Scholar]
- Greifenegger, N.; Jäger, M.; Kunz-Schughart, L.A.; Wolf, H.; Schwarzmann, F. Epstein-Barr virus small RNA (EBER) genes: Differential regulation during lytic viral replication. J. Virol. 1998, 72, 9323–9328. [Google Scholar] [PubMed]
- Clarke, P.A.; Sharp, N.A.; Clemens, M.J. Expression of genes for the Epstein-Barr virus small RNAs EBER-1 and EBER-2 in Daudi Burkitt’s lymphoma cells: Effects of interferon treatment. J. Gen. Virol. 1992, 73, 3169–3175. [Google Scholar] [CrossRef]
- Gilligan, K.; Rajadurai, P.; Resnick, L.; Raab-Traub, N. Epstein-Barr virus small nuclear RNAs are not expressed in permissively infected cells in AIDS-associated leukoplakia. Proc. Natl. Acad. Sci. USA 1990, 87, 8790–8794. [Google Scholar] [CrossRef] [PubMed]
- Wen, S.; Mizugaki, Y.; Shinozaki, F.; Takada, K. Epstein-Barr virus (EBV) infection in salivary gland tumors: Lytic EBV infection in nonmalignant epithelial cells surrounded by EBV-positive T-lymphoma cells. Virology 1997, 227, 484–487. [Google Scholar] [CrossRef] [PubMed]
- Wen, S.; Shimizu, N.; Yoshiyama, H.; Mizugaki, Y.; Shinozaki, F.; Takada, K. Association of Epstein-Barr virus (EBV) with Sjögren’s syndrome: Differential EBV expression between epithelial cells and lymphocytes in salivary glands. Am. J. Pathol. 1996, 149, 1511–1517. [Google Scholar] [PubMed]
- Mizugaki, Y.; Sugawara, Y.; Shinozaki, F.; Takada, K. Detection of Epstein-Barr virus in oral papilloma. Jpn. J. Cancer Res. 1998, 89, 604–607. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Minter, H.A.; Chen, X.; Reynolds, G.M.; Bromley, M.; Arrand, J.R. Heterogeneity of HLA and EBER expression in Epstein-Barr virus-associated nasopharyngeal carcinoma. Int. J. Cancer. 2000, 88, 949–955. [Google Scholar] [CrossRef] [PubMed]
- Howe, J.G.; Steitz, J.A. Localization of Epstein-Barr virus-encoded small RNAs by in situ hybridization. Proc. Natl. Acad. Sci. USA 1986, 83, 9006–9010. [Google Scholar] [CrossRef]
- Schwemmle, M.; Clemens, M.J.; Hilse, K.; Pfeifer, K.; Tröster, H.; Müller, W.E.; Bachmann, M. Localization of Epstein-Barr virus-encoded RNAs EBER-1 and EBER-2 in interphase and mitotic Burkitt lymphoma cells. Proc. Natl. Acad. Sci. USA 1992, 89, 10292–10296. [Google Scholar] [CrossRef] [PubMed]
- Fok, V.; Friend, K.; Steitz, J.A. Epstein-Barr virus noncoding RNAs are confined to the nucleus, whereas their partner, the human La protein, undergoes nuclecytoplasmic shuttling. J. Cell. Biol. 2006, 173, 319–325. [Google Scholar] [CrossRef] [PubMed]
- Clarke, P.A.; Schwemmle, M.; Schickinger, J.; Hilse, K.; Clemens, M.J. Binding of Epstein-Barr virus small RNA EBER-1 to the double-stranded RNA-activated protein kinase DAI. Nucleic Acids Res. 1991, 19, 243–248. [Google Scholar] [CrossRef]
- Lee, N.; Pimienta, G.; Steitz, J.A. AUF1/hnRNP D is a novel protein partner of the EBER1 noncoding RNA of Epstein-Barr virus. RNA 2012, 18, 2073–2082. [Google Scholar]
- Bachmann, M.; Pfeifer, K.; Schröder, H.C.; Müller, W.E. The La antigen shuttles between the nucleus and the cytoplasm in CV-1 cells. Mol. Cell. Biochem. 1989, 85, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Ida, H.; Arima, K.; Nakamura, H.; Aramaki, T.; Fujikawa, K.; Tamai, M.; Kamachi, M.; Kawakami, A.; Yamasaki, H.; et al. La autoantigen translocates to cytoplasm after cleavage during granzyme B-mediated cytotoxicity. Life Sci. 2007, 81, 1461–1466. [Google Scholar]
- Meerovitch, K.; Svitkin, Y.V.; Lee, H.S.; Lejbkowicz, F.; Kenan, D.J.; Chan, E.K.; Agol, V.I.; Keene, J.D.; Sonenberg, N. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J. Virol. 1993, 67, 3798–3807. [Google Scholar] [PubMed]
- Shiroki, K.; Isoyama, T.; Kuge, S.; Ishii, T.; Ohmi, S.; Hata, S.; Suzuki, K.; Takasaki, Y.; Nomoto, A. Intracellular redistribution of truncated La protein produced by poliovirus 3Cpro-mediated cleavage. J. Virol. 1999, 73, 2193–2200. [Google Scholar] [PubMed]
- Akusjärvi, G.; Mathews, M.B.; Andersson, P.; Vennström, B.; Pettersson, U. Structure of genes for virus-associated RNAI and RNAII of adenovirus type 2. Proc. Natl. Acad. Sci. USA 1980, 77, 2424–2428. [Google Scholar] [CrossRef] [PubMed]
- Bhat, R.A.; Thimmappaya, B. Two small RNAs encoded by Epstein-Barr virus can functionally substitute for the virus-associated RNAs in the lytic growth of adenovirus 5. Proc. Natl. Acad. Sci. USA 1983, 80, 4789–4793. [Google Scholar] [CrossRef] [PubMed]
- Meurs, E.; Chong, K.; Galabru, J.; Thomas, N.S.; Kerr, I.M.; Williams, B.R.; Hovanessian, A.G. Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell 1990, 62, 379–390. [Google Scholar] [CrossRef] [PubMed]
- Sharp, T.V.; Schwemmle, M.; Jeffrey, I.; Laing, K.; Mellor, H.; Proud, C.G.; Hilse, K.; Clemens, M.J. Comparative analysis of the regulation of the interferon-inducible protein kinase PKR by Epstein-Barr virus RNAs EBER-1 and EBER-2 and adenovirus VAI RNA. Nucleic Acids Res. 1993, 21, 4483–4490. [Google Scholar] [CrossRef] [PubMed]
- Vuyisich, M.; Spanggord, R.J.; Beal, P.A. The binding site of the RNA-dependent protein kinase (PKR) on EBER1 RNA from Epstein-Barr virus. EMBO Rep. 2000, 3, 622–627. [Google Scholar] [CrossRef]
- Clarke, P.A.; Sharp, N.A.; Clemens, M.J. Translational control by the Epstein-Barr virus small RNA EBER-1. Reversal of the double-stranded RNA-induced inhibition of protein synthesis in reticulocyte lysates. Eur. J. Biochem. 1990, 193, 635–641. [Google Scholar]
- McKenna, S.A.; Lindhout, D.A.; Shimoike, T.; Aitken, C.E.; Puglisis, J.D. Viral dsRNA inhibitors prevent self-association and autophosphorylation of PKR. J. Mol. Biol. 2007, 372, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Toczyski, D.P.; Steitz, J.A. EAP, a highly conserved cellular protein associated with Epstein-Barr virus small RNAs (EBERs). EMBO J. 1991, 10, 459–466. [Google Scholar]
- Toczyski, D.P.; Steitz, J.A. The cellular RNA-binding protein EAP recognizes a conserved stem-loop in the Epstein-Barr virus small RNA EBER 1. Mol. Cell. Biol. 1993, 1, 703–710. [Google Scholar]
- Toczyski, D.P.; Matera, A.G.; Ward, D.C.; Steitz, J.A. The Epstein-Barr virus (EBV) small RNA EBER1 binds and relocalizes ribosomal protein L22 in EBV-infected human B lymphocytes. Proc. Natl. Acad. Sci. USA 1994, 91, 3463–3467. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Tarlé, S.A.; Hajra, A.; Claxton, D.F.; Marlton, P.; Freedman, M.; Siciliano, M.J.; Collins, F.S. Fusion between transcription factor CBF beta/PEBP2 beta and a myosin heavy chain in acute myeloid leukemia. Science 1993, 261, 1041–1044. [Google Scholar] [CrossRef] [PubMed]
- Nucifora, G.; Begy, C.R.; Erickson, P.; Drabkin, H.A.; Rowley, J.D. The 3;21 translocation in myelodysplasia results in a fusion transcript between the AML1 gene and the gene for EAP, a highly conserved protein associated with the Epstein-Barr virus small RNA EBER 1. Proc. Natl. Acad. Sci. USA 1993, 90, 7784–7788. [Google Scholar] [CrossRef] [PubMed]
- Le, S.; Sternglanz, R.; Greider, C.W. Identification of two RNA-binding proteins associated with human telomerase RNA. Mol. Biol. Cell 2000, 11, 999–1010. [Google Scholar] [CrossRef] [PubMed]
- Leopardi, R.; Ward, P.L.; Ogle, W.O.; Roizman, B. Association of herpes simplex virus regulatory protein ICP22 with transcrip-tional complexes containing EAP, ICP4, RNA polymerase II, and viral DNA requires posttranslational modification by the U(L)13 proteinkinase. J. Virol. 1997, 71, 1133–1139. [Google Scholar] [PubMed]
- Wood, J.; Frederickson, R.M.; Fields, S.; Patel, A.H. Hepatitis C virus 3'X region interacts with human ribosomal proteins. J. Virol. 2001, 75, 1348–1358. [Google Scholar] [CrossRef] [PubMed]
- Gregorovic, G.; Bosshard, R.; Karstegl, C.E.; White, R.E.; Pattle, S.; Chiang, A.K.; Dittrich-Breiholz, O.; Kracht, M.; Russ, R.; Farrell, P.J. Cellular gene expression that correlates with EBER expression in Epstein-Barr virus-infected lymphoblastoid cell lines. J. Virol. 2011, 85, 3535–3545. [Google Scholar]
- Dobbelstein, M.; Shenk, T. In vitro selection of RNA ligands for the ribosomal L22 protein associated with Epstein-Barr virus-expressed RNA by using randomized and cDNA-derived RNA libraries. J. Virol. 1995, 69, 8027–8034. [Google Scholar] [PubMed]
- Fok, V.; Mitton-Fry, R.M.; Grech, A.; Steitz, J.A. Multiple domains of EBER 1, an Epstein-Barr virus noncoding RNA, recruit human ribosomal protein L22. RNA 2006, 12, 872–882. [Google Scholar] [CrossRef] [PubMed]
- Elia, A.; Vyas, J.; Laing, K.G.; Clemens, M.J. Ribosomal protein L22 inhibits regulation of cellular activities by the Epstein-Barr virus small RNA EBER-1. Eur. J. Biochem. 2004, 27, 1895–1905. [Google Scholar] [CrossRef]
- Yoneyama, M.; Kikuchi, M.; Natsukawa, T.; Shinobu, N.; Imaizumi, T.; Miyagishi, M.; Taira, K.; Akira, S.; Fujita, T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 2004, 5, 730–737. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.C.; Gopalkrishnan, R.V.; Wu, Q.; Jankowsky, E.; Pyle, A.M.; Fisher, P.B. mda-5: An interferon-inducible putative RNA helicase with double-stranded RNA-dependent ATPase activity and melanoma growth-suppressive properties. Proc. Natl. Acad. Sci. USA 2002, 99, 637–642. [Google Scholar]
- Yoneyama, M.; Fujita, T. Function of RIG-I-like receptors in antiviral innate immunity. J. Biol. Chem. 2007, 282, 15315–15318. [Google Scholar] [CrossRef] [PubMed]
- Meylan, E.; Tschopp, J. Toll-like receptors and RNA helicases: Two parallel ways to trigger antiviral responses. Mol. Cell 2006, 22, 561–569. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Takahashi, K.; Sato, S.; Coban, C.; Kumar, H.; Kato, H.; Ishii, K.J.; Takeuchi, O.; Akira, S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 2005, 11, 1074–1076. [Google Scholar]
- Hornung, V.; Ellegast, J.; Kim, S.; Brzózka, K.; Jung, A.; Kato, H.; Poeck, H.; Akira, S.; Conzelmann, K.K.; Schlee, M.; et al. 5'-Triphosphate RNA is the ligand for RIG-I. Science 2006, 314, 994–997. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.-Y.; Sadri, N.; Schneider, R.J. Endotoxic shock in AUF1 knockout mice mediated by failure to degrade proinflammatory cytokine mRNAs. Gen. Dev. 2006, 20, 3174–3184. [Google Scholar] [CrossRef]
- Gratacós, F.M.; Brewer, G. The role of AUF1 in regulated mRNA decay. Wiley Interdiscip. Rev. RNA 2010, 1, 457–473. [Google Scholar] [CrossRef] [PubMed]
- Lal, A.; Mazan-Mamczarz, K.; Kawai, T.; Yang, X.; Martindale, J.L.; Gorospe, M. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 2004, 23, 3092–3102. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, N.; Tanabe-Tochikura, A.; Kuroiwa, Y.; Takada, K. Isolation of Epstein-Barr virus (EBV)-negative cell clones from the EBV- positive Burkitt’s lymphoma (BL) line Akata: Malignant phenotypes of BL cells are dependent on EBV. J. Virol. 1994, 68, 6069–6073. [Google Scholar]
- Yamamoto, N.; Takizawa, T.; Iwanaga, Y.; Shimizu, N.; Yamamoto, N. Malignant transformation of B lymphoma cell line BJAB by Epstein-Barr virus-encoded small RNAs. FEBS Lett. 2000, 484, 153–158. [Google Scholar] [CrossRef] [PubMed]
- Iwakiri, D.; Takada, K. Role of EBERs in the pathogenesis of EBV infection. Adv. Cancer Res. 2010, 107, 119–136. [Google Scholar]
- Wong, H.L.; Wang, X.; Chang, R.C.; Jin, D.Y.; Feng, H.; Wang, Q.; Lo, K.W.; Huang, D.P.; Yuen, P.W.; Takada, K.; et al. Stable expression of EBERs in immortalized nasopharyngeal epithelial cells confers resistance to apoptotic stress. Mol. Carccing. 2005, 44, 92–101. [Google Scholar] [CrossRef]
- Yoshizaki, T.; Endo, K.; Ren, Q.; Wakisaka, N.; Murono, S.; Kondo, S.; Sato, H.; Furukawa, M. Oncogenic role of Epstein-Barr virus-encoded small RNAs (EBERs) in nasopharyngeal carcinoma. Auris. Nasus. Larynx 2007, 34, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Tsang, C.M.; Zhang, G.; Seto, E.; Takada, K.; Deng, W.; Yip, Y.L.; Man, C.; Hau, P.M.; Chen, H.; Cao, Y.; et al. Epstein-Barr virus infection in immortalized nasopharyngeal epithelial cells: Regulation of infection and phenotypic characterization. Int. J. Cancer 2010, 127, 1570–1583. [Google Scholar] [CrossRef] [PubMed]
- Swaminathan, S.; Tomkinson, B.; Kieff, E. Recombinant Epstein-Barr virus with small RNA (EBER) genes deleted transforms lymphocytes and replicates in vitro. J. Virol. 1991, 66, 5133–5136. [Google Scholar]
- Yajima, M.; Kanda, T.; Takada, K. Critical role of Epstein-Barr Virus (EBV)-encoded RNA in efficient EBV-induced B-lymphocyte growth transformation. J. Virol. 2005, 79, 4298–4307. [Google Scholar]
- Wu, Y.; Maruo, S.; Yajima, M.; Kanda, T.; Takada, K. Epstein-Barr virus (EBV)-encoded RNA 2 (EBER2) but not EBER1 plays a critical role in EBV-induced B-cell growth transformation. J. Virol. 2007, 80, 11236–11245. [Google Scholar] [CrossRef]
- Akira, S.; Takeda, K. Toll-like receptor signaling. Nat. Rev. Immunol. 2004, 4, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Alexopoulou, L.; Holt, A.C.; Medzhitov, R.; Flavell, R.A. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 2001, 413, 732–738. [Google Scholar] [CrossRef] [PubMed]
- Kikuta, H.; Sakiyama, Y.; Matsumoto, S.; Oh-Ishi, T.; Nakano, T.; Nagashima, T.; Oka, T.; Hironaka, T.; Hirai, K. Fatal Epstein-Barr virus-associated hemophagocytic syndrome. Blood 1993, 82, 3259–3264. [Google Scholar] [PubMed]
- Kasahara, Y.; Yachie, A.; Takei, K.; Kanegane, C.; Okada, K.; Ohta, K.; Seki, H.; Igarashi, N.; Maruhashi, K.; Katayama, K.; et al. Differential cellular targets of Epstein-Barr virus (EBV) infection between acute EBV-associated hemophagocytic lymphohistiocytosis and chronic active EBV infection. Blood 2001, 98, 1882–1888. [Google Scholar] [CrossRef] [PubMed]
- Tabiasco, J.; Devêvre, E.; Rufer, N.; Salaun, B.; Cerottini, J.C.; Speiser, D.; Romero, D. Human effector CD8+ T lymphocytes express TLR3 as a functional coreceptor. J. Immunol. 2006, 177, 8708–8713. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, K.N.; Leung, B.; Kwong, M.; Zarember, K.A.; Satyal, S.; Navas, T.A.; Wang, F.; Godowski, P.J. APC-independent activation of NK cells by the Toll-like receptor 3 agonist double-stranded RNA. J. Immunol. 2004, 172, 138–143. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Misawa, N.; Nie, C.; Satou, Y.; Iwakiri, D.; Matsuoka, M.; Takahashi, R.; Kuzushima, K.; Ito, M.; Takada, K.; et al. A novel animal model of Epstein-Barr virus-associated hemophagocytic lymphohistiocytosis in humanized mice. Blood 2011, 117, 5663–5673. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell. Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Pant, S.; Hilton, H.; Burczynski, M.E. The multi-faceted exosome: Biogenesis, role in normal and aberrant cellular function, and frontiers for pharmacological and biomarker opportunities. Biochem. Pharmacol. 2012, 83, 1484–1494. [Google Scholar] [CrossRef] [PubMed]
- Pegtel, D.M.; Cosmopoulos, K.; Thorley-Lawson, D.A.; van Eijndhoven, M.A.; Hopmans, E.S.; Lindenberg, J.L.; de Gruijl, T.D.; Würdinger, T.; Middeldorp, J.M. Functional delivery of viral miRNAs via exosomes. Proc. Natl. Acad. Sci. USA 2010, 107, 6328–6333. [Google Scholar] [CrossRef] [PubMed]
- Kapsogeorgou, E.K.; Abu-Helu, R.F.; Moutsopoulos, H.M.; Manoussakis, M.N. Salivary gland epithelial cell exosomes: A source of autoantigenic ribonucleoproteins. Arthtitis Rheum. 2005, 52, 1517–1521. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Iwakiri, D. Epstein-Barr Virus-Encoded RNAs: Key Molecules in Viral Pathogenesis. Cancers 2014, 6, 1615-1630. https://doi.org/10.3390/cancers6031615
Iwakiri D. Epstein-Barr Virus-Encoded RNAs: Key Molecules in Viral Pathogenesis. Cancers. 2014; 6(3):1615-1630. https://doi.org/10.3390/cancers6031615
Chicago/Turabian StyleIwakiri, Dai. 2014. "Epstein-Barr Virus-Encoded RNAs: Key Molecules in Viral Pathogenesis" Cancers 6, no. 3: 1615-1630. https://doi.org/10.3390/cancers6031615
APA StyleIwakiri, D. (2014). Epstein-Barr Virus-Encoded RNAs: Key Molecules in Viral Pathogenesis. Cancers, 6(3), 1615-1630. https://doi.org/10.3390/cancers6031615



