Exploring the Impact of DNA Methylation on Gene Expression in CRC: A Computational Approach for Identifying Epigenetically Regulated Genes in Multi-Omic Datasets
Simple Summary
Abstract
1. Introduction
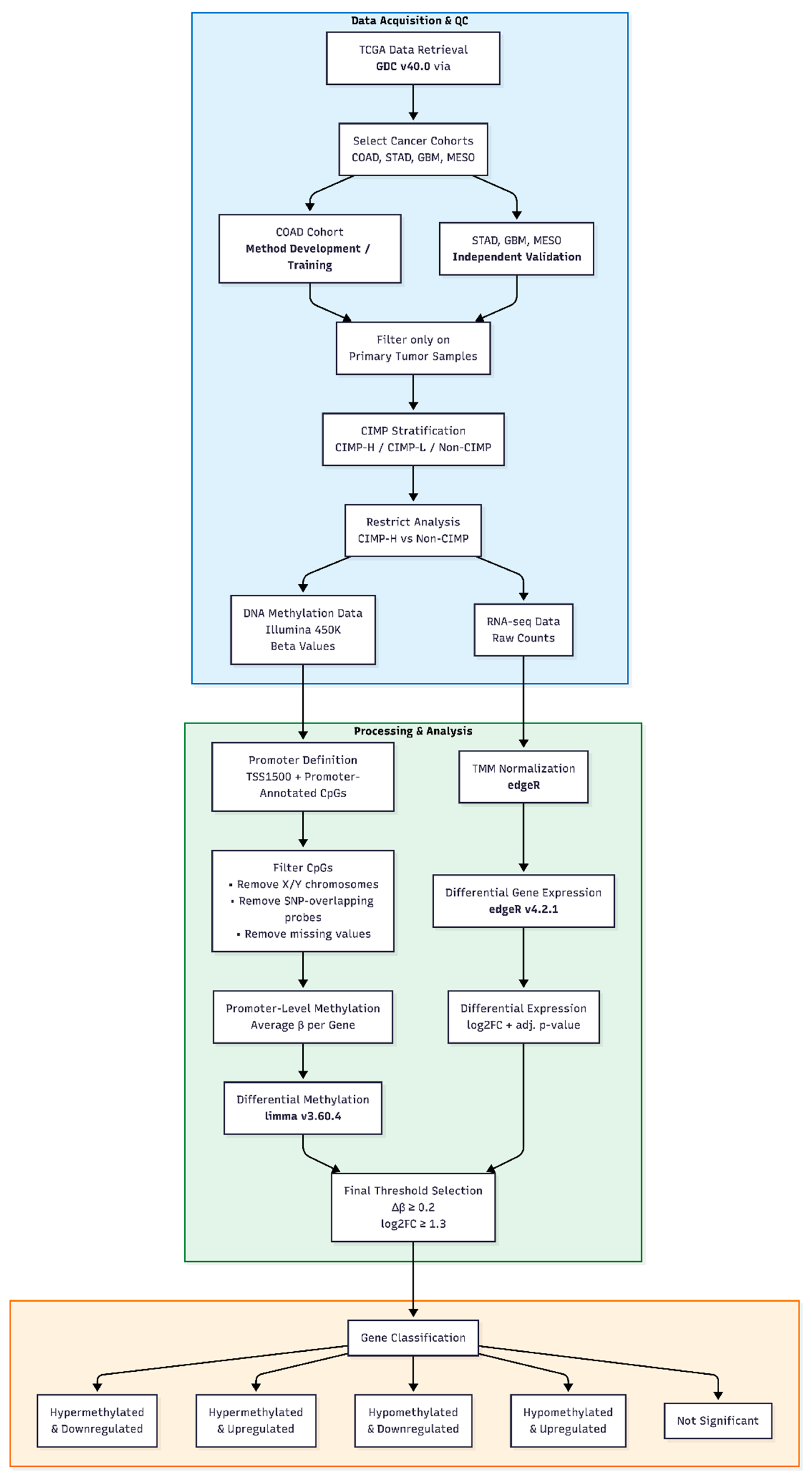
2. Materials and Methods
2.1. Dataset Selection and CIMP-Based Stratification
2.2. Data Preprocessing
2.3. Integrated Analysis of Differential Methylation and Gene Expression
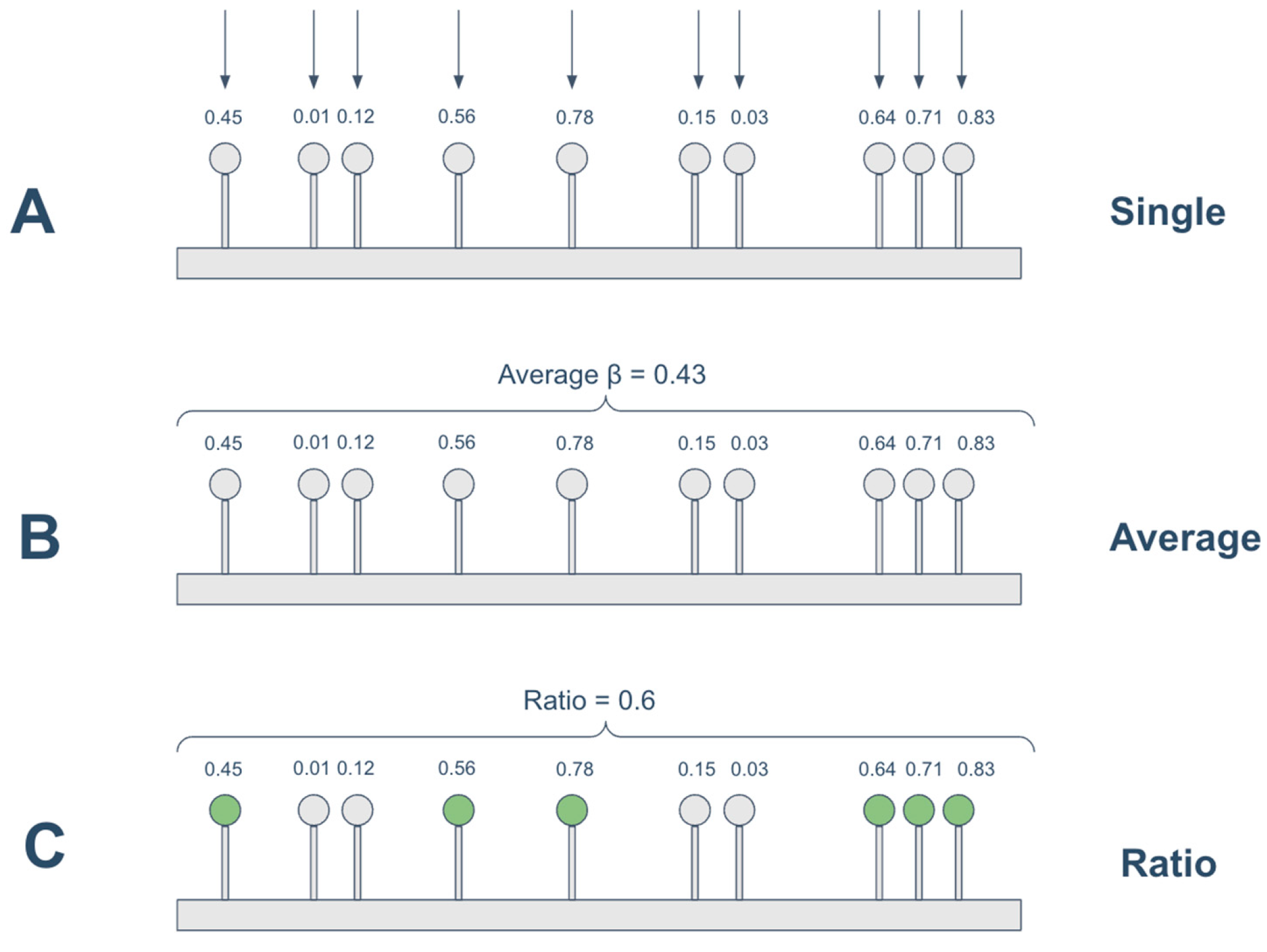
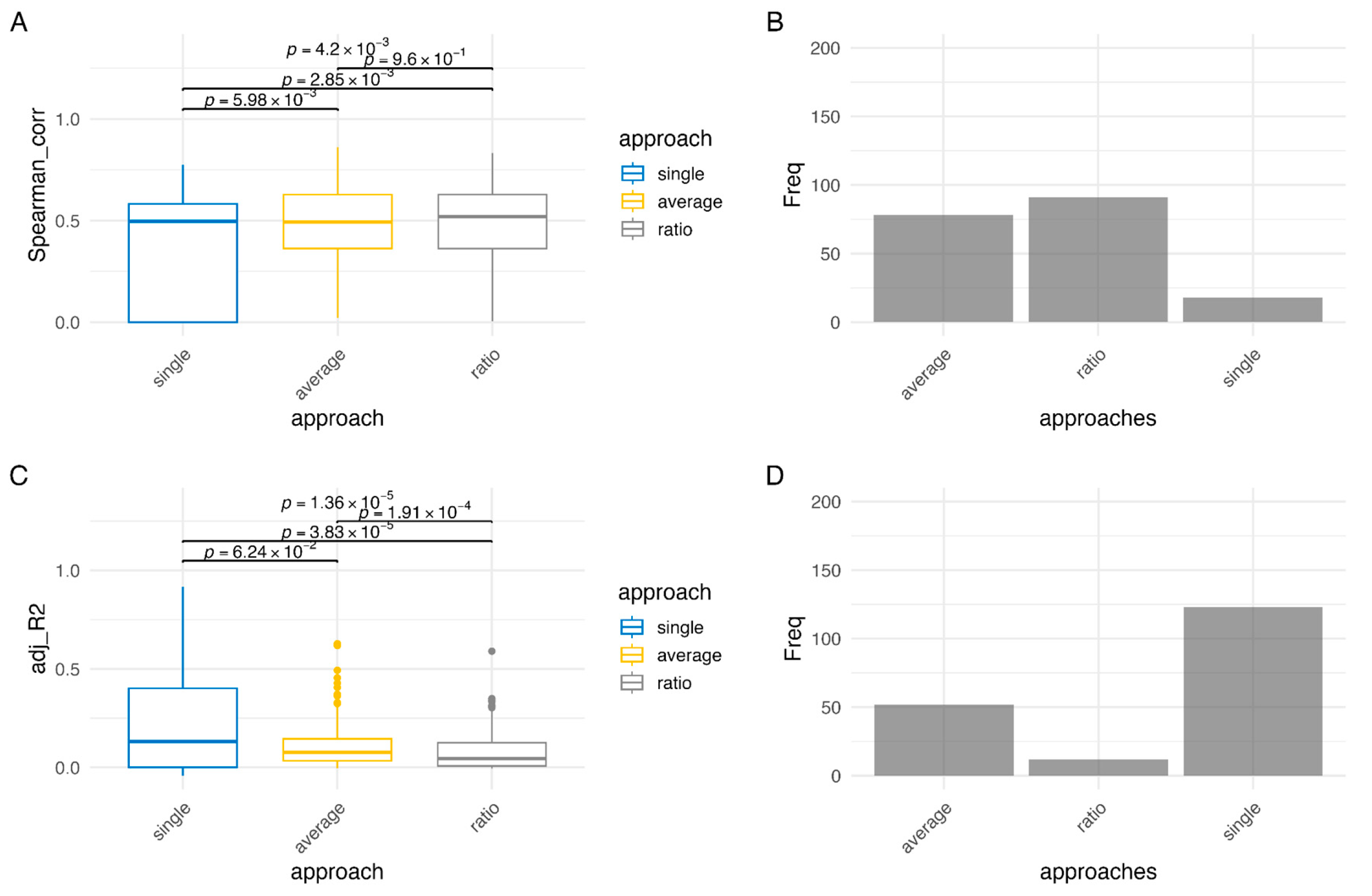
2.4. Methylation-Expression Correlation: Spearman- and Regression-Based Approaches
- •
- single: the unit methylation data is the beta value of each individual CpG site;
- •
- average: the promoter methylation status is calculated as the average of the beta values located on the promoter region;
- •
- ratio: the promoter methylation status was calculated as the ratio of methylated CpGs to the total number of CpGs in the promoter region. A CpG was considered methylated if its beta value was ≥0.3, following previous studies [26,33]. This threshold captures partially methylated CpGs that may influence transcription. More stringent cutoffs (e.g., β ≥ 0.5 or 0.7) could miss biologically relevant intermediate methylation, but future analyses may examine the impact of alternative thresholds.
2.5. Validation Against Independent Studies
2.6. Code Availability
3. Results
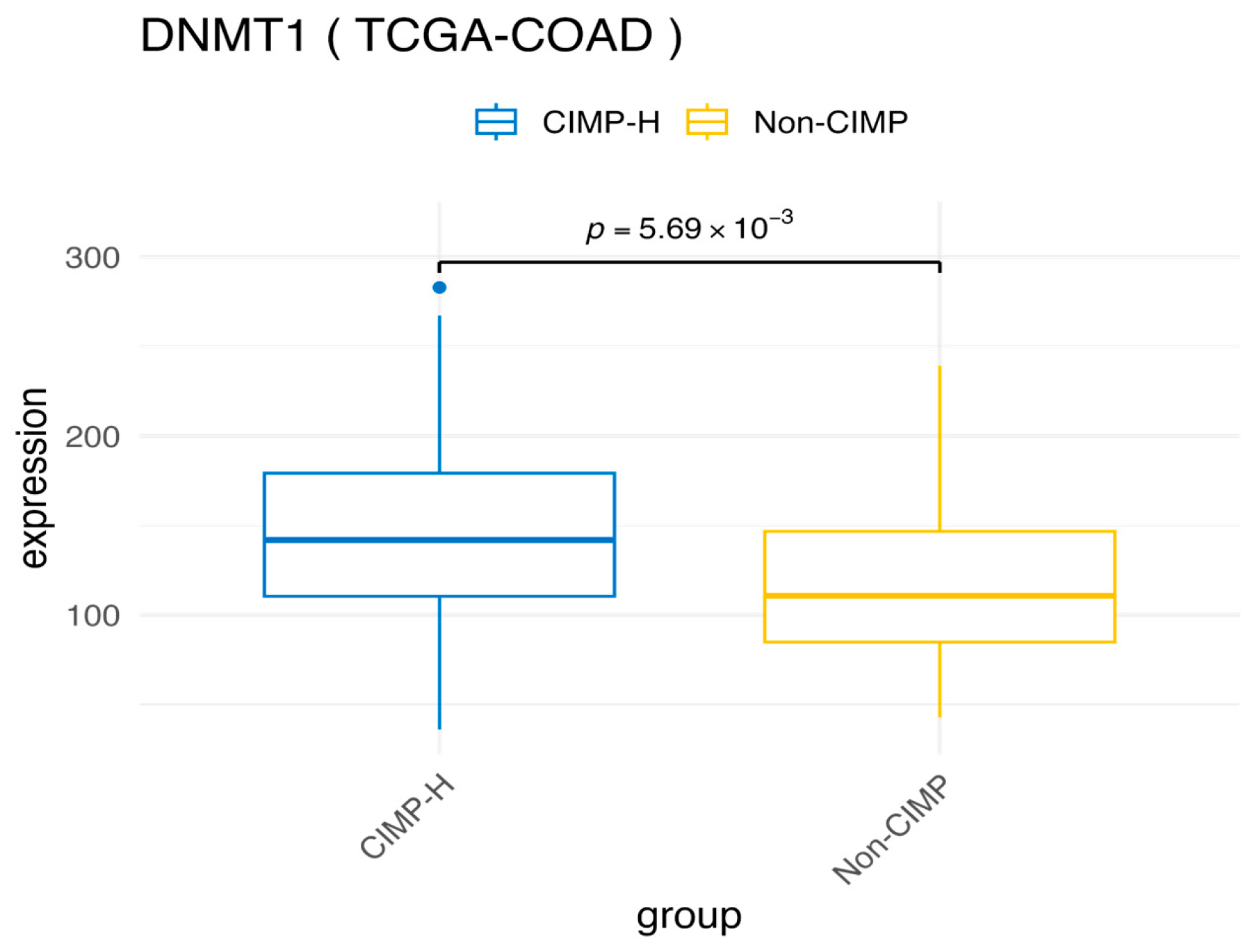
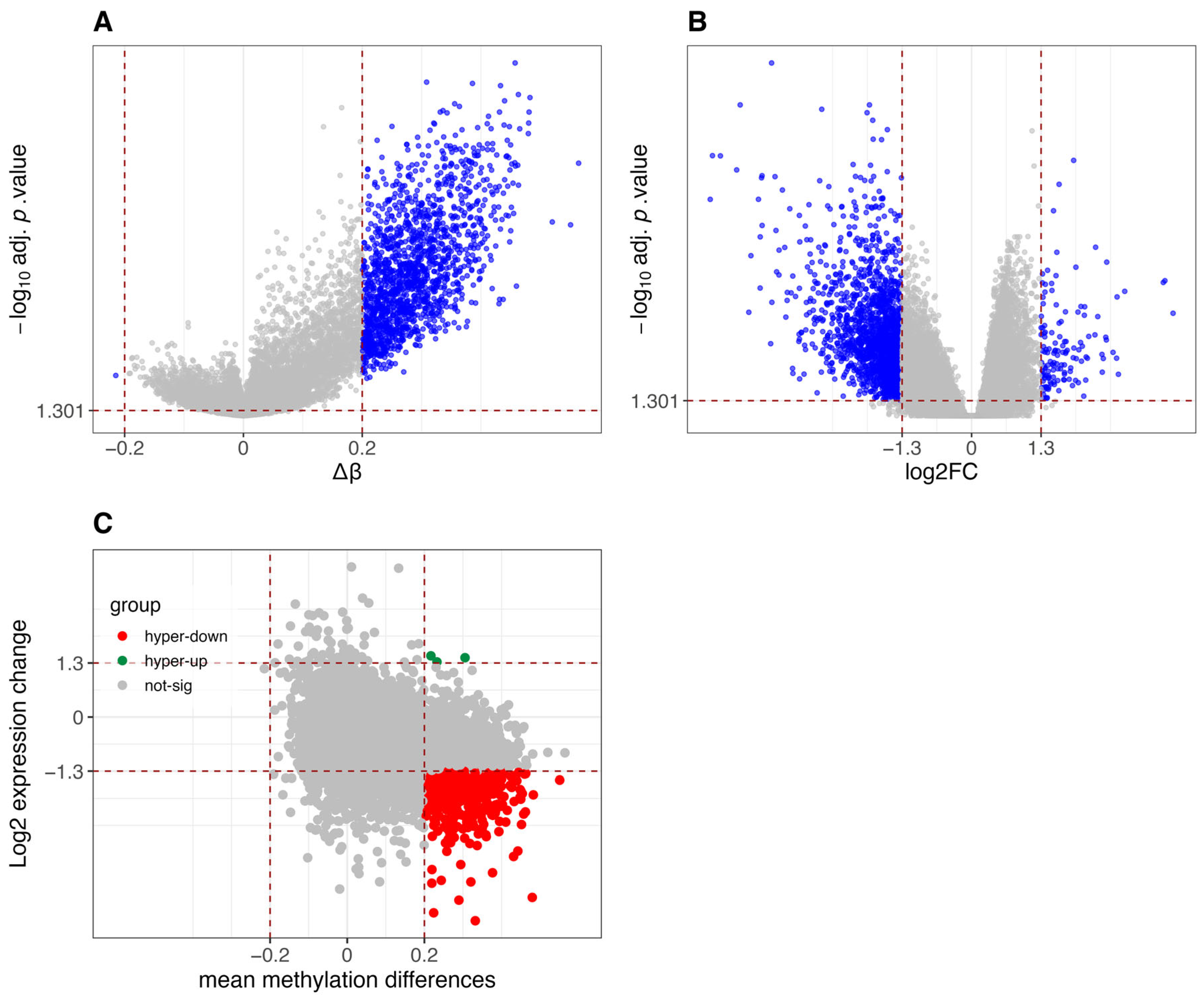
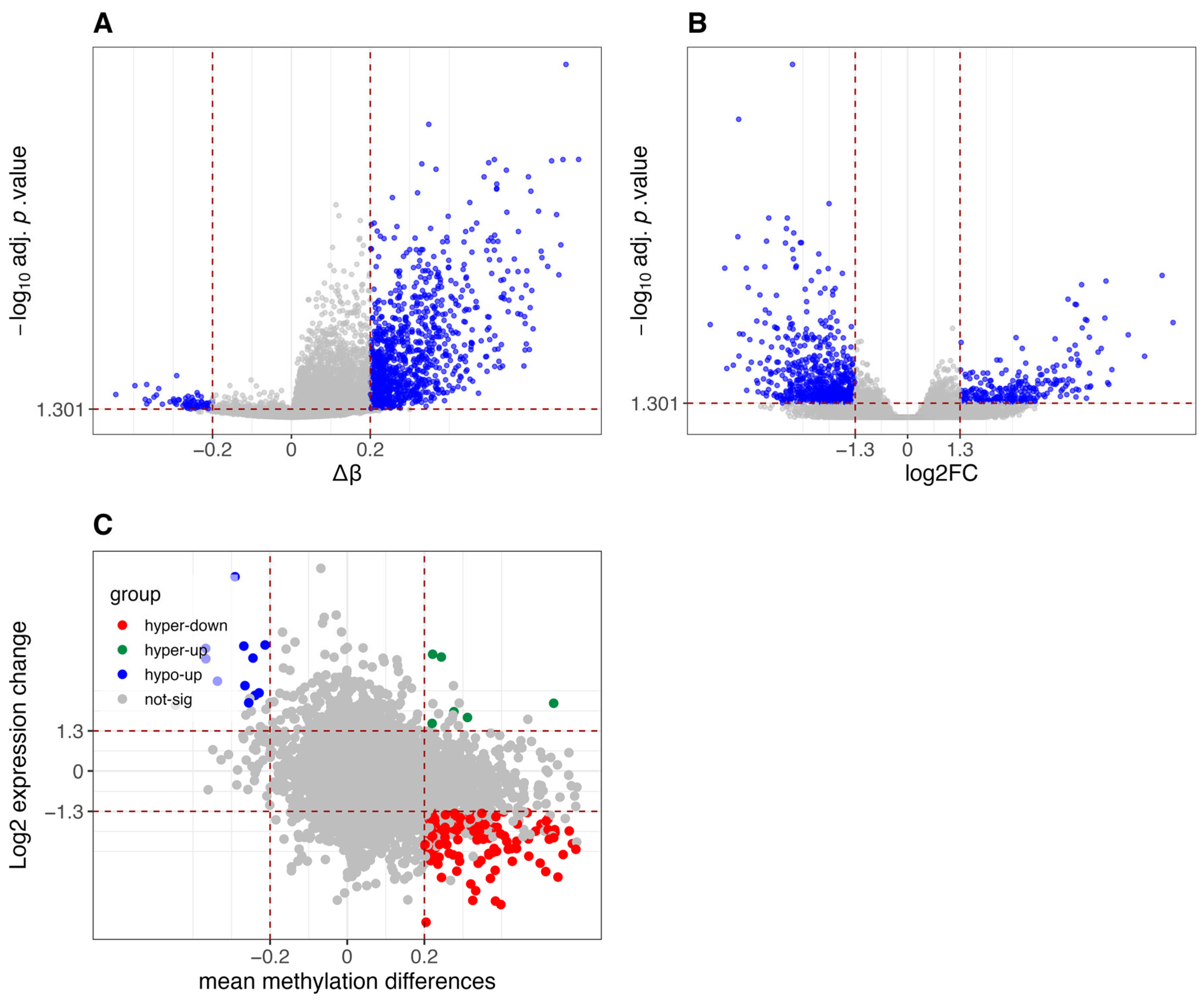
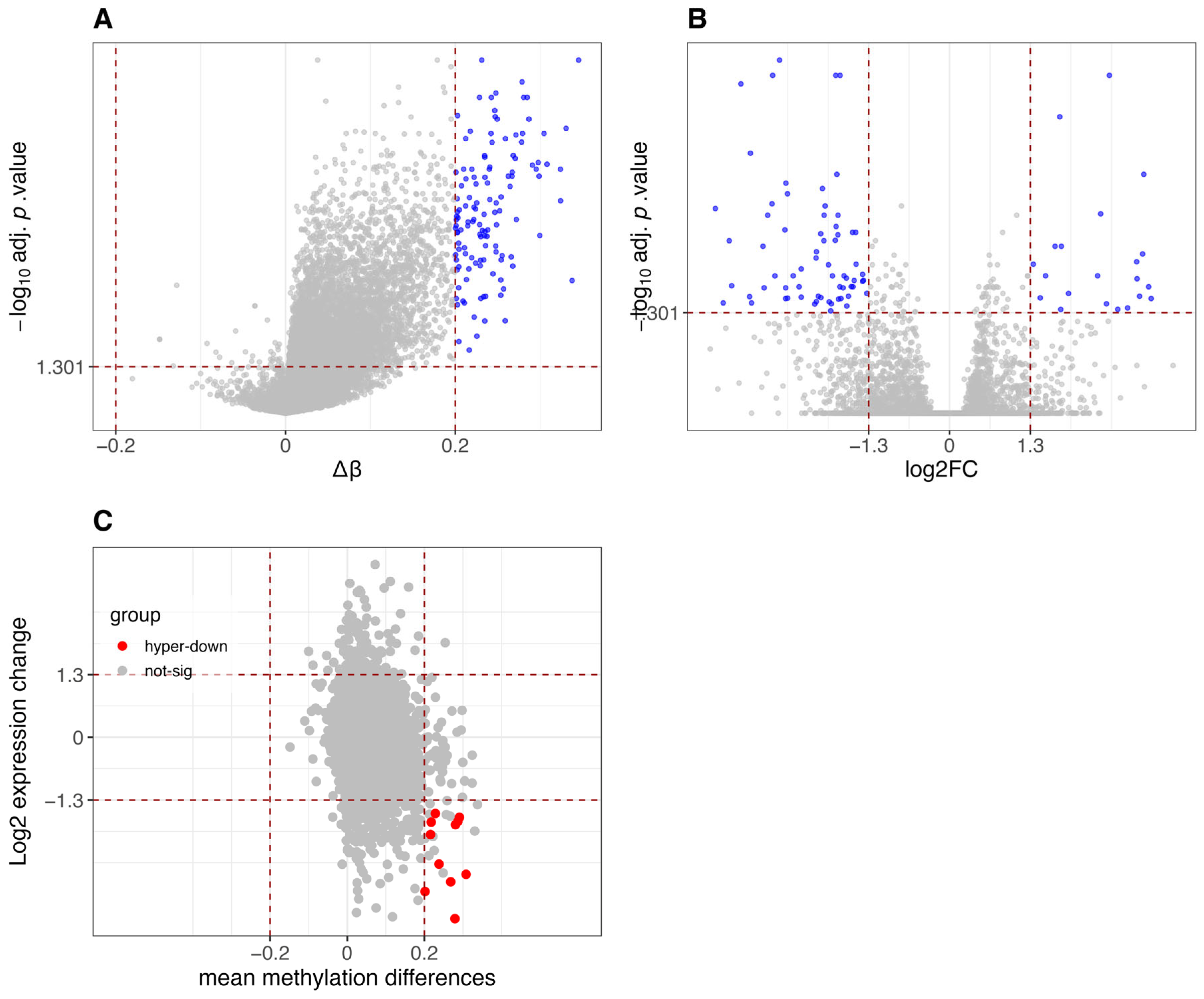
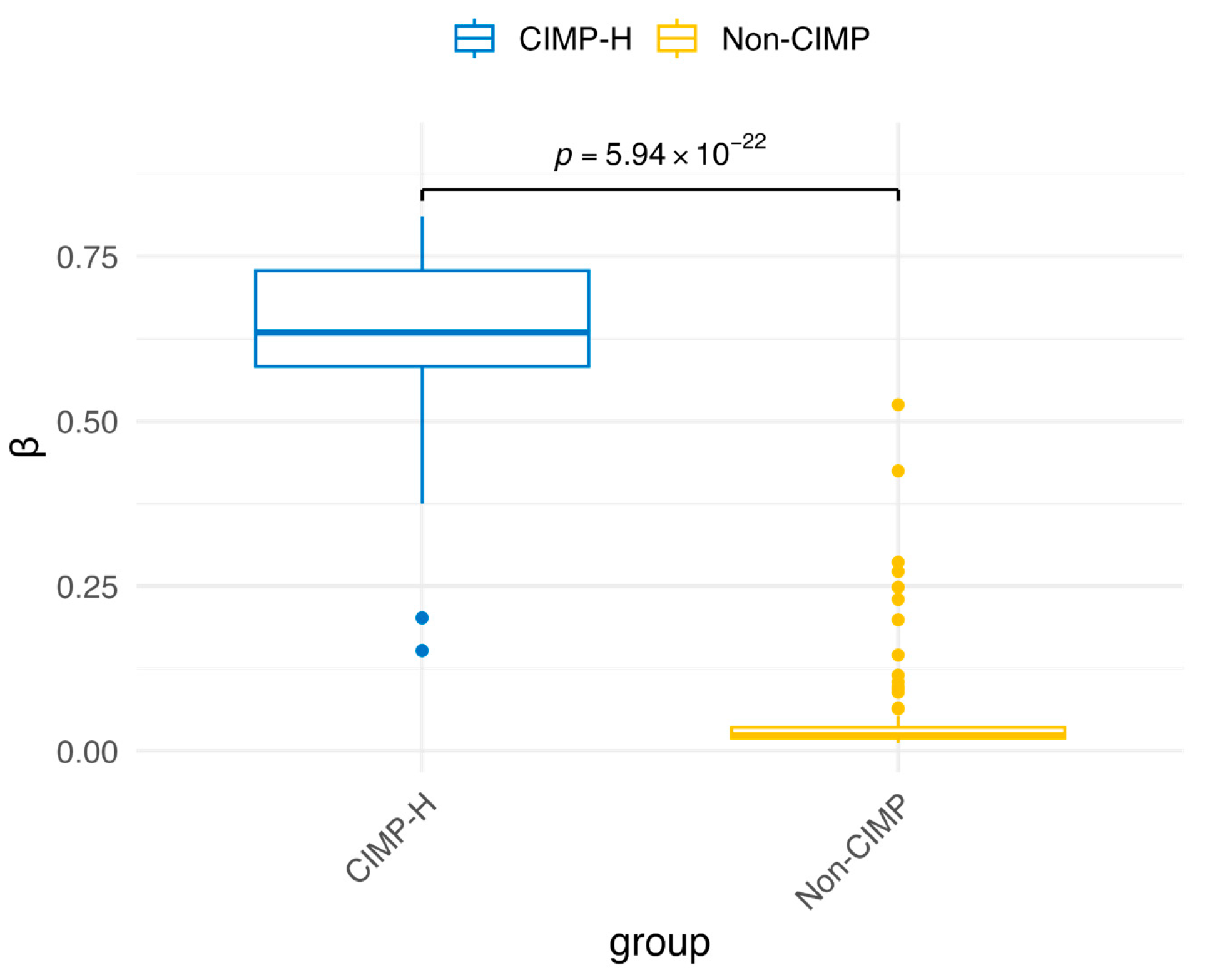
3.1. Comparative Analysis of Differential Methylation and Gene Expression Between CIMP-H and Non-CIMP Groups
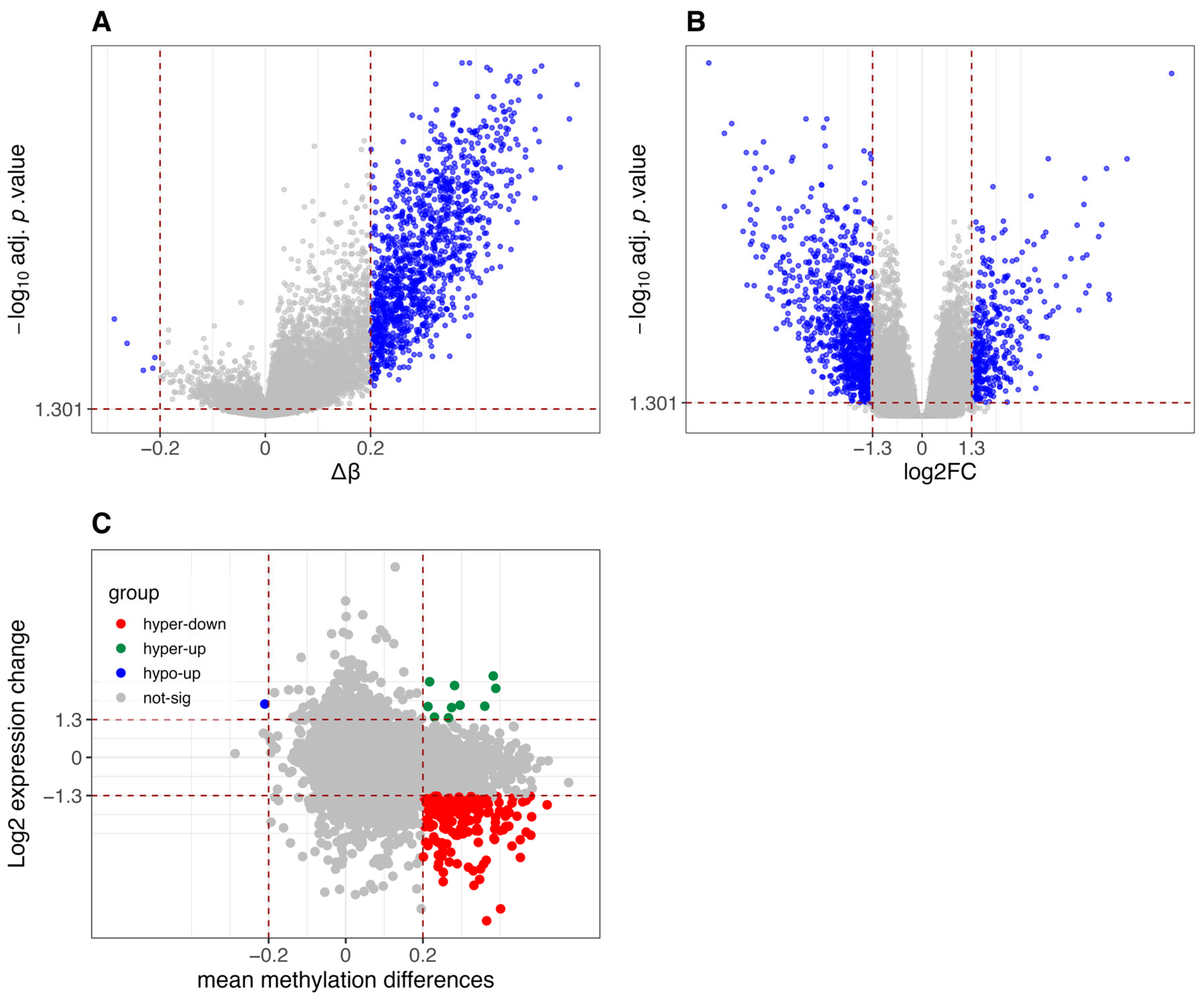
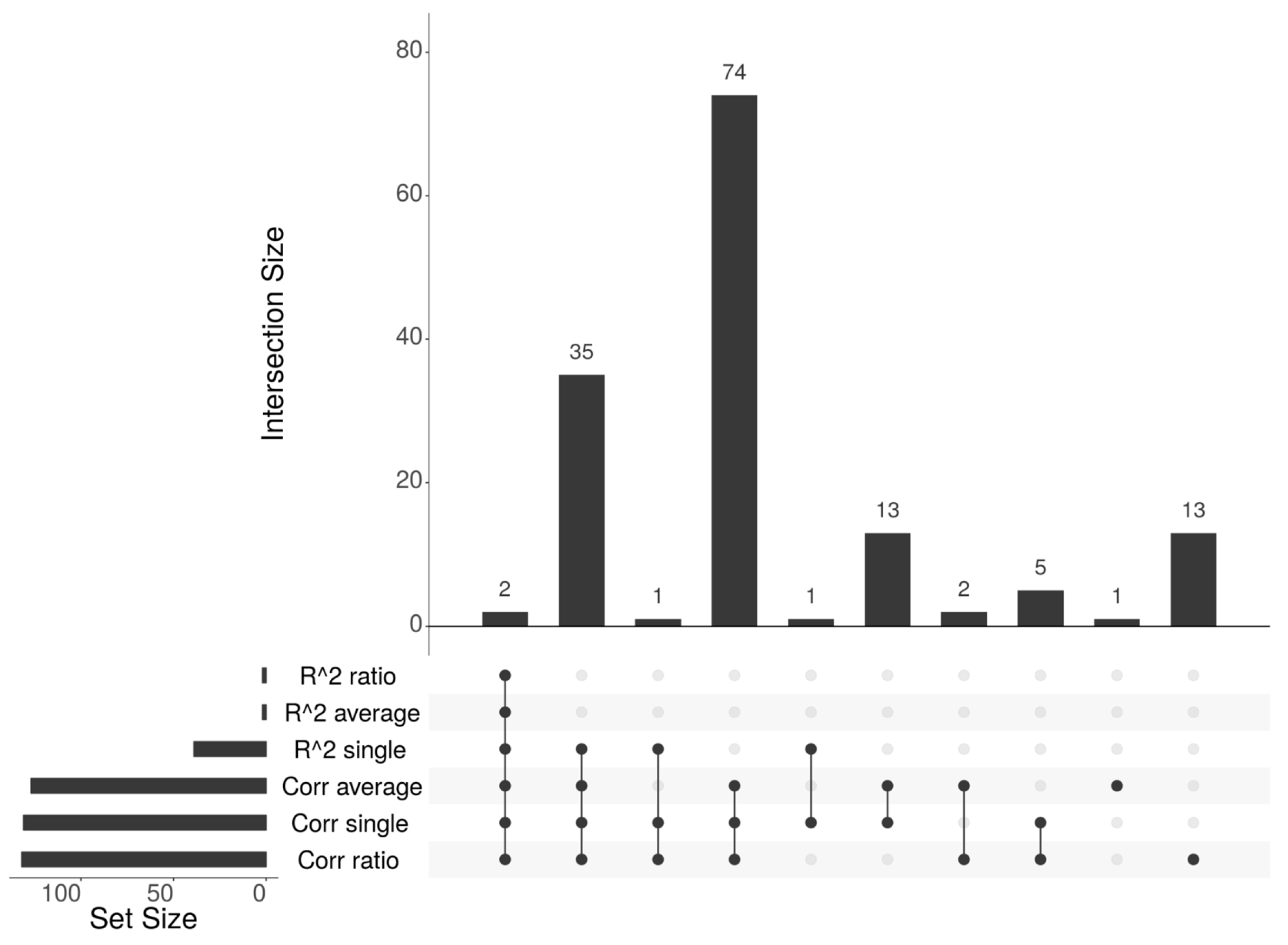
3.2. Results of Methylation-Expression Correlation Methods and Method Selection
3.3. Identified Epigenetically Regulated Genes
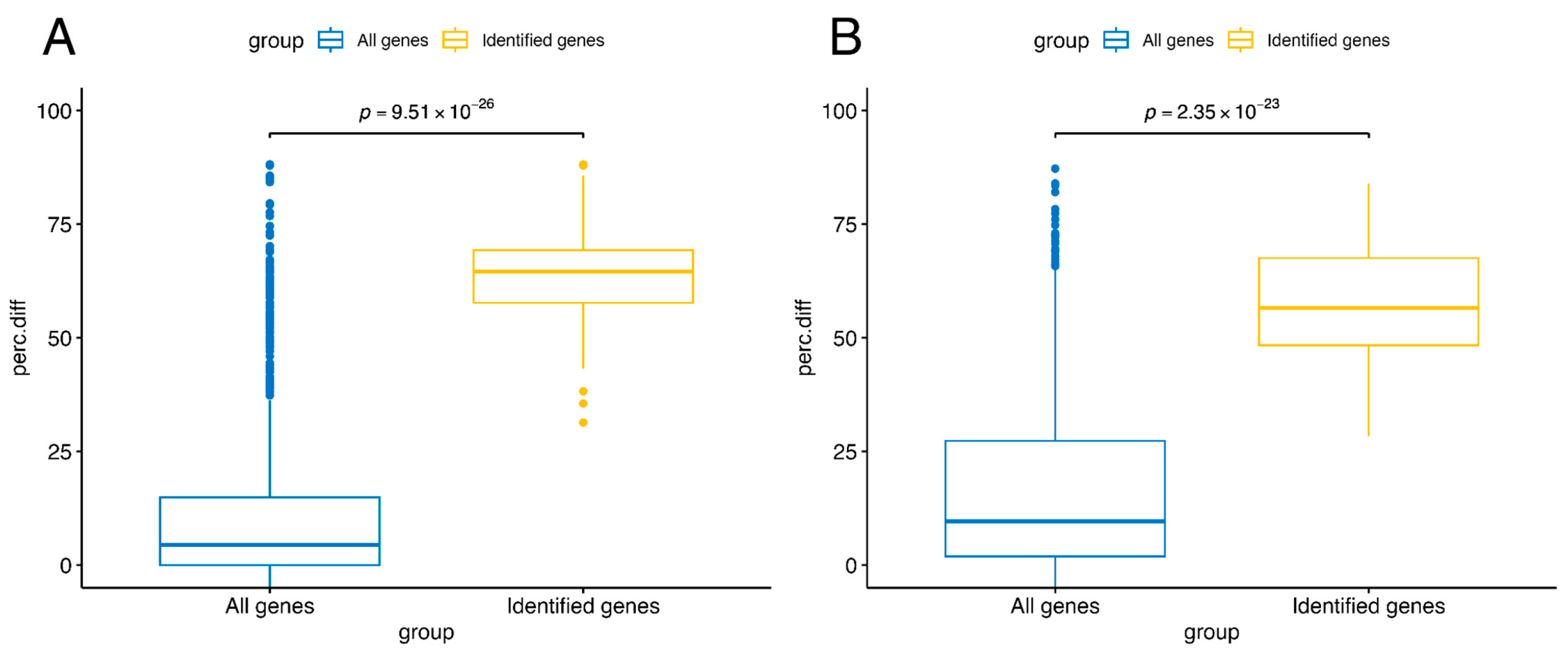
3.4. Validation Against Independent Studies
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CRC | Colorectal Cancer |
| CIMP | CpG Island Methylator Phenotype |
| COAD | Colon Adenocarcinoma |
| DE | Differentially Expressed |
| GBM | Glioblastoma Multiforme |
| GDC | NCI Genomic Data Commons |
| MESO | Mesothelioma |
| SNPs | Single Nucleotide Polymorphisms |
| STAD | Stomach Adenocarcinoma |
| TCGA | The Cancer Genome Atlas |
| TSS | Transcriptional Start Site |
Appendix A
| Diff Methylation Threshold | Diff Expression Threshold (log2FC) |
|---|---|
| log2FC ≥ log2(1.05) or log2FC ≤ log2(0.95) | |log2FC| ≥ 1 |
| |log2FC| ≥ 1.3 | |
| |log2FC| ≥ 1.5 |log2FC| ≥ 2 | |
| |log2FC| ≥ 1.2 | |log2FC| ≥ 1 |log2FC| ≥ 1.3 |log2FC| ≥ 1.5 |
| |log2FC| ≥ 2 | |
| |∆β| ≥ 0.1 | |log2FC| ≥ 1 |log2FC| ≥ 1.3 |log2FC| ≥ 1.5 |
| |log2FC| ≥ 2 | |
| |∆β| ≥ 0.2 | |log2FC| ≥ 1 |log2FC| ≥ 1.3 |log2FC| ≥ 1.5 |
| |log2FC| ≥ 2 | |
| |∆β| ≥ 0.3 | |log2FC| ≥ 1 |log2FC| ≥ 1.3 |log2FC| ≥ 1.5 |log2FC| ≥ 2 |
| Genes | adj-R2 | Model p.Value | Meth Δβ | Expr |log2FC| |
|---|---|---|---|---|
| MLH1 | 0.9160142218 | 9.7573301802 × 10−51 | 0.2696545 | −1.665074 |
| CHFR | 0.9155066294 | 8.0396914024 × 10−60 | 0.269226246 | −1.32773189 |
| TMEM176B | 0.8590336994 | 3.3328801090 × 10−54 | 0.214482 | −1.416893 |
| ZNF350 | 0.8545988847 | 4.4278195945 × 10−60 | 0.2320397 | −1.318949 |
| ZNF570 | 0.8445531222 | 1.2327774024 × 10−50 | 0.271608 | −1.721735 |
| ZNF530 | 0.8400372136 | 5.5140546279 × 10−52 | 0.2465791 | −2.057586 |
| ZNF347 | 0.8114975965 | 5.5301184056 × 10−52 | 0.2658497 | −1.447183 |
| ZNF461 | 0.8051404749 | 3.8759977094 × 10−46 | 0.3696641 | −1.593841 |
| ZNF470 | 0.8011371961 | 1.5487570193 × 10−45 | 0.3167758 | −2.546431 |
| ZNF665 | 0.7960067914 | 5.4817053359 × 10−47 | 0.2725474 | −1.64277 |
| FUZ | 0.7777392954 | 2.5091023605 × 10−40 | 0.2435446 | −1.825009 |
| NHLRC1 | 0.7712743506 | 4.0644414518 × 10−40 | 0.3041523 | −2.612487 |
| ZNF518B | 0.7698917405 | 6.0750372604 × 10−40 | 0.2561586 | −2.0496 |
| ZSCAN18 | 0.7674092534 | 1.9739502642 × 10−38 | 0.2257022 | −1.548055 |
| RAB32 | 0.7542588235 | 4.8471220810 × 10−38 | 0.273973 | −1.748984 |
| EPM2AIP1 | 0.7421457065 | 2.0466792632 × 10−32 | 0.3108179 | −1.668124 |
| ZNF790 | 0.7166895081 | 2.2100181493 × 10−33 | 0.3014525 | −1.802083 |
| GSTM3 | 0.7049150426 | 3.8140509799 × 10−35 | 0.2129139 | −1.305608 |
| PCDHGC3 | 0.7017538942 | 5.1118072493 × 10−33 | 0.2672849 | −1.390975 |
| LARP6 | 0.6748827652 | 2.9597049278 × 10−34 | 0.3334754 | −1.758083 |
| BBS5 | 0.6656853753 | 4.8494375654 × 10−32 | 0.2283376 | −1.345157 |
| GNG4 | 0.6557856995 | 5.0993101991 × 10−26 | 0.3651545 | −5.601045 |
| PAX9 | 0.6531760303 | 6.1897381747 × 10−31 | 0.3822773 | 2.796667 |
| ZNF287 | 0.648190007 | 9.0622211349 × 10−29 | 0.3056667 | −1.320021 |
| MYEF2 | 0.6400660127 | 3.2151261728 × 10−29 | 0.4526309 | −3.425723 |
| ZNF345 | 0.6273470281 | 4.2866553672 × 10−27 | 0.3128804 | −1.834955 |
| SCRN1 | 0.6178338195 | 1.9406897476 × 10−27 | 0.2349142 | −1.310881 |
| TTC9B | 0.6120338869 | 7.9436566855 × 10−29 | 0.3603467 | 1.759013 |
| ZNF256 | 0.608153717 | 3.8447277982 × 10−25 | 0.2595887 | −1.940782 |
| DNM3 | 0.6032257626 | 8.6872374039 × 10−26 | 0.2889181 | −1.577103 |
| VANGL2 | 0.5803117683 | 1.1544112232 × 10−24 | 0.2977 | −2.432072 |
| TMEM176A | 0.5797805563 | 4.1558335632 × 10−24 | 0.251388 | −1.813204 |
| KLF7 | 0.5782472007 | 1.6506221588 × 10−23 | 0.2988387 | −1.478293 |
| STC2 | 0.5533759868 | 6.4931066518 × 10−24 | 0.3305407 | −1.505171 |
| GAL | 0.5355766068 | 3.4449879148 × 10−22 | 0.2289686815 | −2.82493938 |
| TRMT12 | 0.5256366293 | 2.6472947155 × 10−23 | 0.2417141 | −1.468805 |
| ACSL6 | 0.5238291627 | 5.0859492373 × 10−20 | 0.3321547 | −4.38583 |
| ADAM32 | 0.5231521371 | 1.6271688295 × 10−22 | 0.3019306 | −2.247536 |
| DENND2C | 0.5085058292 | 1.4737898361 × 10−17 | 0.2506021 | −1.629142 |
| Genes | adj-R2 | Model p.Value | Meth Δβ | Expr |log2FC| |
|---|---|---|---|---|
| MLH1 | 0.8625194402 | 5.1115944567 × 10−82 | 0.3309603 | −1.313101 |
| SPAG16 | 0.8108002593 | 7.9047197457 × 10−82 | 0.2190329 | −1.335292 |
| ZNF549 | 0.7659061963 | 2.8651608949 × 10−73 | 0.2224513 | −1.352792 |
| ZNF530 | 0.7521410701 | 9.5033608340 × 10−69 | 0.2278235 | −1.364519 |
| EPM2AIP1 | 0.7326966892 | 1.2653387700 × 10−59 | 0.3128086 | −1.575874 |
| FUZ | 0.7183519805 | 1.6965220867 × 10−59 | 0.2771977 | −1.806915 |
| PCDHGC3 | 0.690208358 | 5.1966558535 × 10−58 | 0.271416 | −1.424485 |
| ZNF415 | 0.6898570341 | 6.8339031726 × 10−63 | 0.2125039 | −1.356719 |
| ZNF518B | 0.6557446401 | 1.1716878367 × 10−50 | 0.2012544 | −1.572526 |
| TTC9B | 0.6534714454 | 6.1507217211 × 10−53 | 0.2325077 | 1.326743 |
| STOX2 | 0.6262812014 | 9.8970559076 × 10−50 | 0.3389031 | −2.172084 |
| PPP1R9A | 0.6216226523 | 1.7206102252 × 10−46 | 0.2289913 | −2.537084 |
| PYGO1 | 0.6045898697 | 4.5360754050 × 10−49 | 0.21845 | −1.49573 |
| ZNF512B | 0.5981651401 | 2.0369709415 × 10−47 | 0.2650061 | −1.691932 |
| TUB | 0.5694345938 | 1.5470821262 × 10−40 | 0.2091746743 | −1.72417821 |
| VANGL2 | 0.5676130373 | 3.6114328641 × 10−42 | 0.3398634 | −2.803349 |
| LARP6 | 0.5645034498 | 1.7903179607 × 10−42 | 0.2753438 | −1.619616 |
| NUP210 | 0.5544244614 | 1.2961432358 × 10−40 | 0.23946828 | −1.7079703 |
| ZSCAN18 | 0.5467536543 | 2.3698137358 × 10−36 | 0.2450219 | −1.674229 |
| NHLRC1 | 0.5453508891 | 3.1074756177 × 10−37 | 0.262541 | −2.077767 |
| PAIP2B | 0.534069801 | 5.2728482658 × 10−36 | 0.2455338 | −1.795764 |
| ZNF300 | 0.5334687427 | 1.4050379317 × 10−39 | 0.2397008 | −1.31001 |
| CABYR | 0.5072583955 | 8.0060346562 × 10−35 | 0.2526839 | −1.592365 |
| Genes | adj-R2 | Model p.Value | Meth Δβ | Expr |log2FC| |
|---|---|---|---|---|
| VILL | 0.9262673517 | 2.4725768581 × 10−17 | 0.3397819 | −1.525788 |
| FAM50B | 0.9094371166 | 1.0201226544 × 10−12 | 0.2101957 | −1.867682 |
| TRIP4 | 0.9025516512 | 2.5642870743 × 10−22 | 0.3142913 | −1.61957 |
| FBXO17 | 0.8961476435 | 1.7337920747 × 10−22 | 0.3254043 | −4.190574 |
| EMP3 | 0.8679583888 | 2.2357782662 × 10−19 | 0.3466201 | −2.898656 |
| FCHSD1 | 0.8678618202 | 2.2724422625 × 10−19 | 0.2259016 | −1.523468 |
| KHNYN | 0.8342158227 | 7.6944683535 × 10−15 | 0.5379234 | −1.894583 |
| TUBA1C | 0.8262239337 | 1.9594887247 × 10−13 | 0.2737355 | −1.856427 |
| ZDHHC12 | 0.8093786572 | 7.7184376089 × 10−16 | 0.2317068 | −1.426678 |
| TSTD1 | 0.7886016084 | 7.6354783070 × 10−12 | 0.3710361 | −3.477444 |
| RAB34 | 0.7870253404 | 3.6418158146 × 10−13 | 0.367253 | −2.712978 |
| XKR8 | 0.7818698663 | 1.3668922651 × 10−11 | 0.3775083 | −2.632459 |
| MARVELD1 | 0.7664193197 | 6.8408254916 × 10−14 | 0.4048824 | −1.322334 |
| KCNB1 | 0.7546708328 | 6.3690196282 × 10−12 | 0.5351048 | 2.197043 |
| TOM1L1 | 0.7536948059 | 6.8998771422 × 10−12 | 0.5146205 | −3.254208 |
| CLIC1 | 0.7321082475 | 6.8432347298 × 10−9 | 0.3569989 | −1.734158 |
| LRRC61 | 0.7236492858 | 9.4667143224 × 10−8 | 0.2163285 | −2.899631 |
| ALDH7A1 | 0.7176096928 | 3.3559684485 × 10−13 | 0.339439 | −2.984188 |
| B3GNT5 | 0.7117808817 | 6.0406915694 × 10−11 | 0.5755067 | −1.936565 |
| PPCS | 0.7024967074 | 1.1598064690 × 10−10 | 0.2597928 | −1.438017 |
| FABP5 | 0.700038623 | 4.7278840357 × 10−9 | 0.2886457 | −2.75996 |
| TTC12 | 0.6936032628 | 5.4736520843 × 10−10 | 0.5353078 | −2.154691 |
| PYROXD2 | 0.6820653714 | 1.8500042218 × 10−11 | 0.2564855 | −1.939967 |
| MIR155HG | 0.6767940524 | 8.3669349604 × 10−11 | 0.407702 | −2.100055 |
| B3GNT7 | 0.6724731901 | 4.8141644041 × 10−9 | 0.2946536 | −1.592759 |
| ECHDC2 | 0.670169979 | 4.2148141764 × 10−11 | 0.4173006 | −2.530786 |
| EID3 | 0.6485458465 | 3.4989560072 × 10−9 | 0.592721 | −2.531831 |
| OSMR | 0.641828216 | 8.1542530517 × 10−11 | 0.2419406 | −1.97559 |
| RBP1 | 0.6248659311 | 2.1109179887 × 10−9 | 0.3841675936 | −4.21122843 |
| FERMT1 | 0.6111895937 | 1.6592323584 × 10−9 | −0.3364869 | 2.915307 |
| LRRC34 | 0.6099437671 | 6.3478581723 × 10−8 | 0.2528083 | −1.948628 |
| TCEA3 | 0.5991307379 | 1.0867475374 × 10−9 | 0.5596619 | −2.702318 |
| STEAP3 | 0.5970572542 | 9.8376572230 × 10−9 | 0.3717444 | −2.009174 |
| CBLN3 | 0.5849646858 | 2.1146152728 × 10−7 | 0.5835179 | −2.35043 |
| NIPAL2 | 0.5834731801 | 2.0029059943 × 10−8 | 0.2036887 | −1.565651 |
| PDLIM4 | 0.5379376026 | 1.6548089798 × 10−6 | 0.3202394 | −3.657253 |
| ZIC5 | 0.5184717764 | 4.3845145105 × 10−7 | 0.2867333 | −2.217452 |
| CMYA5 | 0.5093551602 | 5.1501871268 × 10−6 | 0.6127386 | −2.663765 |
| CD109 | 0.5007389969 | 1.6346569625 × 10−7 | 0.3198041 | −1.770341 |
| Genes | adj-R2 | Model p.Value | Meth Δβ | Expr |log2FC| |
|---|---|---|---|---|
| NMNAT3 | 0.71194155 | 6.6224383300 × 10−12 | 0.2379173 | −2.629047 |
| TMEM220 | 0.65779601 | 8.0332932199 × 10−11 | 0.2176805 | −1.757845 |
| RNF208 | 0.59085732 | 7.4437667235 × 10−8 | 0.2284904 | −1.577897 |
| OCIAD2 | 0.56425239 | 1.1039000636 × 10−6 | 0.2867297 | −1.744734 |
| CMBL | 0.55763349 | 2.2651777226 × 10−8 | 0.268212 | −2.997392 |
| MSI-H | MSI-L | MSS | NA | |
|---|---|---|---|---|
| CIMP-H | 30 | 4 | 9 | 0 |
| Non-CIMP | 9 | 14 | 78 | 1 |
| MSI-H | MSI-L | MSS | |
|---|---|---|---|
| CIMP-H | 36 | 7 | 9 |
| Non-CIMP | 9 | 20 | 168 |
References
- Jones, P.A.; Baylin, S.B. The Fundamental Role of Epigenetic Events in Cancer. Nat. Rev. Genet. 2002, 3, 415–428. [Google Scholar] [CrossRef]
- Davalos, V.; Esteller, M. Cancer Epigenetics in Clinical Practice. CA Cancer J. Clin. 2023, 73, 376–424. [Google Scholar] [CrossRef]
- Liu, R.; Zhao, E.; Yu, H.; Yuan, C.; Abbas, M.N.; Cui, H. Methylation across the Central Dogma in Health and Diseases: New Therapeutic Strategies. Signal Transduct. Target. Ther. 2023, 8, 310. [Google Scholar] [CrossRef]
- Herman, J.G.; Baylin, S.B. Gene Silencing in Cancer in Association with Promoter Hypermethylation. N. Engl. J. Med. 2003, 349, 2042–2054. [Google Scholar] [CrossRef] [PubMed]
- Issa, J.-P. CpG Island Methylator Phenotype in Cancer. Nat. Rev. Cancer 2004, 4, 988–993. [Google Scholar] [CrossRef] [PubMed]
- Nazemalhosseini Mojarad, E.; Kuppen, P.J.; Aghdaei, H.A.; Zali, M.R. The CpG Island Methylator Phenotype (CIMP) in Colorectal Cancer. Gastroenterol. Hepatol. Bed Bench 2013, 6, 120–128. [Google Scholar]
- Toyota, M.; Ahuja, N.; Suzuki, H.; Itoh, F.; Ohe-Toyota, M.; Imai, K.; Baylin, S.B.; Issa, J.P. Aberrant Methylation in Gastric Cancer Associated with the CpG Island Methylator Phenotype. Cancer Res. 1999, 59, 5438–5442. [Google Scholar]
- Malta, T.M.; de Souza, C.F.; Sabedot, T.S.; Silva, T.C.; Mosella, M.S.; Kalkanis, S.N.; Snyder, J.; Castro, A.V.B.; Noushmehr, H. Glioma CpG Island Methylator Phenotype (G-CIMP): Biological and Clinical Implications. Neuro-Oncology 2018, 20, 608–620. [Google Scholar] [CrossRef]
- Curtin, K.; Slattery, M.L.; Samowitz, W.S. CpG Island Methylation in Colorectal Cancer: Past, Present and Future. Pathol. Res. Int. 2011, 2011, 902674. [Google Scholar] [CrossRef]
- Ma, Y.; Li, J.; Zhao, X.; Ji, C.; Hu, W.; Ma, Y.; Qu, F.; Sun, Y.; Zhang, X. Multi-Omics Cluster Defines the Subtypes of CRC with Distinct Prognosis and Tumor Microenvironment. Eur. J. Med. Res. 2024, 29, 207. [Google Scholar] [CrossRef] [PubMed]
- Müller, D.; Győrffy, B. DNA Methylation-Based Diagnostic, Prognostic, and Predictive Biomarkers in Colorectal Cancer. Biochim. Biophys. Acta BBA—Rev. Cancer 2022, 1877, 188722. [Google Scholar] [CrossRef]
- Fatemi, N.; Tierling, S.; Es, H.A.; Varkiani, M.; Mojarad, E.N.; Aghdaei, H.A.; Walter, J.; Totonchi, M. DNA Methylation Biomarkers in Colorectal Cancer: Clinical Applications for Precision Medicine. Int. J. Cancer 2022, 151, 2068–2081. [Google Scholar] [CrossRef] [PubMed]
- Baião, A.R.; Cai, Z.; Poulos, R.C.; Robinson, P.J.; Reddel, R.R.; Zhong, Q.; Vinga, S.; Gonçalves, E. A Technical Review of Multi-Omics Data Integration Methods: From Classical Statistical to Deep Generative Approaches. Brief. Bioinform. 2025, 26, bbaf355. [Google Scholar] [CrossRef] [PubMed]
- Sibilio, P.; De Smaele, E.; Paci, P.; Conte, F. Integrating Multi-Omics Data: Methods and Applications in Human Complex Diseases. Biotechnol. Rep. 2025, 48, e00938. [Google Scholar] [CrossRef] [PubMed]
- Majumdar, K.; Jaffrézic, F.; Rau, A.; Gormley, I.C.; Murphy, T.B. Integrated Differential Analysis of Multi-Omics Data Using a Joint Mixture Model: Idiffomix. arXiv 2024, arXiv:2412.17511. [Google Scholar] [CrossRef]
- Singh, A.; Shannon, C.P.; Gautier, B.; Rohart, F.; Vacher, M.; Tebbutt, S.J.; Lê Cao, K.-A. DIABLO: An Integrative Approach for Identifying Key Molecular Drivers from Multi-Omics Assays. Bioinformatics 2019, 35, 3055–3062. [Google Scholar] [CrossRef]
- Cedoz, P.-L.; Prunello, M.; Brennan, K.; Gevaert, O. MethylMix 2.0: An R Package for Identifying DNA Methylation Genes. Bioinformatics 2018, 34, 3044–3046. [Google Scholar] [CrossRef]
- Huang, H.; Fu, J.; Zhang, L.; Xu, J.; Li, D.; Onwuka, J.U.; Zhang, D.; Zhao, L.; Sun, S.; Zhu, L.; et al. Integrative Analysis of Identifying Methylation-Driven Genes Signature Predicts Prognosis in Colorectal Carcinoma. Front. Oncol. 2021, 11, 629860. [Google Scholar] [CrossRef]
- Silva, T.C.; Coetzee, S.G.; Gull, N.; Yao, L.; Hazelett, D.J.; Noushmehr, H.; Lin, D.-C.; Berman, B.P. ELMER v.2: An R/Bioconductor Package to Reconstruct Gene Regulatory Networks from DNA Methylation and Transcriptome Profiles. Bioinformatics 2019, 35, 1974–1977. [Google Scholar] [CrossRef]
- Ruiz-Arenas, C.; González, J.R. Redundancy Analysis Allows Improved Detection of Methylation Changes in Large Genomic Regions. BMC Bioinform. 2017, 18, 553. [Google Scholar] [CrossRef]
- Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.M.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer Analysis Project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef]
- Colaprico, A.; Silva, T.C.; Olsen, C.; Garofano, L.; Cava, C.; Garolini, D.; Sabedot, T.S.; Malta, T.M.; Pagnotta, S.M.; Castiglioni, I.; et al. TCGAbiolinks: An R/Bioconductor Package for Integrative Analysis of TCGA Data. Nucleic Acids Res. 2016, 44, e71. [Google Scholar] [CrossRef]
- Bibikova, M.; Barnes, B.; Tsan, C.; Ho, V.; Klotzle, B.; Le, J.M.; Delano, D.; Zhang, L.; Schroth, G.P.; Gunderson, K.L.; et al. High Density DNA Methylation Array with Single CpG Site Resolution. Genomics 2011, 98, 288–295. [Google Scholar] [CrossRef]
- Liu, Y.; Sethi, N.S.; Hinoue, T.; Schneider, B.G.; Cherniack, A.D.; Sanchez-Vega, F.; Seoane, J.A.; Farshidfar, F.; Bowlby, R.; Islam, M.; et al. Comparative Molecular Analysis of Gastrointestinal Adenocarcinomas. Cancer Cell 2018, 33, 721–735.e8. [Google Scholar] [CrossRef]
- Brennan, C.W.; Verhaak, R.G.W.; McKenna, A.; Campos, B.; Noushmehr, H.; Salama, S.R.; Zheng, S.; Chakravarty, D.; Sanborn, J.Z.; Berman, S.H.; et al. The Somatic Genomic Landscape of Glioblastoma. Cell 2013, 155, 462–477. [Google Scholar] [CrossRef]
- Mangiante, L.; Alcala, N.; Sexton-Oates, A.; Di Genova, A.; Gonzalez-Perez, A.; Khandekar, A.; Bergstrom, E.N.; Kim, J.; Liu, X.; Blazquez-Encinas, R.; et al. Multiomic Analysis of Malignant Pleural Mesothelioma Identifies Molecular Axes and Specialized Tumor Profiles Driving Intertumor Heterogeneity. Nat. Genet. 2023, 55, 607–618. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Tricarico, R.; Madzo, J.; Scher, G.; Cohen, M.; Jelinek, J.; Maegawa, S.; Nagarathinam, R.; Scher, C.; Chang, W.-C.; Nicolas, E.; et al. TET1 and TDG Suppress Inflammatory Response in Intestinal Tumorigenesis: Implications for Colorectal Tumors with the CpG Island Methylator Phenotype. Gastroenterology 2023, 164, 921–936.e1. [Google Scholar] [CrossRef]
- Zhang, R.; Chang, C.; Jin, Y.; Xu, L.; Jiang, P.; Wei, K.; Xu, L.; Guo, S.; Sun, S.; He, D. Identification of DNA Methylation-Regulated Differentially Expressed Genes in RA by Integrated Analysis of DNA Methylation and RNA-Seq Data. J. Transl. Med. 2022, 20, 481. [Google Scholar] [CrossRef]
- Xu, W.; Xu, M.; Wang, L.; Zhou, W.; Xiang, R.; Shi, Y.; Zhang, Y.; Piao, Y. Integrative Analysis of DNA Methylation and Gene Expression Identified Cervical Cancer-Specific Diagnostic Biomarkers. Signal Transduct. Target. Ther. 2019, 4, 55. [Google Scholar] [CrossRef]
- Gu, Y.; Zou, Y.M.; Lei, D.; Huang, Y.; Li, W.; Mo, Z.; Hu, Y. Promoter DNA Methylation Analysis Reveals a Novel Diagnostic CpG-Based Biomarker and RAB25 Hypermethylation in Clear Cell Renel Cell Carcinoma. Sci. Rep. 2017, 7, 14200. [Google Scholar] [CrossRef] [PubMed]
- Lakis, V.; Lawlor, R.T.; Newell, F.; Patch, A.-M.; Mafficini, A.; Sadanandam, A.; Koufariotis, L.T.; Johnston, R.L.; Leonard, C.; Wood, S.; et al. DNA Methylation Patterns Identify Subgroups of Pancreatic Neuroendocrine Tumors with Clinical Association. Commun. Biol. 2021, 4, 155. [Google Scholar] [CrossRef]
- Levine, A.J.; Phipps, A.I.; Baron, J.A.; Buchanan, D.D.; Ahnen, D.J.; Cohen, S.A.; Lindor, N.M.; Newcomb, P.A.; Rosty, C.; Haile, R.W.; et al. Clinicopathological Risk Factor Distributions for MLH1 Promoter Region Methylation in CIMP Positive Tumors. Cancer Epidemiol. Biomark. Prev. 2016, 25, 68–75. [Google Scholar] [CrossRef]
- Svedružić, Ž.M. Chapter 6—Dnmt1: Structure and Function. In Progress in Molecular Biology and Translational Science; Cheng, X., Blumenthal, R.M., Eds.; Modifications of Nuclear DNA and Its Regulatory Proteins; Academic Press: Cambridge, UK, 2011; Volume 101, pp. 221–254. [Google Scholar] [CrossRef]
- Liu, J.; Xiao, Q.; Xiao, J.; Niu, C.; Li, Y.; Zhang, X.; Zhou, Z.; Shu, G.; Yin, G. Wnt/β-Catenin Signalling: Function, Biological Mechanisms, and Therapeutic Opportunities. Signal Transduct. Target. Ther. 2022, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Shima, K.; Nosho, K.; Baba, Y.; Cantor, M.; Meyerhardt, J.A.; Giovannucci, E.L.; Fuchs, C.S.; Ogino, S. Prognostic Significance of CDKN2A (P16) Promoter Methylation and Loss of Expression in 902 Colorectal Cancers: Cohort Study and Literature Review. Int. J. Cancer 2011, 128, 1080–1094. [Google Scholar] [CrossRef]
- Jiangzhou, H.; Zhang, H.; Sun, R.; Fahira, A.; Wang, K.; Li, Z.; Shi, Y.; Wang, Z. Integrative Omics Analysis Reveals Effective Stratification and Potential Prognosis Markers of Pan-Gastrointestinal Cancers. iScience 2021, 24, 102824. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.S.; Lin, X.; Chan, T.-F.; Chan, H.Y.E. Pan-Cancer Investigation Reveals Mechanistic Insights of Planar Cell Polarity Gene Fuz in Carcinogenesis. Aging 2021, 13, 7259–7283. [Google Scholar] [CrossRef]
- Cubiella, T.; Celada, L.; San-Juan-Guardado, J.; Rodríguez-Aguilar, R.; Suárez-Priede, Á.; Poch, M.; Dominguez, F.; Fernández-Vega, I.; Montero-Pavón, P.; Fraga, M.F.; et al. PCDHGC3 Hypermethylation as a Potential Biomarker of Intestinal Neuroendocrine Carcinomas. J. Pathol. 2024, 263, 418–428. [Google Scholar] [CrossRef]
- Vega-Benedetti, A.F.; Loi, E.; Moi, L.; Blois, S.; Fadda, A.; Antonelli, M.; Arcella, A.; Badiali, M.; Giangaspero, F.; Morra, I.; et al. Clustered Protocadherins Methylation Alterations in Cancer. Clin. Epigenet. 2019, 11, 100. [Google Scholar] [CrossRef]
- Liu, S.; Liu, X.; Lin, X.; Chen, H. Zinc Finger Proteins in the War on Gastric Cancer: Molecular Mechanism and Clinical Potential. Cells 2023, 12, 1314. [Google Scholar] [CrossRef]
- Jin, Z.; Cheng, Y.; Gu, W.; Zheng, Y.; Sato, F.; Mori, Y.; Olaru, A.V.; Paun, B.C.; Yang, J.; Kan, T.; et al. A Multicenter, Double-Blinded Validation Study of Methylation Biomarkers for Progression Prediction in Barrett’s Esophagus. Cancer Res. 2009, 69, 4112–4115. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gao, L.; Guo, X.; Lian, W.; Deng, K.; Xing, B. Development and Validation of a Novel DNA Methylation-Driven Gene Based Molecular Classification and Predictive Model for Overall Survival and Immunotherapy Response in Patients with Glioblastoma: A Multiomic Analysis. Front. Cell Dev. Biol. 2020, 8, 576996. [Google Scholar] [CrossRef]
- Wang, N.; Song, Q.; Yu, H.; Bao, G. Overexpression of FBXO17 Promotes the Proliferation, Migration and Invasion of Glioma Cells Through the Akt/GSK-3β/Snail Pathway. Cell Transplant. 2021, 30, 9636897211007395. [Google Scholar] [CrossRef]
- Du, D.; Yuan, J.; Ma, W.; Ning, J.; Weinstein, J.N.; Yuan, X.; Fuller, G.N.; Liu, Y. Clinical Significance of FBXO17 Gene Expression in High-Grade Glioma. BMC Cancer 2018, 18, 773. [Google Scholar] [CrossRef]
- Li, L.; Xia, S.; Zhao, Z.; Deng, L.; Wang, H.; Yang, D.; Hu, Y.; Ji, J.; Huang, D.; Xin, T. EMP3 as a Prognostic Biomarker Correlates with EMT in GBM. BMC Cancer 2024, 24, 89. [Google Scholar] [CrossRef]
- Zhu, H.; Hu, X.; Gu, L.; Jian, Z.; Li, L.; Hu, S.; Qiu, S.; Xiong, X. TUBA1C Is a Prognostic Marker in Low-Grade Glioma and Correlates with Immune Cell Infiltration in the Tumor Microenvironment. Front. Genet. 2021, 12, 759953. [Google Scholar] [CrossRef]
- Xu, S.; Wang, Z.; Ye, J.; Mei, S.; Zhang, J. Identification of Iron Metabolism-Related Genes as Prognostic Indicators for Lower-Grade Glioma. Front. Oncol. 2021, 11, 729103. [Google Scholar] [CrossRef] [PubMed]
- Lu, F.; Shen, S.-H.; Wu, S.; Zheng, P.; Lin, K.; Liao, J.; Jiang, X.; Zeng, G.; Wei, D. Hypomethylation-Induced Prognostic Marker Zinc Finger DHHC-Type Palmitoyltransferase 12 Contributes to Glioblastoma Progression. Ann. Transl. Med. 2022, 10, 334. [Google Scholar] [CrossRef]
- Ansar, M.; Thu, L.T.A.; Hung, C.-S.; Su, C.-M.; Huang, M.-H.; Liao, L.-M.; Chung, Y.-M.; Lin, R.-K. Promoter Hypomethylation and Overexpression of TSTD1 Mediate Poor Treatment Response in Breast Cancer. Front. Oncol. 2022, 12, 1004261. [Google Scholar] [CrossRef] [PubMed]
| Dataset | Primary Tumors | CIMP-H | CIMP-L | Non-CIMP |
|---|---|---|---|---|
| TCGA-COAD | 292 | 43 | 116 | 102 |
| TCGA-STAD | 373 | 52 | 63 | 197 |
| TCGA-GBM | 60 | 3 | 0 | 48 |
| TCGA-MESO | 87 | 23 | 24 | 26 |
| Gene | adj-R2 | Model p.Value | Meth Δβ | Meth adj.p.Value | Expr |log2FC| | Expr adj.p.Value |
|---|---|---|---|---|---|---|
| MLH1 | 0.9160 | 9.757 × 10−51 | 0.2696 | 1.005 × 10−18 | −1.6650 | 4.159 × 10−21 |
| CHFR | 0.9155 | 8.039 × 10−60 | 0.2692 | 4.361 × 10−17 | −1.3277 | 9.634 × 10−14 |
| TMEM176B | 0.8590 | 3.332 × 10−54 | 0.2144 | 2.907 × 10−26 | −1.4168 | 4.478 × 10−16 |
| ZNF350 | 0.8545 | 4.427 × 10−60 | 0.2320 | 1.861 × 10−18 | −1.3189 | 4.834 × 10−13 |
| ZNF570 | 0.8445 | 1.232 × 10−50 | 0.2716 | 5.133 × 10−32 | −1.7217 | 9.649 × 10−20 |
| ZNF530 | 0.8400 | 5.514 × 10−52 | 0.2465 | 1.043 × 10−21 | −2.0575 | 9.957 × 10−20 |
| ZNF347 | 0.8114 | 5.530 × 10−52 | 0.2658 | 4.999 × 10−17 | −1.4471 | 9.699 × 10−12 |
| ZNF461 | 0.8051 | 3.875 × 10−46 | 0.3696 | 4.018 × 10−39 | −1.5938 | 7.787 × 10−18 |
| ZNF470 | 0.8011 | 1.548 × 10−45 | 0.3167 | 5.709 × 10−31 | −2.5464 | 1.020 × 10−27 |
| ZNF665 | 0.7960 | 5.481 × 10−47 | 0.2725 | 2.512 × 10−13 | −1.6427 | 7.221 × 10−10 |
| Gene | adj-R2 | Model p.Value | Meth Δβ | Meth adj.p.Value | Expr |log2FC| | Expr adj.p.Value |
|---|---|---|---|---|---|---|
| MLH1 | 0.8625 | 5.111 × 10−82 | 0.3309 | 1.735 × 10−38 | −1.3131 | 2.522 × 10−21 |
| SPAG16 | 0.8108 | 7.904 × 10−82 | 0.2190 | 1.693 × 10−22 | −1.3352 | 6.162 × 10−9 |
| ZNF549 | 0.7659 | 2.865 × 10−73 | 0.2224 | 1.639 × 10−18 | −1.3527 | 2.983 × 10−11 |
| ZNF530 | 0.7521 | 9.503 × 10−69 | 0.2278 | 5.345 × 10−34 | −1.3645 | 2.023 × 10−11 |
| EPM2AIP1 | 0.7326 | 1.265 × 10−59 | 0.3128 | 5.515 × 10−39 | −1.5758 | 1.698 × 10−25 |
| FUZ | 0.7183 | 1.696 × 10−59 | 0.2771 | 1.017 × 10−32 | −1.8069 | 8.646 × 10−19 |
| PCDHGC3 | 0.6902 | 5.196 × 10−58 | 0.2714 | 9.279 × 10−34 | −1.4244 | 2.100 × 10−1 |
| ZNF415 | 0.6898 | 6.833 × 10−63 | 0.2125 | 6.708 × 10−24 | −1.3567 | 7.768 × 10−10 |
| ZNF518B | 0.6557 | 1.171 × 10−50 | 0.2012 | 7.820 × 10−20 | −1.5725 | 1.583 × 10−13 |
| TTC9B | 0.6534 | 6.150 × 10−53 | 0.2325 | 2.510 × 10−30 | 1.3267 | 5.290 × 10−13 |
| Gene | adj-R2 | Model p.Value | Meth Δβ | Meth adj.p.Value | Expr |log2FC| | Expr adj.p.Value |
|---|---|---|---|---|---|---|
| VILL | 0.9262 | 2.472 × 10−17 | 0.3397 | 6.303 × 10−17 | −1.5257 | 2.000 × 10−4 |
| FAM50B | 0.9094 | 1.020 × 10−12 | 0.2101 | 9.718 × 10−8 | −1.8676 | 4.300 × 10−3 |
| TRIP4 | 0.9025 | 2.564 × 10−22 | 0.3142 | 4.801 × 10−20 | −1.6195 | 3.990 × 10−14 |
| FBXO17 | 0.8961 | 1.733 × 10−22 | 0.3254 | 5.730 × 10−26 | −4.1905 | 1.744 × 10−28 |
| EMP3 | 0.8679 | 2.235 × 10−19 | 0.3466 | 2.387 × 10−24 | −2.8986 | 2.188 × 10−9 |
| FCHSD1 | 0.8678 | 2.272 × 10−19 | 0.2259 | 1.471 × 10−6 | −1.5234 | 4.983 × 10−6 |
| KHNYN | 0.8342 | 7.694 × 10−15 | 0.5379 | 6.496 × 10−23 | −1.8945 | 4.784 × 10−9 |
| TUBA1C | 0.8262 | 1.959 × 10−13 | 0.2737 | 1.811 × 10−23 | −1.8564 | 4.579 × 10−7 |
| ZDHHC12 | 0.8093 | 7.718 × 10−16 | 0.2317 | 2.483 × 10−7 | −1.4266 | 4.424 × 10−5 |
| TSTD1 | 0.7886 | 7.635 × 10−12 | 0.3710 | 3.538 × 10−6 | −3.4774 | 4.892 × 10−6 |
| Gene | adj-R2 | Model p.Value | Meth Δβ | Meth adj.p.Value | Expr |log2FC| | Expr adj.p.Value |
|---|---|---|---|---|---|---|
| NMNAT3 | 0.7119 | 6.622 × 10−12 | 0.2379 | 3.593 × 10−7 | −2.6290 | 1.200 × 10−3 |
| TMEM220 | 0.6577 | 8.033 × 10−11 | 0.2176 | 1.281 × 10−8 | −1.7578 | 4.226 × 10−5 |
| RNF208 | 0.5908 | 7.443 × 10−8 | 0.2284 | 1.447 × 10−9 | −1.577 | 2.290 × 10−2 |
| OCIAD2 | 0.5642 | 1.103 × 10−6 | 0.2867 | 5.852 × 10−9 | −1.744 | 3.270 × 10−2 |
| CMBL | 0.5576 | 2.265 × 10−8 | 0.2682 | 6.128 × 10−8 | −2.9973 | 6.900 × 10−3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Blindu, A.S.; Berardelli, S.; De Paoli, F.; Manai, F.; Tricarico, R.; Zucca, S.; Magni, P. Exploring the Impact of DNA Methylation on Gene Expression in CRC: A Computational Approach for Identifying Epigenetically Regulated Genes in Multi-Omic Datasets. Cancers 2026, 18, 211. https://doi.org/10.3390/cancers18020211
Blindu AS, Berardelli S, De Paoli F, Manai F, Tricarico R, Zucca S, Magni P. Exploring the Impact of DNA Methylation on Gene Expression in CRC: A Computational Approach for Identifying Epigenetically Regulated Genes in Multi-Omic Datasets. Cancers. 2026; 18(2):211. https://doi.org/10.3390/cancers18020211
Chicago/Turabian StyleBlindu, Andrei Stefan, Silvia Berardelli, Federica De Paoli, Federico Manai, Rossella Tricarico, Susanna Zucca, and Paolo Magni. 2026. "Exploring the Impact of DNA Methylation on Gene Expression in CRC: A Computational Approach for Identifying Epigenetically Regulated Genes in Multi-Omic Datasets" Cancers 18, no. 2: 211. https://doi.org/10.3390/cancers18020211
APA StyleBlindu, A. S., Berardelli, S., De Paoli, F., Manai, F., Tricarico, R., Zucca, S., & Magni, P. (2026). Exploring the Impact of DNA Methylation on Gene Expression in CRC: A Computational Approach for Identifying Epigenetically Regulated Genes in Multi-Omic Datasets. Cancers, 18(2), 211. https://doi.org/10.3390/cancers18020211








