Computer-Aided Detection (CADe) of Small Metastatic Prostate Cancer Lesions on 3D PSMA PET Volumes Using Multi-Angle Maximum Intensity Projections
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Dataset Specifications
- Histologically proven PCa with BCR after initial curative therapy with radical prostatectomy, with a PSA > 0.4 ng/mL and an additional PSA measurement confirming an increase.
- Histologically proven PCa with BCR after initial curative radiotherapy, with a PSA > 2 ng/mL after therapy [12].
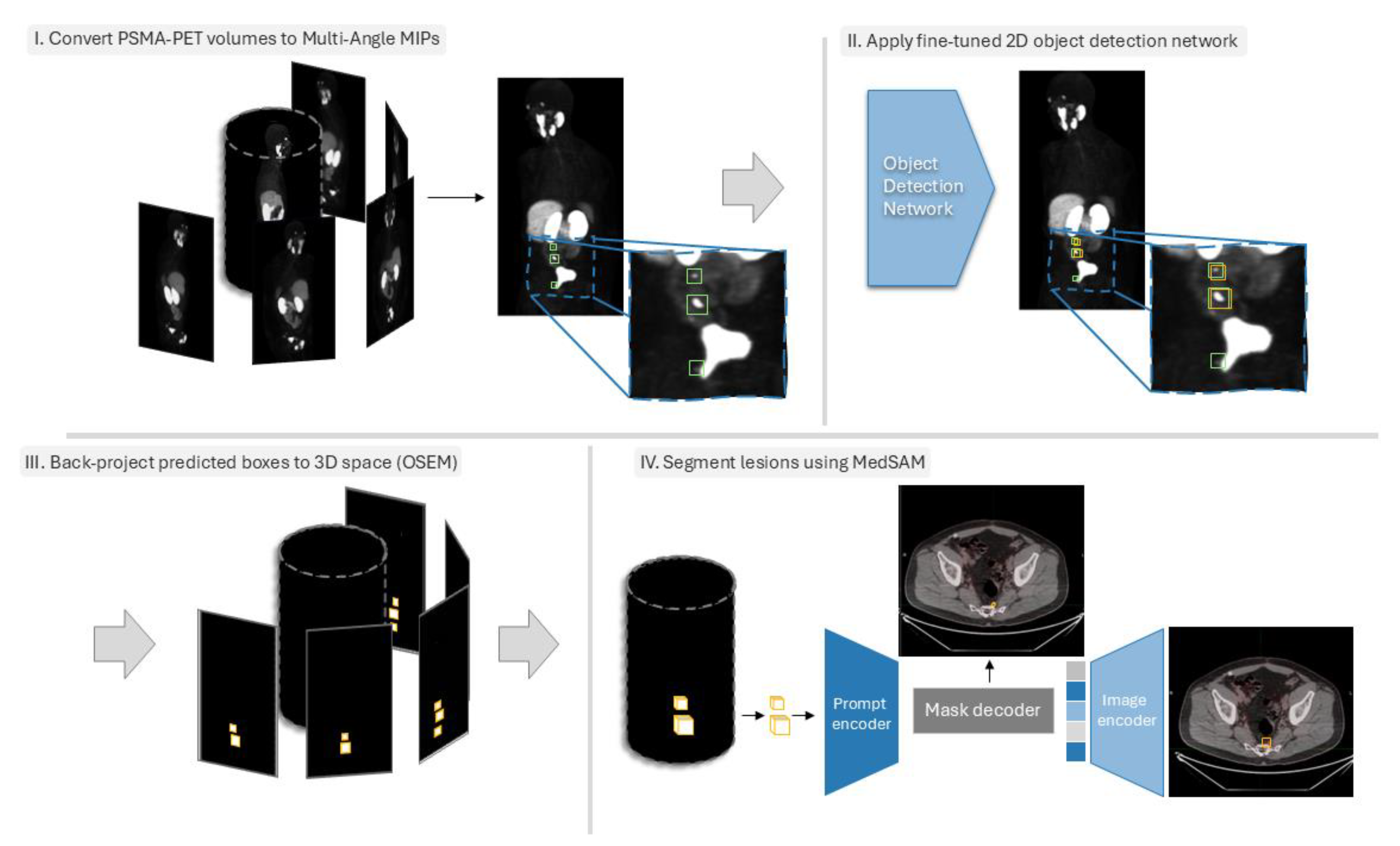
2.2. Data Preprocessing
2.3. Object Detection Networks Training
2.4. Back-Projecting Predicted Bounding Boxes Using OSEM
2.5. Lesion Segmentation Within 3D Bounding Boxes Using MedSAM
2.6. Experimental Details
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Soldatov, A.; von Klot, C.A.; Walacides, D.; Derlin, T.; Bengel, F.M.; Ross, T.L.; Wester, H.-J.; Derlin, K.; Kuczyk, M.A.; Christiansen, H.; et al. Patterns of Progression After 68Ga-PSMA-Ligand PET/CT-Guided Radiation Therapy for Recurrent Prostate Cancer. Int. J. Radiat. Oncol. 2018, 103, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Shore, N.D.; Moul, J.W.; Pienta, K.J.; Czernin, J.; King, M.T.; Freedland, S.J. Biochemical recurrence in patients with prostate cancer after primary definitive therapy: Treatment based on risk stratification. Prostate Cancer Prostatic Dis. 2023, 27, 192–201. [Google Scholar] [CrossRef] [PubMed]
- Freedland, S.J.; Presti, J.C.; Amling, C.L.; Kane, C.J.; Aronson, W.J.; Dorey, F.; Terris, M.K. Time trends in biochemical recurrence after radical prostatectomy: Results of the SEARCH database. Urology 2003, 61, 736–741. [Google Scholar] [CrossRef] [PubMed]
- Bubendorf, L.; Schöpfer, A.; Wagner, U.; Sauter, G.; Moch, H.; Willi, N.; Gasser, T.C.; Mihatsch, M.J. Metastatic patterns of prostate cancer: An autopsy study of 1589 patients. Hum. Pathol. 2000, 31, 578–583. [Google Scholar] [CrossRef]
- Slart, R.H. FDG-PET/CT (A) imaging in large vessel vasculitis and polymyalgia rheumatica: Joint procedural recommen-dation of the EANM, SNMMI, and the PET Interest Group (PIG), and endorsed by the ASNC. Eur. J. Nucl. Med. Mol. Imaging 2018, 45, 1250–1269. [Google Scholar] [CrossRef]
- Rousseau, E.; Wilson, D.; Lacroix-Poisson, F.; Krauze, A.; Chi, K.; Gleave, M.; McKenzie, M.; Tyldesley, S.; Goldenberg, S.L.; Bénard, F. A Prospective Study on 18F-DCFPyL PSMA PET/CT Imaging in Biochemical Recurrence of Prostate Cancer. J. Nucl. Med. 2019, 60, 1587–1593. [Google Scholar] [CrossRef]
- Ma, K.; Harmon, S.A.; Klyuzhin, I.S.; Rahmim, A.; Turkbey, B. Clinical Application of Artificial Intelligence in Positron Emission Tomography: Imaging of Prostate Cancer. PET Clin. 2022, 17, 137–143. [Google Scholar] [CrossRef]
- Iqbal, A.; Sharif, M.; Yasmin, M.; Raza, M.; Aftab, S. Generative adversarial networks and its applications in the biomedical image segmentation: A comprehensive survey. Int. J. Multimedia Inf. Retr. 2022, 11, 333–368. [Google Scholar] [CrossRef]
- Gadosey, P.K.; Li, Y.; Agyekum, E.A.; Zhang, T.; Liu, Z.; Yamak, P.T.; Essaf, F. SD-UNet: Stripping down U-Net for Segmentation of Biomedical Images on Platforms with Low Computational Budgets. Diagnostics 2020, 10, 110. [Google Scholar] [CrossRef]
- Van Opbroek, A.; Ikram, M.A.; Vernooij, M.W.; De Bruijne, M. Transfer learning improves supervised image segmen-tation across imaging protocols. IEEE Trans. Med. Imaging 2014, 34, 1018–1030. [Google Scholar] [CrossRef]
- Sureshbabu, W.; Mawlawi, O. PET/CT imaging artifacts. J. Nucl. Med. Technol. 2005, 33, 156. [Google Scholar] [PubMed]
- Harsini, S.; Wilson, D.; Saprunoff, H.; Allan, H.; Gleave, M.; Goldenberg, L.; Chi, K.N.; Kim-Sing, C.; Tyldesley, S.; Bénard, F. Outcome of patients with biochemical recurrence of prostate cancer after PSMA PET/CT-directed radiotherapy or surgery without systemic therapy. Cancer Imaging 2023, 23, 27. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.-Y.; Maire, M.; Belongie, S.; Hays, J.; Perona, P.; Ramanan, D.; Dollár, P.; Zitnick, C.L. Microsoft coco: Common objects in context. In Proceedings of the Computer Vision–ECCV 2014: 13th European Conference, Zurich, Switzerland, 6–12 September 2014; Proceedings, Part V 13. pp. 740–755. [Google Scholar]
- Cai, Z.; Vasconcelos, N. Cascade R-CNN: High Quality Object Detection and Instance Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2019, 43, 1483–1498. [Google Scholar] [CrossRef]
- Sun, P.; Zhang, R.; Jiang, Y.; Kong, T.; Xu, C.; Zhan, W.; Tomizuka, M.; Li, L.; Yuan, Z.; Wang, C.; et al. Sparse r-cnn: End-to-end object detection with learnable proposals. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 14454–14463. [Google Scholar]
- Zhu, X.; Hu, H.; Lin, S.; Dai, J. Deformable convnets v2: More deformable, better results. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 9308–9316. [Google Scholar]
- Li, Y.; Chen, Y.; Wang, N.; Zhang, Z. Scale-aware trident networks for object detection. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Republic of Korea, 27 October–2 November 2019; pp. 6054–6063. [Google Scholar]
- Zhang, S.; Chi, C.; Yao, Y.; Lei, Z.; Li, S.Z. Bridging the gap between anchor-based and anchor-free detection via adaptive training sample selection. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seoul, Republic of Korea, 16–18 June 2020; pp. 9759–9768. [Google Scholar]
- Zhang, X.; Wan, F.; Liu, C.; Ji, R.; Ye, Q. Freeanchor: Learning to match anchors for visual object detection. Adv. Neural Inf. Process. Syst. 2019, 32. [Google Scholar]
- Kim, K.; Lee, H.S. Probabilistic Anchor Assignment with IoU Prediction for Object Detection. In Proceedings of the European Conference on Computer Vision (ECCV), Glasgow, UK, 23–28 August 2020; pp. 355–371. [Google Scholar] [CrossRef]
- Li, X.; Wang, W.; Wu, L.; Chen, S.; Hu, X.; Li, J.; Tang, J.; Yang, J. Generalized focal loss: Learning qualified and distributed bounding boxes for dense object detection. Adv. Neural Inf. Process. Syst. 2020, 33, 21002–21012. [Google Scholar]
- Zhang, H.; Wang, Y.; Dayoub, F.; Sunderhauf, N. Varifocalnet: An iou-aware dense object detector. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 8514–8523. [Google Scholar]
- Chen, Z.; Yang, C.; Li, Q.; Zhao, F.; Zha, Z.-J.; Wu, F. Disentangle Your Dense Object Detector. In Proceedings of the 29th ACM International Conference on Multimedia, Chengdu, China, 20–24 October 2021; pp. 4939–4948. [Google Scholar]
- Feng, C.; Zhong, Y.; Gao, Y.; Scott, M.R.; Huang, W. Tood: Task-aligned one-stage object detection. In Proceedings of the 2021 IEEE/CVF International Conference on Computer Vision (ICCV), Montreal, QC, Canada, 10–17 October 2021; pp. 3490–3499. [Google Scholar]
- Yang, Z.; Liu, S.; Hu, H.; Wang, L.; Lin, S. Reppoints: Point set representation for object detection. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Republic of Korea, 27 October–2 November 2019; pp. 9657–9666. [Google Scholar]
- Zhu, C.; He, Y.; Savvides, M. Feature selective anchor-free module for single-shot object detection. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 840–849. [Google Scholar]
- Tian, Z.; Shen, C.; Chen, H.; He, T. Fcos: Fully convolutional one-stage object detection. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Republic of Korea, 27 October–2 November 2019; pp. 9627–9636. [Google Scholar]
- Dong, Z.; Li, G.; Liao, Y.; Wang, F.; Ren, P.; Qian, C. Centripetalnet: Pursuing high-quality keypoint pairs for object de-tection. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 10519–10528. [Google Scholar]
- Zhu, X.; Su, W.; Lu, L.; Li, B.; Wang, X.; Dai, J. Deformable detr: Deformable transformers for end-to-end object detection. arXiv 2020, arXiv:2010.04159. [Google Scholar]
- Hudson, H.M.; Larkin, R.S. Accelerated image reconstruction using ordered subsets of projection data. IEEE Trans. Med. Imaging 1994, 13, 601–609. [Google Scholar] [CrossRef]
- Ma, J.; He, Y.; Li, F.; Han, L.; You, C.; Wang, B. Segment anything in medical images. Nat. Commun. 2024, 15, 654. [Google Scholar] [CrossRef]
- Chen, K.; Wang, J.; Pang, J.; Cao, Y.; Xiong, Y.; Li, X.; Sun, S.; Feng, W.; Liu, Z.; Xu, J.; et al. MMDetection: Open mmlab detection toolbox and benchmark. arXiv 2019, arXiv:1906.07155. [Google Scholar]
- Kostyszyn, D.; Fechter, T.; Bartl, N.; Grosu, A.L.; Gratzke, C.; Sigle, A.; Mix, M.; Ruf, J.; Fassbender, T.F.; Kiefer, S.; et al. Intraprostatic Tumor Segmentation on PSMA PET Images in Patients with Primary Prostate Cancer with a Convolutional Neural Network. J. Nucl. Med. 2021, 62, 823–828. [Google Scholar] [CrossRef]
- Jafari, E.; Zarei, A.; Dadgar, H.; Keshavarz, A.; Manafi-Farid, R.; Rostami, H.; Assadi, M. A convolutional neural network–based system for fully automatic segmentation of whole-body [68Ga]Ga-PSMA PET images in prostate cancer. Eur. J. Nucl. Med. 2023, 51, 1476–1487. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Klyuzhin, I.; Harsini, S.; Ortiz, A.; Zhang, S.; Bénard, F.; Dodhia, R.; Uribe, C.F.; Rahmim, A.; Ferres, J.L. Automatic segmentation of prostate cancer metastases in PSMA PET/CT images using deep neural networks with weighted batch-wise dice loss. Comput. Biol. Med. 2023, 158, 106882. [Google Scholar] [CrossRef]


| Characteristic | Entire Cohort (N = 317) |
|---|---|
| Age at diagnosis (years), median (range) | 71 (45–91) |
| Initial Gleason score | |
| 6 | 31 (9.6%) |
| 3 + 4 | 81 (25.3%) |
| 4 + 3 | 108 (33.7%) |
| 8 | 36 (11.3%) |
| 9 | 58 (18.1%) |
| Primary tumor classification | |
| p/cT1 | 38 (11.9%) |
| p/cT2 | 144 (45%) |
| p/cT3 | 129 (40.3%) |
| p/cT4 | 3 (0.9%) |
| p/cTx | 3 (0.9%) |
| Primary nodal status | |
| p/cN0 | 223 (69.7%) |
| p/cN1 | 24 (7.5%) |
| p/cNx | 72 (22.5%) |
| D’Amico Score | |
| High risk | 241 (75.31%) |
| Intermediate risk | 63 (19.68%) |
| Low risk | 18 (5.62%) |
| PSA at PET/CT (ng/mL), median (range) | 3.2 (0.06–53.3) |
| Number of lesions at PET/CT | |
| 1 | 170 (53.1%) |
| 2 | 66 (20.6%) |
| 3–5 | 85 (26.6%) |
| Site of lesion | |
| Local recurrence | 162 (50.6%) |
| Regional lymph node | 130 (40.6%) |
| Distant lymph node | 41 (12.8%) |
| Bone | 59 (18.4%) |
| Other | 11 (3.4%) |
| SUVmax (g/mL), median (range) | 7.51 (0.97–97.39) |
| TMTV (mL), median (range) | 4.02 (0.18–92) |
| TLA (g), median (range) | 16.02 (0.35–716.87) |
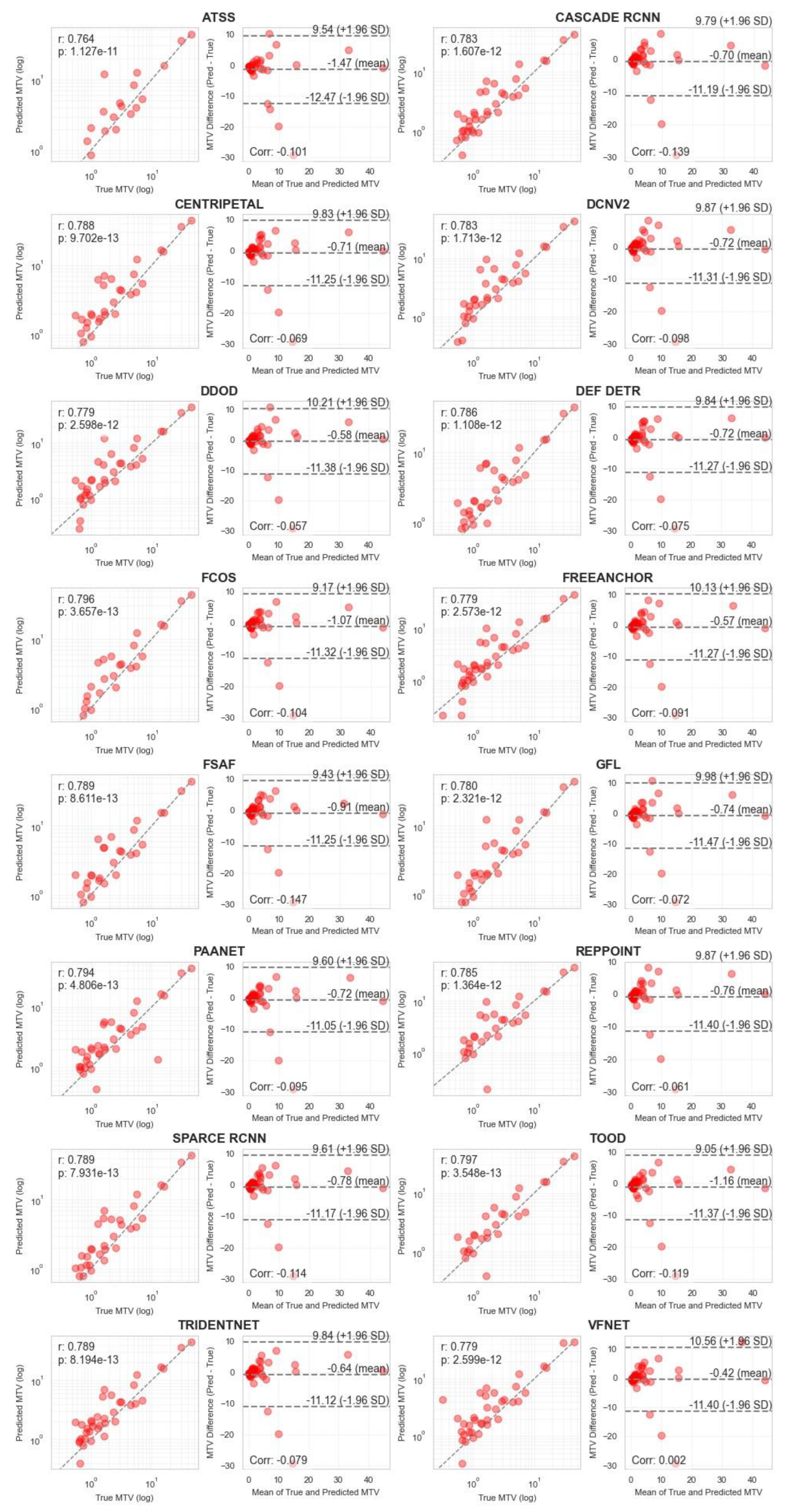
| Object Detection (OD) Network | Model Type | Backbone | AP |
|---|---|---|---|
| Cascade R-CNN [14] | Multi-stage | R-50 | 26.6 |
| Sparse R-CNN [15] | Multi-stage | R-101 | 28.3 |
| Deformable ConvNets v2 (DCNv2) [16] | Multi-stage | R-101-DCN | 27.8 |
| Trident-Net [17] | Multi-stage | R-101-DCN | 28 |
| Adaptive Training Sample Selection (ATSS) [18] | Single stage | R-101 | 26.1 |
| FreeAnchor [19] | Single stage | X-101 | 26.8 |
| Probabilistic Anchor Assignment with IoU Pred. [20] | Single stage | R-101-DCN | 27.9 |
| Generalized Focal Loss (GFL) [21] | Single stage | X-101-DCN | 29.2 |
| Varifocal-Net [22] | Single stage | X-101-DCN | 30.7 |
| Disentangle Your Dense Object Detector (DDOD) [23] | Single stage | R-50 | 27.2 |
| Task-aligned One-stage Object Detection (TOOD) [24] | Single stage | R-101-DCN | 30.5 |
| RepPoints [25] | Anchor-free | R-101-DCN | 30.3 |
| Feature Selective Anchor-Free (FSAF) [26] | Anchor-free | X-101 | 26.6 |
| Fully Convolutional One-Stage Object Detection [27] | Anchor-free | X-101 | 27.6 |
| Centripetal Net [28] | Anchor-free | H-104 | 29 |
| Deformable DETR [29] | Query-based | R-50 | 27.7 |
| Networks | TP | FP | FN | Precision | Recall | F1-Score | ||
|---|---|---|---|---|---|---|---|---|
| Detection Models | Multi-Stage | Cascade R-CNN | 38 | 31 | 16 | 0.55 | 0.70 | 0.62 |
| Sparse R-CNN | 32 | 12 | 22 | 0.73 | 0.59 | 0.65 | ||
| DCNv2 | 32 | 20 | 22 | 0.62 | 0.59 | 0.60 | ||
| Trident-Net | 38 | 34 | 16 | 0.53 | 0.70 | 0.60 | ||
| Single-Stage | ATSS | 18 | 3 | 36 | 0.86 | 0.33 | 0.48 | |
| FreeAnchor | 40 | 22 | 14 | 0.65 | 0.74 | 0.69 | ||
| PAA Net | 33 | 25 | 21 | 0.57 | 0.61 | 0.59 | ||
| GFL | 30 | 8 | 24 | 0.79 | 0.56 | 0.65 | ||
| Varifocal-Net | 35 | 32 | 19 | 0.52 | 0.65 | 0.58 | ||
| DDOD | 35 | 28 | 19 | 0.56 | 0.65 | 0.60 | ||
| TOOD | 25 | 8 | 29 | 0.76 | 0.46 | 0.57 | ||
| Anchor-Free | RepPoints | 29 | 12 | 25 | 0.71 | 0.54 | 0.61 | |
| FSAF | 25 | 21 | 29 | 0.54 | 0.46 | 0.50 | ||
| FCOS | 25 | 7 | 29 | 0.78 | 0.46 | 0.58 | ||
| Centripetal Net | 23 | 6 | 31 | 0.79 | 0.43 | 0.55 | ||
| Query-based | Deformable DETR | 35 | 16 | 19 | 0.69 | 0.65 | 0.67 | |
| 3D | nnDetection | 38 | 28 | 16 | 0.58 | 0.70 | 0.63 | |
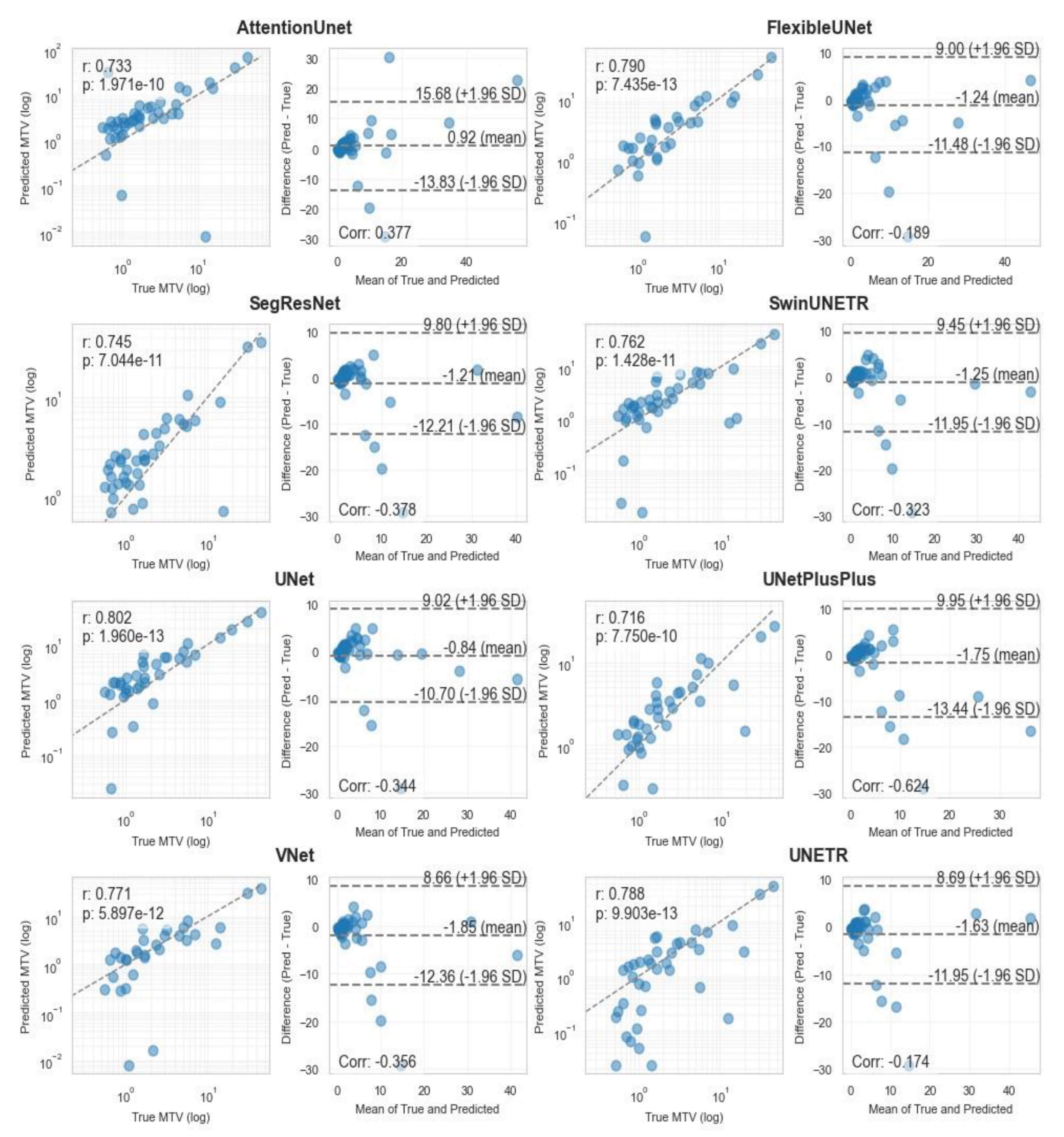
| Segmentation Models | 3D | AttentionUnet | 39 | 193 | 15 | 0.17 | 0.72 | 0.27 |
| FlexibleUNet | 30 | 19 | 24 | 0.61 | 0.56 | 0.58 | ||
| SegResNet | 39 | 160 | 15 | 0.20 | 0.72 | 0.31 | ||
| SwinUNETR | 36 | 81 | 18 | 0.31 | 0.67 | 0.42 | ||
| UNet | 35 | 57 | 19 | 0.38 | 0.65 | 0.48 | ||
| UNetPlusPlus | 35 | 68 | 19 | 0.34 | 0.65 | 0.45 | ||
| Vnet | 29 | 19 | 25 | 0.60 | 0.54 | 0.57 | ||
| UNETR | 33 | 193 | 21 | 0.15 | 0.61 | 0.24 | ||
| nnUNet | 39 | 16 | 15 | 0.71 | 0.72 | 0.72 |
| All Lesions | Local Relapse | Regional Lymph Nodes | Distant Lymph Nodes | Bone Metastasis | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Networks | TP | FP | FN | Recall | F1 | TP | FN | Recall | TP | FN | Recall | TP | FN | Recall | TP | FN | Recall | ||
| Detection Models | Multi-Stage | Cascade R-CNN | 38 | 31 | 16 | 0.70 | 0.62 | 9 | 2 | 0.82 | 21 | 9 | 0.70 | 2 | 1 | 0.67 | 6 | 4 | 0.60 |
| Sparse R-CNN | 32 | 12 | 22 | 0.59 | 0.65 | 8 | 3 | 0.73 | 19 | 11 | 0.63 | 1 | 2 | 0.33 | 5 | 5 | 0.50 | ||
| DCNv2 | 32 | 20 | 22 | 0.59 | 0.60 | 8 | 3 | 0.73 | 18 | 12 | 0.60 | 2 | 1 | 0.67 | 5 | 5 | 0.50 | ||
| Trident-Net | 38 | 34 | 16 | 0.70 | 0.60 | 8 | 3 | 0.73 | 21 | 9 | 0.70 | 2 | 1 | 0.67 | 8 | 2 | 0.80 | ||
| Single-Stage | ATSS | 18 | 3 | 36 | 0.33 | 0.48 | 7 | 4 | 0.64 | 8 | 22 | 0.27 | 1 | 2 | 0.33 | 5 | 5 | 0.50 | |
| FreeAnchor | 40 | 22 | 14 | 0.74 | 0.69 | 9 | 2 | 0.82 | 21 | 9 | 0.70 | 2 | 1 | 0.67 | 8 | 2 | 0.80 | ||
| PAA Net | 33 | 25 | 21 | 0.61 | 0.59 | 10 | 1 | 0.91 | 20 | 10 | 0.67 | 1 | 2 | 0.33 | 7 | 3 | 0.70 | ||
| GFL | 30 | 8 | 24 | 0.56 | 0.65 | 8 | 3 | 0.73 | 18 | 12 | 0.60 | 1 | 2 | 0.33 | 5 | 5 | 0.50 | ||
| Varifocal-Net | 35 | 32 | 19 | 0.65 | 0.58 | 9 | 2 | 0.82 | 19 | 11 | 0.63 | 2 | 1 | 0.67 | 8 | 2 | 0.80 | ||
| DDOD | 35 | 28 | 19 | 0.65 | 0.60 | 8 | 3 | 0.73 | 19 | 11 | 0.63 | 2 | 1 | 0.67 | 6 | 4 | 0.60 | ||
| TOOD | 25 | 8 | 29 | 0.46 | 0.57 | 7 | 4 | 0.64 | 15 | 15 | 0.50 | 2 | 1 | 0.67 | 4 | 6 | 0.40 | ||
| Anchor-Free | RepPoints | 29 | 12 | 25 | 0.54 | 0.61 | 8 | 3 | 0.73 | 16 | 14 | 0.53 | 1 | 2 | 0.33 | 4 | 6 | 0.40 | |
| FSAF | 25 | 21 | 29 | 0.46 | 0.50 | 7 | 4 | 0.64 | 16 | 14 | 0.53 | 1 | 2 | 0.33 | 5 | 5 | 0.50 | ||
| FCOS | 25 | 7 | 29 | 0.46 | 0.58 | 8 | 3 | 0.73 | 12 | 18 | 0.40 | 1 | 2 | 0.33 | 4 | 6 | 0.40 | ||
| Centripetal Net | 23 | 6 | 31 | 0.43 | 0.55 | 8 | 3 | 0.73 | 16 | 14 | 0.53 | 1 | 2 | 0.33 | 6 | 4 | 0.60 | ||
| Query-based | Deformable DETR | 35 | 16 | 19 | 0.61 | 0.59 | 8 | 3 | 0.73 | 18 | 12 | 0.60 | 1 | 2 | 0.33 | 6 | 4 | 0.60 | |
| 3D | nnDetection | 38 | 28 | 16 | 0.70 | 0.63 | 10 | 1 | 0.91 | 20 | 10 | 0.67 | 2 | 1 | 0.67 | 6 | 4 | 0.60 | |
| Segmentation Models | 3D | AttentionUnet | 39 | 193 | 15 | 0.72 | 0.27 | 10 | 1 | 0.91 | 21 | 9 | 0.70 | 2 | 1 | 0.67 | 6 | 4 | 0.60 |
| FlexibleUNet | 30 | 19 | 24 | 0.56 | 0.58 | 9 | 2 | 0.82 | 15 | 15 | 0.50 | 1 | 2 | 0.33 | 5 | 5 | 0.50 | ||
| SegResNet | 39 | 160 | 15 | 0.72 | 0.31 | 9 | 2 | 0.82 | 21 | 9 | 0.70 | 2 | 1 | 0.67 | 7 | 3 | 0.70 | ||
| SwinUNETR | 36 | 81 | 18 | 0.67 | 0.42 | 11 | 0 | 1.00 | 19 | 11 | 0.63 | 1 | 2 | 0.33 | 5 | 5 | 0.50 | ||
| UNet | 35 | 57 | 19 | 0.65 | 0.48 | 9 | 2 | 0.82 | 19 | 11 | 0.63 | 0 | 3 | 0.00 | 6 | 4 | 0.60 | ||
| UNetPlusPlus | 35 | 68 | 19 | 0.65 | 0.45 | 9 | 2 | 0.82 | 19 | 11 | 0.63 | 0 | 3 | 0.00 | 6 | 4 | 0.60 | ||
| Vnet | 29 | 19 | 25 | 0.54 | 0.57 | 9 | 2 | 0.82 | 14 | 16 | 0.47 | 0 | 3 | 0.00 | 6 | 4 | 0.60 | ||
| UNETR | 33 | 193 | 21 | 0.61 | 0.24 | 10 | 1 | 0.91 | 16 | 14 | 0.53 | 0 | 3 | 0.00 | 6 | 4 | 0.60 | ||
| nnUNet | 39 | 16 | 15 | 0.72 | 0.72 | 9 | 2 | 0.82 | 20 | 10 | 0.67 | 2 | 1 | 0.67 | 7 | 3 | 0.70 | ||
| Networks | Dice↑ | HD95↓ | Vol Err.↓ | Sens.↑ | |||
|---|---|---|---|---|---|---|---|
| Detection Models | Multi-Stage | Cascade R-CNN | 0.45 | 29.68 | 47.72 | 0.33 | MedSAM Segmentation model |
| Sparse R-CNN | 0.48 | 19.42 | 48.27 | 0.35 | |||
| DCNv2 | 0.42 | 21.11 | 47.19 | 0.30 | |||
| Trident-Net | 0.45 | 20.34 | 50.66 | 0.33 | |||
| Single-Stage | ATSS | 0.29 | 36.81 | 65.05 | 0.22 | ||
| FreeAnchor | 0.45 | 24.24 | 47.98 | 0.32 | |||
| PAA Net | 0.43 | 24.26 | 45.58 | 0.32 | |||
| GFL | 0.45 | 20.08 | 46.91 | 0.34 | |||
| Varifocal-Net | 0.44 | 33.71 | 48.34 | 0.32 | |||
| DDOD | 0.44 | 30.15 | 47.15 | 0.32 | |||
| TOOD | 0.40 | 9.30 | 51.19 | 0.30 | |||
| Anchor-Free | RepPoints | 0.41 | 19.32 | 52.57 | 0.31 | ||
| FSAF | 0.38 | 18.20 | 51.67 | 0.28 | |||
| FCOS | 0.39 | 17.83 | 48.90 | 0.30 | |||
| Centripetal Net | 0.38 | 18.95 | 57.52 | 0.29 | |||
| Query-based | Deformable DETR | 0.47 | 20.53 | 39.36 | 0.35 | ||
| 3D | nnDetection | 0.45 | 31.23 | 47.46 | 0.32 | ||
| Segmentation Models | 3D | AttentionUnet | 0.35 | 167.24 | 66.89 | 0.24 | Task-specific Segmentation models |
| FlexibleUNet | 0.43 | 23.53 | 50.28 | 0.32 | |||
| SegResNet | 0.39 | 94.18 | 57.05 | 0.27 | |||
| SwinUNETR | 0.46 | 18.2 | 40.64 | 0.34 | |||
| UNet | 0.42 | 56.02 | 49.16 | 0.30 | |||
| UNetPlusPlus | 0.40 | 79.21 | 52.95 | 0.28 | |||
| Vnet | 0.42 | 30.26 | 53.62 | 0.32 | |||
| UNETR | 0.36 | 80.6 | 49.61 | 0.26 | |||
| nnUNet | 0.47 | 30.74 | 49.90 | 0.35 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toosi, A.; Harsini, S.; Divband, G.; Bénard, F.; Uribe, C.F.; Oviedo, F.; Dodhia, R.; Weeks, W.B.; Lavista Ferres, J.M.; Rahmim, A. Computer-Aided Detection (CADe) of Small Metastatic Prostate Cancer Lesions on 3D PSMA PET Volumes Using Multi-Angle Maximum Intensity Projections. Cancers 2025, 17, 1563. https://doi.org/10.3390/cancers17091563
Toosi A, Harsini S, Divband G, Bénard F, Uribe CF, Oviedo F, Dodhia R, Weeks WB, Lavista Ferres JM, Rahmim A. Computer-Aided Detection (CADe) of Small Metastatic Prostate Cancer Lesions on 3D PSMA PET Volumes Using Multi-Angle Maximum Intensity Projections. Cancers. 2025; 17(9):1563. https://doi.org/10.3390/cancers17091563
Chicago/Turabian StyleToosi, Amirhosein, Sara Harsini, Ghasemali Divband, François Bénard, Carlos F. Uribe, Felipe Oviedo, Rahul Dodhia, William B. Weeks, Juan M. Lavista Ferres, and Arman Rahmim. 2025. "Computer-Aided Detection (CADe) of Small Metastatic Prostate Cancer Lesions on 3D PSMA PET Volumes Using Multi-Angle Maximum Intensity Projections" Cancers 17, no. 9: 1563. https://doi.org/10.3390/cancers17091563
APA StyleToosi, A., Harsini, S., Divband, G., Bénard, F., Uribe, C. F., Oviedo, F., Dodhia, R., Weeks, W. B., Lavista Ferres, J. M., & Rahmim, A. (2025). Computer-Aided Detection (CADe) of Small Metastatic Prostate Cancer Lesions on 3D PSMA PET Volumes Using Multi-Angle Maximum Intensity Projections. Cancers, 17(9), 1563. https://doi.org/10.3390/cancers17091563








