Molecular Profiling of Nasopharyngeal Carcinoma Using the AACR Project GENIE Repository
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Patient Demographics of Nasopharyngeal Carcinoma
3.2. Most Common Somatic Mutations and Copy Number Alterations
3.3. Genetic Differences by Race and Sex
3.4. Co-Occurrence and Mutual Exclusivity of Mutations
3.5. Primary vs. Metastatic Mutations
4. Discussion
4.1. Subgroups and Mutational Landscape
4.2. Commonly-Mutated Genes and Known Pathways
4.3. p53 Pathway
4.4. NF-κB Pathway
4.5. PI3K Pathway
4.6. Co-Occurrence Patterns and Functional Implications
4.7. Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
Appendix A
| Gene | Protein Change | Mutation Type |
|---|---|---|
| KMT2D | M4159Cfs*2 | FS del |
| S2431Lfs*53 | FS del | |
| A2119Lfs*25 | FS del | |
| L3542Vfs*13 | FS del | |
| C5226Afs*16 | FS del | |
| P648Tfs*2 | FS ins | |
| L2331Pfs*46 | FS ins | |
| Q3863del | IF del | |
| E2393K | Missense | |
| D32N | Missense | |
| R4420W | Missense | |
| I3065M | Missense | |
| K2288N | Missense | |
| L894F | Missense | |
| Q4249H | Missense | |
| D1724Y | Missense | |
| A2925V | Missense | |
| R1615 * | Nonsense | |
| Q3441 * | Nonsense | |
| W4987 * | Nonsense | |
| Q2416 * | Nonsense | |
| E4662 * | Nonsense | |
| Q4609 * | Nonsense | |
| S3239 * | Nonsense | |
| X4882_splice | Splice | |
| TP53 | K382Nfs*40 | FS del |
| L257Gfs*6 | FS del | |
| P71Rfs*87 | FS ins | |
| Y220C | Missense | |
| Y220C | Missense | |
| R248Q | Missense | |
| R273C | Missense | |
| Y163C | Missense | |
| E285K | Missense | |
| P278S | Missense | |
| V157F | Missense | |
| V157F | Missense | |
| E271K | Missense | |
| C238G | Missense | |
| G245A | Missense | |
| E349K | Missense | |
| R213 * | Nonsense | |
| Q165 * | Nonsense | |
| R196 * | Nonsense | |
| X307_splice | Splice | |
| CYLD | E585Rfs*2 | FS del |
| P910Qfs*3 | FS del | |
| Q443Pfs*4 | FS ins | |
| I161Efs*43 | FS ins | |
| D681N | Missense | |
| Q898 * | Nonsense | |
| Q554 * | Nonsense | |
| S371 * | Nonsense | |
| S371 * | Nonsense | |
| E626 * | Nonsense | |
| Q823 * | Nonsense | |
| X896_splice | Splice | |
| NFKBIA | R264Pfs*21 | FS ins |
| D100Rfs*29 | FS ins | |
| K238Efs*5 | FS ins | |
| E14Gfs*72 | FS ins | |
| NFKBIA intragenic | Fusion | |
| NFKBIA intragenic | Fusion | |
| NFKBIA intragenic | Fusion | |
| A158_N182del | IF del | |
| G161E | Missense | |
| X113_splice | Splice | |
| X154_splice | Splice |
References
- Chen, Y.-P.; Chan, A.T.C.; Le, Q.-T.; Blanchard, P.; Sun, Y.; Ma, J. Nasopharyngeal Carcinoma. Lancet 2019, 394, 64–80. [Google Scholar] [CrossRef] [PubMed]
- Badoual, C. Update from the 5th Edition of the World Health Organization Classification of Head and Neck Tumors: Oropharynx and Nasopharynx. Head Neck Pathol. 2022, 16, 19–30. [Google Scholar] [CrossRef]
- Sinha, S.; Winters, R.; Gajra, A. Nasopharyngeal Cancer. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2025. [Google Scholar]
- Liu, K.; Wang, J. Developing a Nomogram Model and Prognostic Analysis of Nasopharyngeal Squamous Cell Carcinoma Patients: A Population-Based Study. J. Cancer Res. Clin. Oncol. 2023, 149, 12165–12175. [Google Scholar] [CrossRef]
- Zhang, Y.; Rumgay, H.; Li, M.; Cao, S.; Chen, W. Nasopharyngeal Cancer Incidence and Mortality in 185 Countries in 2020 and the Projected Burden in 2040: Population-Based Global Epidemiological Profiling. JMIR Public Health Surveill. 2023, 9, e49968. [Google Scholar] [CrossRef]
- Jia, W.-H.; Qin, H.-D. Non-Viral Environmental Risk Factors for Nasopharyngeal Carcinoma: A Systematic Review. Semin. Cancer Biol. 2012, 22, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Chang, E.T.; Ye, W.; Zeng, Y.-X.; Adami, H.-O. The Evolving Epidemiology of Nasopharyngeal Carcinoma. Cancer Epidemiol. Biomark. Prev. 2021, 30, 1035–1047. [Google Scholar] [CrossRef]
- Abdullah, B.; Alias, A.; Hassan, S. Challenges in the Management of Nasopharyngeal Carcinoma: A Review. Malays. J. Med. Sci. MJMS 2009, 16, 50–54. [Google Scholar] [PubMed]
- Juarez-Vignon Whaley, J.J.; Afkhami, M.; Onyshchenko, M.; Massarelli, E.; Sampath, S.; Amini, A.; Bell, D.; Villaflor, V.M. Recurrent/Metastatic Nasopharyngeal Carcinoma Treatment from Present to Future: Where Are We and Where Are We Heading? Curr. Treat. Options Oncol. 2023, 24, 1138–1166. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, L.; Hu, G.-Q.; Zhang, N.; Zhu, X.-D.; Yang, K.-Y.; Jin, F.; Shi, M.; Chen, Y.-P.; Hu, W.-H.; et al. Final Overall Survival Analysis of Gemcitabine and Cisplatin Induction Chemotherapy in Nasopharyngeal Carcinoma: A Multicenter, Randomized Phase III Trial. J. Clin. Oncol. 2022, 40, 2420–2425. [Google Scholar] [CrossRef]
- Lin, J.-C.; Wang, W.-Y.; Chen, K.Y.; Wei, Y.-H.; Liang, W.-M.; Jan, J.-S.; Jiang, R.-S. Quantification of Plasma Epstein–Barr Virus DNA in Patients with Advanced Nasopharyngeal Carcinoma. N. Engl. J. Med. 2004, 350, 2461–2470. [Google Scholar] [CrossRef]
- Krishnan, M.; Babu, S. Biomarkers in Nasopharyngeal Carcinoma (NPC): Clinical Relevance and Prognostic Potential. Oral Oncol. Rep. 2024, 11, 100640. [Google Scholar] [CrossRef]
- Tang, L.-Q.; Li, C.-F.; Li, J.; Chen, W.-H.; Chen, Q.-Y.; Yuan, L.-X.; Lai, X.-P.; He, Y.; Xu, Y.-X.-X.; Hu, D.-P.; et al. Establishment and Validation of Prognostic Nomograms for Endemic Nasopharyngeal Carcinoma. J. Natl. Cancer Inst. 2016, 108, djv291. [Google Scholar] [CrossRef] [PubMed]
- Sykes, E.A.; Weisbrod, N.; Rival, E.; Haque, A.; Fu, R.; Eskander, A. Methods, Detection Rates, and Survival Outcomes of Screening for Head and Neck Cancers: A Systematic Review. JAMA Otolaryngol. Neck Surg. 2023, 149, 1047. [Google Scholar] [CrossRef]
- Arter, Z.; Shieh, K.; Nagasaka, M.; Ou, S.-H. Comprehensive Survey of AACR GENIE Database of Tumor Mutation Burden (TMB) Among All Three Classes (I, II, III) of BRAF Mutated (BRAF+) NSCLC. Lung Cancer Targets Ther. 2025, 16, 1–9. [Google Scholar] [CrossRef]
- Lee, A.W.; Sou, A.; Patel, M.; Guzman, S.; Liu, L. Early Onset of Nasopharyngeal Cancer in Asian/Pacific Islander Americans Revealed by Age-Specific Analysis. Ann. Epidemiol. 2023, 80, 25–29. [Google Scholar] [CrossRef]
- Zhou, X.; Cui, J.; Macias, V.; Kajdacsy-Balla, A.A.; Ye, H.; Wang, J.; Rao, P.N. The Progress on Genetic Analysis of Nasopharyngeal Carcinoma. Comp. Funct. Genom. 2007, 2007, 57513. [Google Scholar] [CrossRef] [PubMed]
- Xiong, G.; Zhang, B.; Huang, M.; Zhou, H.; Chen, L.; Feng, Q.; Luo, X.; Lin, H.; Zeng, Y. Epstein-Barr Virus (EBV) Infection in Chinese Children: A Retrospective Study of Age-Specific Prevalence. PLoS ONE 2014, 9, e99857. [Google Scholar] [CrossRef]
- Liu, X.; Deng, Y.; Huang, Y.; Ye, J.; Xie, S.; He, Q.; Chen, Y.; Lin, Y.; Liang, R.; Wei, J.; et al. Nasopharyngeal Carcinoma Progression: Accumulating Genomic Instability and Persistent Epstein–Barr Virus Infection. Curr. Oncol. 2022, 29, 6035–6052. [Google Scholar] [CrossRef]
- Zhou, Z.; Li, P.; Zhang, X.; Xu, J.; Xu, J.; Yu, S.; Wang, D.; Dong, W.; Cao, X.; Yan, H.; et al. Mutational Landscape of Nasopharyngeal Carcinoma Based on Targeted Next-Generation Sequencing: Implications for Predicting Clinical Outcomes. Mol. Med. Camb. Mass 2022, 28, 55. [Google Scholar] [CrossRef]
- Lin, D.-C.; Meng, X.; Hazawa, M.; Nagata, Y.; Varela, A.M.; Xu, L.; Sato, Y.; Liu, L.-Z.; Ding, L.-W.; Sharma, A.; et al. The Genomic Landscape of Nasopharyngeal Carcinoma. Nat. Genet. 2014, 46, 866–871. [Google Scholar] [CrossRef]
- Na, D.; Chae, J.; Cho, S.-Y.; Kang, W.; Lee, A.; Min, S.; Kang, J.; Kim, M.J.; Choi, J.; Lee, W.; et al. Predictive Biomarkers for 5-Fluorouracil and Oxaliplatin-Based Chemotherapy in Gastric Cancers via Profiling of Patient-Derived Xenografts. Nat. Commun. 2021, 12, 4840. [Google Scholar] [CrossRef]
- Petitjean, A.; Mathe, E.; Kato, S.; Ishioka, C.; Tavtigian, S.V.; Hainaut, P.; Olivier, M. Impact of Mutant P53 Functional Properties on TP53 Mutation Patterns and Tumor Phenotype: Lessons from Recent Developments in the IARC TP53 Database. Hum. Mutat. 2007, 28, 622–629. [Google Scholar] [CrossRef]
- Armstrong, M.B.; Bian, X.; Liu, Y.; Subramanian, C.; Ratanaproeksa, A.B.; Shao, F.; Yu, V.C.; Kwok, R.P.S.; Opipari, A.W.; Castle, V.P. Signaling from P53 to NF-κB Determines the Chemotherapy Responsiveness of Neuroblastoma. Neoplasia 2006, 8, 964–974. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, Y.; Zhou, X.; Zhu, S.; Kaya, N.A.; Chan, Y.S.; Ma, L.; Xu, M.; Zhai, W. An Integrative Analysis of Nasopharyngeal Carcinoma Genomes Unraveled Unique Processes Driving a Viral-Positive Cancer. Cancers 2023, 15, 1243. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Feng, R.; Liu, Z.; Zhou, X.; Chen, Y.; Cao, Y.; Valeri, L.; Li, Z.; Liu, Z.; Cao, S.-M.; et al. Host Genetic Variants, Epstein-Barr Virus Subtypes, and the Risk of Nasopharyngeal Carcinoma: Assessment of Interaction and Mediation. Cell Genom. 2024, 4, 100474. [Google Scholar] [CrossRef] [PubMed]
- Wong, K.C.W.; Hui, E.P.; Lo, K.-W.; Lam, W.K.J.; Johnson, D.; Li, L.; Tao, Q.; Chan, K.C.A.; To, K.-F.; King, A.D.; et al. Nasopharyngeal Carcinoma: An Evolving Paradigm. Nat. Rev. Clin. Oncol. 2021, 18, 679–695. [Google Scholar] [CrossRef]
- Dai, W.; Chung, D.L.-S.; Chow, L.K.-Y.; Yu, V.Z.; Lei, L.C.; Leong, M.M.-L.; Chan, C.K.-C.; Ko, J.M.-Y.; Lung, M.L. Clinical Outcome–Related Mutational Signatures Identified by Integrative Genomic Analysis in Nasopharyngeal Carcinoma. Clin. Cancer Res. 2020, 26, 6494–6504. [Google Scholar] [CrossRef]
- Tsang, C.M.; Lui, V.W.Y.; Bruce, J.P.; Pugh, T.J.; Lo, K.W. Translational Genomics of Nasopharyngeal Cancer. Semin. Cancer Biol. 2020, 61, 84–100. [Google Scholar] [CrossRef]
- Lin, M.; Zhang, X.-L.; You, R.; Liu, Y.-P.; Cai, H.-M.; Liu, L.-Z.; Liu, X.-F.; Zou, X.; Xie, Y.-L.; Zou, R.-H.; et al. Evolutionary Route of Nasopharyngeal Carcinoma Metastasis and Its Clinical Significance. Nat. Commun. 2023, 14, 610. [Google Scholar] [CrossRef]
- Bruce, J.P.; To, K.-F.; Lui, V.W.Y.; Chung, G.T.Y.; Chan, Y.-Y.; Tsang, C.M.; Yip, K.Y.; Ma, B.B.Y.; Woo, J.K.S.; Hui, E.P.; et al. Whole-Genome Profiling of Nasopharyngeal Carcinoma Reveals Viral-Host Co-Operation in Inflammatory NF-κB Activation and Immune Escape. Nat. Commun. 2021, 12, 4193. [Google Scholar] [CrossRef]
- Chung, A.; OuYang, C.; Liu, H.; Chao, M.; Luo, J.; Lee, C.; Lu, Y.; Chung, I.; Chen, L.; Wu, S.; et al. Targeted Sequencing of Cancer-related Genes in Nasopharyngeal Carcinoma Identifies Mutations in the TGF-β Pathway. Cancer Med. 2019, 8, 5116–5127. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Dai, W.; Cheung, A.K.L.; Ko, J.M.Y.; Kan, R.; Wong, B.W.Y.; Leong, M.M.L.; Deng, M.; Kwok, T.C.T.; Chan, J.Y.-W.; et al. Whole-Exome Sequencing Identifies Multiple Loss-of-Function Mutations of NF-κB Pathway Regulators in Nasopharyngeal Carcinoma. Proc. Natl. Acad. Sci. USA 2016, 113, 11283–11288. [Google Scholar] [CrossRef]
- Chow, Y.P.; Tan, L.P.; Chai, S.J.; Abdul Aziz, N.; Choo, S.W.; Lim, P.V.H.; Pathmanathan, R.; Mohd Kornain, N.K.; Lum, C.L.; Pua, K.C.; et al. Exome Sequencing Identifies Potentially Druggable Mutations in Nasopharyngeal Carcinoma. Sci. Rep. 2017, 7, 42980. [Google Scholar] [CrossRef] [PubMed]
- You, R.; Liu, Y.-P.; Lin, D.-C.; Li, Q.; Yu, T.; Zou, X.; Lin, M.; Zhang, X.-L.; He, G.-P.; Yang, Q.; et al. Clonal Mutations Activate the NF-κB Pathway to Promote Recurrence of Nasopharyngeal Carcinoma. Cancer Res. 2019, 79, 5930–5943. [Google Scholar] [CrossRef]
- Li, J.; Guo, M.; Chen, L.; Chen, Z.; Fu, Y.; Chen, Y. Amyloid Aggregates Induced by the P53-R280T Mutation Lead to Loss of P53 Function in Nasopharyngeal Carcinoma. Cell Death Dis. 2024, 15, 35. [Google Scholar] [CrossRef] [PubMed]
- EAR, E.N.S.; Irekeola, A.A.; Yean Yean, C. Diagnostic and Prognostic Indications of Nasopharyngeal Carcinoma. Diagnostics 2020, 10, 611. [Google Scholar] [CrossRef]
- Wang, S.; Zhao, Y.; Aguilar, A.; Bernard, D.; Yang, C.-Y. Targeting the MDM2–P53 Protein–Protein Interaction for New Cancer Therapy: Progress and Challenges. Cold Spring Harb. Perspect. Med. 2017, 7, a026245. [Google Scholar] [CrossRef] [PubMed]
- National Cancer Institute. Phase 1/2 Study of APR-246 in Combination with Pembrolizumab in Subjects with Solid Tumor Malignancies. Available online: https://www.cancer.gov/research/participate/clinical-trials-search/v?id=NCI-2020-05550 (accessed on 15 March 2025).
- ClinicalTrials.gov. P53 Gene Combined with Radio- and Chemo-therapy in Treatment of Unresectable Locally Advanced Head and Neck Cancer; NCT02429037. Available online: https://clinicaltrials.gov/study/NCT02429037 (accessed on 15 March 2025).
- National Cancer Institute. A Novel MDM2 Inhibitor (APG-115) for the Treatment of p53 Wild-Type Salivary Gland Cancer. Available online: https://www.cancer.gov/research/participate/clinical-trials-search/v?id=NCI-2022-00506 (accessed on 15 March 2025).
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.-C. NF-κB Signaling in Inflammation. Signal Transduct. Target. Ther. 2017, 2, 17023. [Google Scholar] [CrossRef]
- Li, Y.Y.; Chung, G.T.Y.; Lui, V.W.Y.; To, K.-F.; Ma, B.B.Y.; Chow, C.; Woo, J.K.S.; Yip, K.Y.; Seo, J.; Hui, E.P.; et al. Exome and Genome Sequencing of Nasopharynx Cancer Identifies NF-κB Pathway Activating Mutations. Nat. Commun. 2017, 8, 14121. [Google Scholar] [CrossRef]
- Or, Y.Y.; Hui, A.B.; To, K.; Lam, C.N.; Lo, K. PIK3CA Mutations in Nasopharyngeal Carcinoma. Int. J. Cancer 2006, 118, 1065–1067. [Google Scholar] [CrossRef]
- Chou, C.-C.; Chou, M.-J.; Tzen, C.-Y. PIK3CA Mutation Occurs in Nasopharyngeal Carcinoma but Does Not Significantly Influence the Disease-Specific Survival. Med. Oncol. 2009, 26, 322–326. [Google Scholar] [CrossRef] [PubMed]
- Venot, Q.; Blanc, T.; Rabia, S.H.; Berteloot, L.; Ladraa, S.; Duong, J.-P.; Blanc, E.; Johnson, S.C.; Hoguin, C.; Boccara, O.; et al. Targeted Therapy in Patients with PIK3CA-Related Overgrowth Syndrome. Nature 2018, 558, 540–546. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.; Ni, J.; Guo, H.; Luu, V.; Wang, Y.; Zhao, J.J.; Roberts, T.M. The Role of the PIK3CA Gene in the Development and Aging of the Brain. Sci. Rep. 2021, 11, 291. [Google Scholar] [CrossRef] [PubMed]
- Alqahtani, A.; Ayesh, H.S.K.; Halawani, H. PIK3CA Gene Mutations in Solid Malignancies: Association with Clinicopathological Parameters and Prognosis. Cancers 2019, 12, 93. [Google Scholar] [CrossRef]
- Migliaccio, I.; Paoli, M.; Risi, E.; Biagioni, C.; Biganzoli, L.; Benelli, M.; Malorni, L. PIK3CA Co-Occurring Mutations and Copy-Number Gain in Hormone Receptor Positive and HER2 Negative Breast Cancer. NPJ Breast Cancer 2022, 8, 24. [Google Scholar] [CrossRef]
- Rasti, A.R.; Guimaraes-Young, A.; Datko, F.; Borges, V.F.; Aisner, D.L.; Shagisultanova, E. PIK3CA Mutations Drive Therapeutic Resistance in Human Epidermal Growth Factor Receptor 2-Positive Breast Cancer. JCO Precis. Oncol. 2022, 6, e2100370. [Google Scholar] [CrossRef]
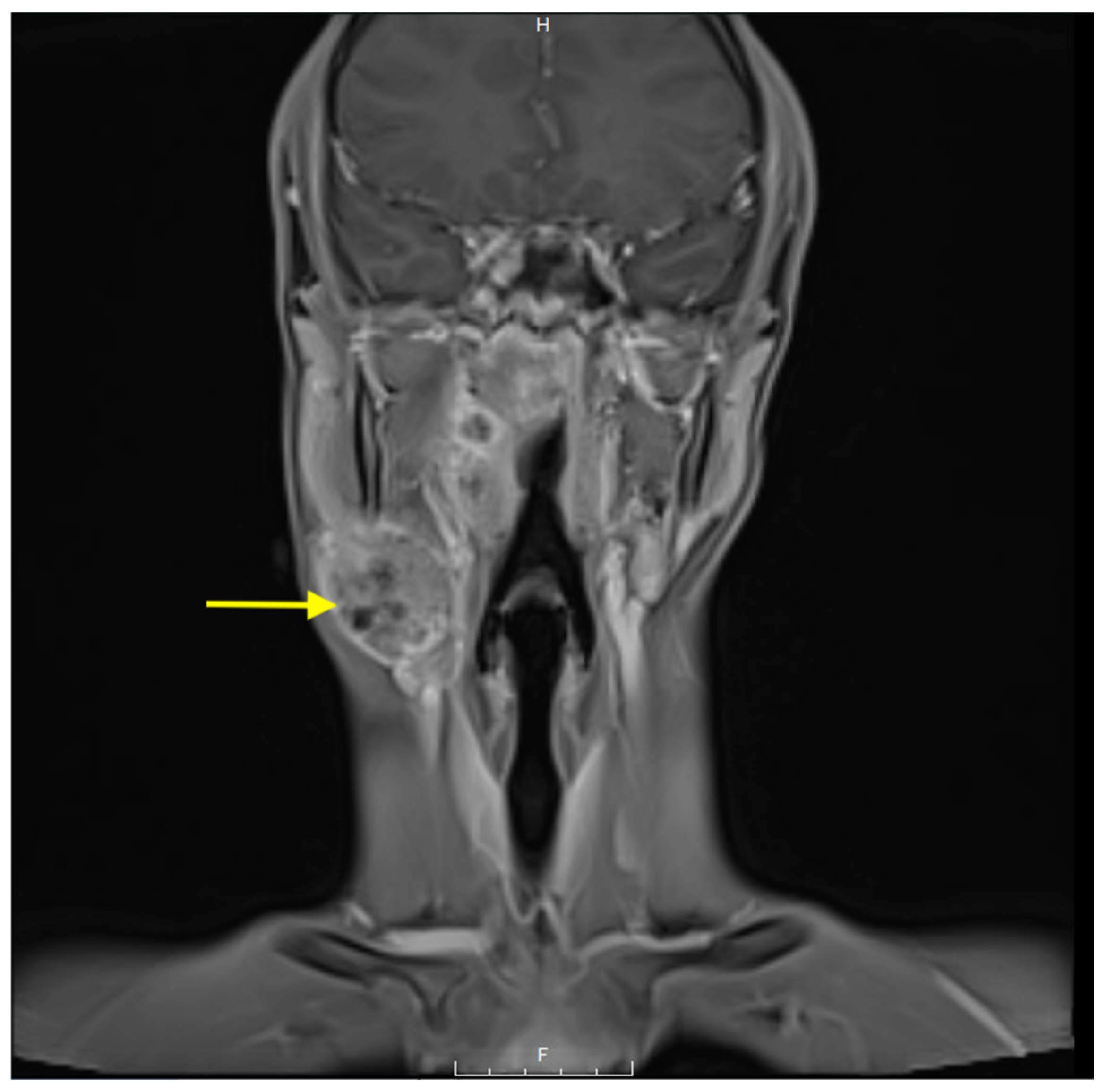
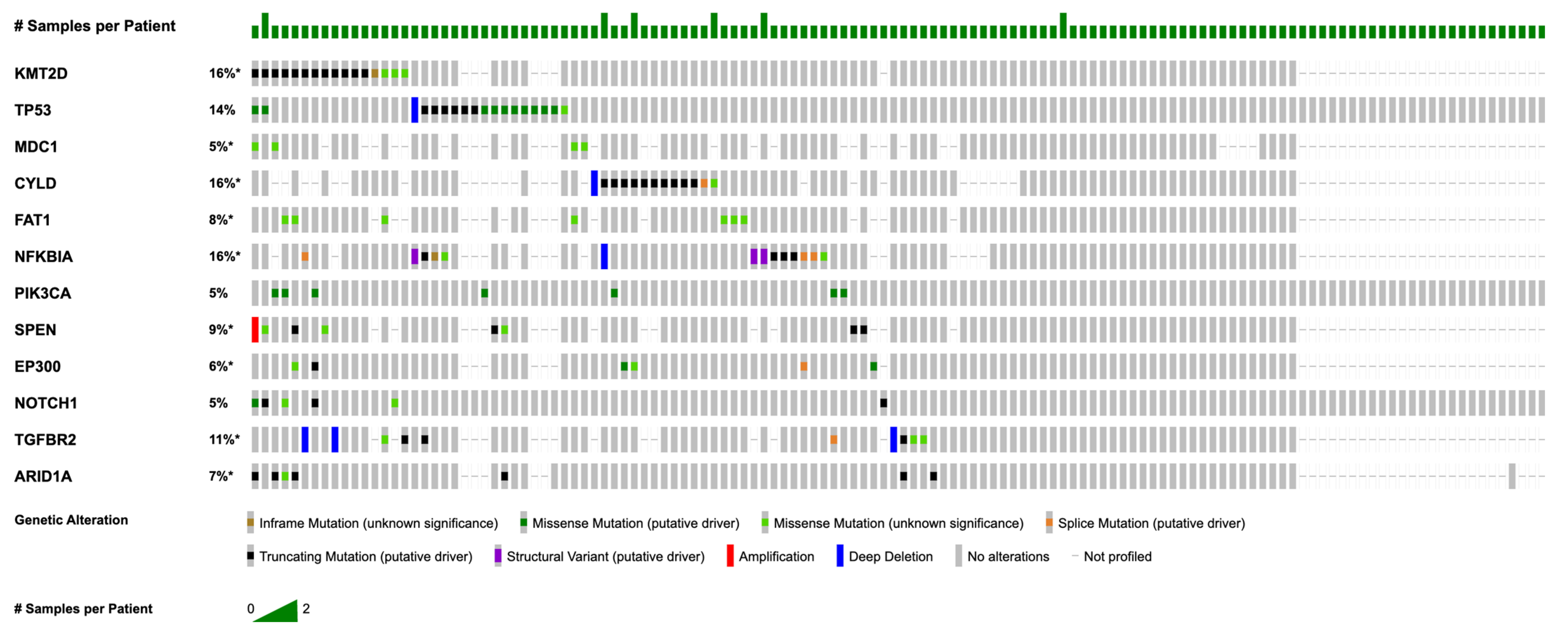
| Demographics | Category | n (%) |
|---|---|---|
| Sex | Male | 83 (69.7) |
| Female | 42 (35.5) | |
| Age category | Adult | 121 (96.6) |
| Pediatric | 4 (3.4) | |
| Ethnicity | Non-Hispanic | 71 (59.7) |
| Unknown/Not Collected | 22 (18.5) | |
| Hispanic | 9 (7.6) | |
| Race | Asian | 51 (42.9) |
| White | 33 (27.7) | |
| Black | 15 (12.6) | |
| Other | 8 (6.7) | |
| Unknown | 5 (4.2) | |
| Sample Type | Primary | 48 (40.3) |
| Metastasis | 67 (56.3) | |
| Not Collected | 6 (5.0) |
| Gene (Chi-Squared) | Asian, n (%) | Non-Asian, n (%) | p Value |
|---|---|---|---|
| TP53 | 3 (5.9) | 15 (12.0) | p = 0.0361 |
| CCND2 | 0 (0.0) | 6 (4.8) | p = 0.0272 |
| KDM5A | 0 (0.0) | 6 (4.8) | p = 0.0259 |
| ITPKB | 0 (0.0) | 1 (0.8) | p = 0.0192 |
| CD40 | 0 (0.0) | 1 (0.8) | p = 0.0192 |
| CHD7 | 0 (0.0) | 1 (0.8) | p = 0.0192 |
| ASNS | 0 (0.0) | 1 (0.8) | p = 0.0192 |
| Male, n (%) | Female, n (%) | ||
| KDM5A | 1 (0.8) | 5 (4.0) | p = 0.0137 |
| CCND2 | 1 (0.8) | 5 (4.0) | p = 0.0149 |
| PIK3C2G | 0 (0.0) | 5 (4.0) | p = 0.0018 |
| ETV6 | 0 (0.0) | 4 (3.2) | p = 0.0093 |
| CDKN1B | 0 (0.0) | 4 (3.2) | p = 0.0099 |
| ASNS | 0 (0.0) | 1 (0.8) | p = 0.0147 |
| CD40 | 0 (0.0) | 1 (0.8) | p = 0.0147 |
| CHD7 | 0 (0.0) | 1 (0.8) | p = 0.0147 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hsia, B.; Sure, A.; Dongre, R.; Jo, N.; Kuzniar, J.; Bitar, G.; Alshaka, S.A.; Kim, J.D.; Valencia-Sanchez, B.A.; Brandel, M.G.; et al. Molecular Profiling of Nasopharyngeal Carcinoma Using the AACR Project GENIE Repository. Cancers 2025, 17, 1544. https://doi.org/10.3390/cancers17091544
Hsia B, Sure A, Dongre R, Jo N, Kuzniar J, Bitar G, Alshaka SA, Kim JD, Valencia-Sanchez BA, Brandel MG, et al. Molecular Profiling of Nasopharyngeal Carcinoma Using the AACR Project GENIE Repository. Cancers. 2025; 17(9):1544. https://doi.org/10.3390/cancers17091544
Chicago/Turabian StyleHsia, Beau, Asritha Sure, Roshan Dongre, Nicolas Jo, Julia Kuzniar, Gabriel Bitar, Saif A. Alshaka, Jeeho D. Kim, Bastien A. Valencia-Sanchez, Michael G. Brandel, and et al. 2025. "Molecular Profiling of Nasopharyngeal Carcinoma Using the AACR Project GENIE Repository" Cancers 17, no. 9: 1544. https://doi.org/10.3390/cancers17091544
APA StyleHsia, B., Sure, A., Dongre, R., Jo, N., Kuzniar, J., Bitar, G., Alshaka, S. A., Kim, J. D., Valencia-Sanchez, B. A., Brandel, M. G., Sato, M., Crawford, J. R., Levy, M. L., Polster, S. P., & Patel, V. A. (2025). Molecular Profiling of Nasopharyngeal Carcinoma Using the AACR Project GENIE Repository. Cancers, 17(9), 1544. https://doi.org/10.3390/cancers17091544






