The Co-Expression and Cellular Location of HER Family Members, EGFRvIII, Putative Cancer Stem Cell Biomarkers CD44 and CD109 in Patients with Glioblastoma, and Their Impacts on Prognosis
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Information
2.2. Immunohistochemical Staining
2.3. Statistical Analysis
3. Results
3.1. Patient Clinicopathological Features
3.2. The Immunohistochemical Expression of HER-Family Members and EGFRvIII in Patients with Brain Cancer
3.3. Immunohistochemical Expression of CD44 and CD109 in Patient with Brain Tumour
3.4. The Association Between the Clinicopathological Parameters and the Expression of HER-Family Members in Patients with Brain Cancer
4. Discussion
| Study | Antibody Used/Scoring System | Number of Specimens Examined (% of Positive Cases) | Summary |
|---|---|---|---|
| (Agosti et al., 1992) [60] | Mouse anti-EGFR (not known). | 103 astrocytic tumours, Glioblastoma (37%) | A close correlation was found between the presence of EGFR gene amplification and over-expression of receptor protein. |
| (Zhu et al., 1996) [61] | Mouse anti-EGFR (Sigma), EGFR positive when >0 of tumour cells with staining | 55 Glioblastoma, 16 anaplastic astrocytoma (69%) | The percentage of EGFR positive cells in patients with astrocytic gliomas was associated significantly reduced overall survival (p = 0.0434). |
| (Korshunov et al., 2000) [62] | EGFR (clone H-11, Dako, catalogue No. M3563)/ ≥5% expression was considered positive | 88 Ependymomas (43%) | (EGFR) revealed its predominantly membranous staining in 38 tumours with staining of 40–80% of the component cells. In addition, no differences in survival time between patients with different grades of EGFR reactivity when tumours with more than 50% of immunostained cells when compared with those with less then 50% or absence of expression. |
| (Andersson et al., 2004) [63] | Mouse anti-EGFR 31G7 (Zymed Laboratories)/ low (<20%), moderate (20–40%) or high (>40%) | 44 Gliomas and 26 Meningiomas (58%) | The significantly higher EGFR protein expression in high-grade tumours than in low-grade tumours (p = 0.004) was correlated with a shorter overall survival compared to patients with low or no protein expression (p < 0.001). The antibody recognizes both wtEGFR and EGFRvIII. |
| (Nishikawa et al., 2004) [64] | EGFR 113 Novocastra (Newcastle, UK) low (<20%), moderate (20–40%) or high (>40%) | 53 Glioblastoma (55%) | No prognostic significance of EGFR was studied. Also, EGFR 113 is not wt EGFR specific and cross react with the EGFRvIII |
| (Varela et al., 2004) [53] | EGFR (M3563, Dako Corporation, Glostrup, Denmark)/EGFR positive at >10% | 103 malignant glioma (22%) | EGFR expression was significantly associated with worse overall survival (p < 0.01). |
| (Chakravarti et al., 2005) [54] | Mouse anti-EGFR 31G7, Zymed Laboratories)/ Immuno program scoring system | 155 Glioblastoma multiforme TMAs (25%) | The antibody recognizes both wtEGFR and EGFRvIII. The EGFR expression was not associated with overall survival or PFS. The antibody recognizes both wtEGFR and EGFRvIII. |
| (Heimberger et al., 2005) [65] | Mouse anti-EGFR 31G7, Zymed Laboratories)/ >10% expression was considered positive tumour cells | 196 Glioblastoma (54%) | Neither the overexpressed wild-type EGFR nor EGFRvIII was an independent predictor of median overall survival in this selected cohort of patients who underwent extensive tumour resection. The antibody recognizes both wtEGFR and EGFRvIII. |
| (Mendrzyk et al., 2006) [66] | EGFR (rabbit polyclonal, clone sc-03 Biotechnology, Santa Cruz, CA, USA) ≥10% expression was considered positive | 163 ependymomas (59%) | EGFR overexpression was associated with a poor prognosis in patients with intracranial grade II tumours (p = 0.002). |
| (Umesh et al., 2009) [67] | EGFR (monoclonal E-30; EGFR (Clone H-11, monoclonal) A cut-off value of >20% was considered positive | 54 supratentorial glioblastoma (35.2%) | Over-expression of EGFR was a significant predictor of poor outcome on multivariate analysis |
| (Nabika et al., 2010) [68] | Mouse anti-HER2 (Novocastra),/scored by counting the numbering of positive cells per 1000 tumour cells and producing labelling index (i.e., <30% as negative, >30% as positive). | 59 High grade astrocytoma EGFR (67.8%) | High expression of EGFR was associated with poor prognosis (p = 0.017). |
| (Senetta et al., 2011) [69] | Mouse anti EGFR mAb, clone 31G7, Zymed A cut-off value of >20% was considered positive | 22 supratentorial glioblastoma (45%) | By univariate analysis, histological grade (p = 0.018) and EGFR (p = 0.014) expression significantly correlated with overall survival. The antibody recognizes both wtEGFR and EGFRvIII. |
| (Hobbs et al., 2012) [56] | EGFR primary antibody (Ventana 790-2988/3C6/prediluted\ Sem quantitative scoring (Negative, weak, intermediate, strong) | 532 glioblastomata (92%) | No significant association between EGFR expression and overall survival in univariate analysis (p = 0.59. The median survival was 39% longer in the high-amplifier group (EGFR: chromosome 7 ratio > 20) versus non-amplified GBMs (p = 0.03). |
| (Choi et al., 2013) [55] | Mouse anti-EGFR Zymed A cut-off value of >10% was considered positive | 33 Glioblastoma (76.7%) | The antibody recognizes both wtEGFR and EGFRvIII. Survival in EGFR expressing GBM patients was significantly less than that in non-expressing patients (median survival: 12.5 versus 17.5 months, p = 0.013). |
| (Lee et al., 2013) [70] | EGFR, Dako, Camarillo, A cut off value of <5% was considered positive | 150 Glioblastoma (62.6%) | No association between the main prognostic factors in glioblastoma. |
| (Michaelsen et al., 2013) [71] | EGFR DAKO, Glostrup, Denmark A cut off value of >10%, was considered positive | 225 Glioblastoma multiforme (64%) | No association between the main prognostic factors in glioblastoma multiforme. |
| (Saha et al., 2014) [72] | Mouse anti EGFR EP38Y A cut off value of >21.7% tumour was considered positive | 57 Glioblastoma (41%) Anaplastic astrocytoma’s (17%) | Distribution of age, EGFR and Ki-67 labelling index expressed strong positive (≥0.5) correlation with the grade of tumours. |
| (Montgomery et al., 2015) [73] | EGFR Dako, K4061 A cut off value of >25% was considered positive | 36 Glioblastoma (28%) | The correlation between the expression of MDM2 and that of the wild variant of EGFR was positive (p-value = 0.04). |
| (Tini et al., 2015) [74] | Mouse anti EGFR clone 31G7, Zymed, Milan, Italy Scored based on intensity and % of tumour positively stained cells (i.e., total score 0–2 as a Low and 3–5 as a high. | 144 Glioblastoma (64.6%) | The antibody recognizes both wtEGFR and EGFRvIII. Patients with a high EGFR expression seemed to present worse clinic neurological status and radiological features of tumour aggressiveness. The antibody recognizes both wtEGFR and EGFRvIII. |
| (Tripathy et al., 2017) [75] | Anti EGFR pan kit; Biogenex, Hyderabad, India >20% expression was considered positive | 52 Glioblastoma multiforme (58%) | EGFR negative patients respond better to therapy along with longer duration of survival as compared to EGFR positive patient. |
| (Abdulghani et al., 2019) [57] | EGFR antibody, clone (EP38Y) Abcam >5% expression was considered positive | 44 Astrocytic tumours (38.8%) | Expression of EGFR was restricted to only grade IV glioblastoma patients and no expression was found in astrocytoma patients grades I, II and III. |
| (Amirpour et al., 2020) [76] | EGFR antibody (Dako, Denmark) >5% expression was considered positive | 70 Glioblastoma (61.4%) | GBM tumour was associated with a poor prognosis and a low survival rate. It was also found that the expression of the EGFR gene did not affect the survival rate of patients with GBM |
| (Miratashi Yazdi et al., 2022) [77] | Monoclonal anti-EGFR antibody clone EP22; Master Diagnóstica, Spain). >10% expression was considered positive | 30 Glioblastoma (56.6%) | EGFR expression and tumour characteristics showed no significant association. |
| Current Study | Mouse anti-human wt EGFR mAb Clone: DAK-H1-WT Isotype: IgG1, kappa ≥5% expression was considered positive | 80 Glioblastoma 46% (≥5%) 40% (≥10%) 34% (≥20%) 24% (≥50%) | No significant correlation was demonstrated between EGFR and other HER family members. |
| Study | Antibody Used/Scoring System | Number of Specimens Examined (% of Positive Cases) | Summary |
|---|---|---|---|
| (A) HER2 | |||
| (Haapasalo et al., 1996) [87] | Anti HER2 monoclonal MAb1 antibody TA250 | 94 Glioblastoma (6.4%) | No significant prognostic value and that HER2 oncogene amplification is not seen in the few cases in which there is overexpression. |
| (Koka et al., 2003) [88] | HER2 (DAKO Diagnostics) | 149 Glioblastoma 15.4% | Age, performance status, smoking history, and treatment, logistic regression analysis (with a survival of <3 months as the dependent variable) revealed that Her-2/neu overexpression significantly (p < 0.01) increased the odds of early mortality (<3 months). |
| (Mineo et al., 2007) [81] | Anti-HER2 antibody (Novocastra, clone CB11) 0 = no staining; 1+ = faint, incomplete membranous pattern; 2+ = moderate, complete membranous pattern; 3+ = strong membranous pattern | 57 Glioblastoma (82.5%) | Survival time was significantly longer when HER2 expression was low p = 0.04). The patterns of HER2 expression were similar between grade III gliomas and secondary GBM. |
| (Meurer et al., 2008) [80] | Mouse anti-HER2, Neu F-11 (Santa Cruz Biotechnology) >20% expression was considered positive tumour cells | 40 Medulloblastoma (57.5%) | HER2 was positive in 23 (57.5%) of the samples and did not show statistical association with survival (p = 0.07). |
| (Nabika et al., 2010) [68] | Mouse anti-HER2 (Novocastra),/scored by counting the numbering of positive cells per 1000 tumour cells and producing labelling index (i.e., < 30% as negative, >30% as positive). | 59 High grade astrocytoma HER2 (28.8%) | No significant association between HER2 overexpression and prognosis |
| (Ramezani et al., 2020) [79] | HER2 (DAKO Diagnostics, Polyclonal Rabbit Anti-Human A0485) >10% expression was considered positive tumour cells | 107 Malignant brain tumours (42.1%) | The type of brain tumours can impact on HER2 expression that high HER2 expression in High grade glioma may be helpful for therapeutics. |
| Mulliqi et al., 2025 Current study | Mouse anti-human HER-2 mAb Clone: 3B5 Isotype: IgG1, kappa ≥5% expression was considered positive tumour cells | 80 Glioblastoma 75% (≥5%) 55% (≥10%) 43.8% (≥20%) 20.% (≥50%) | Co expressions with low HER2 intensity there was a statistically significant association on increase of patient overall survival. HER2 positivity was found to be an independent prognostic marker in multivariate analysis. Furthermore, expression of HER2 positivity with cytoplasmic staining was also associated with significantly increase of overall survival in patients with glioblastoma (p = 0.022). |
| (B) HER3 and HER4 | |||
| (Nabika et al., 2010) [68] | Mouse anti-HER3 (Novocastra), and HER4 (Lab vision) antibodies/scored by counting the numbering of positive cells per 1000 tumour cells and producing labelling index (i.e., < 30% as negative, >30% as positive). | 59 High grade astrocytoma HER3 (5.1%) HER4 (75%) | High expression of HER4 was associated with a poor prognosis (p = 0.004) but no association between HER3 and prognosis |
| (Donoghue et al., 2018) [89] | Anti HER4 antibody (not known) <50% as negative and >50% as positive | 53 Glioblastoma HER4 (11%) | high p-ERBB4 in 11% of archived GBM samples, independent of p-EGFR, was associated with shorter patient survival (12.0 ± 3.2 months) than was no p-ERBB4 (22.5 ± 9.5 months) |
| (Arnli et al., 2019) [90] | HER3 RTJ-1 IgM, mouse monoclonal HER4 HFR1 IgG2b, mouse Monoclonal, Dako >10% expression was considered positive | 186 Meningiomas TMA HER3 (98.4%) HER4 (100%) | Meningiomas of all grades were shown to widely express both HER3 and HER4 receptors however neither HER3 nor HER4 expressions to be of prognostic significance. |
| (Kusuhara et al., 2022) [82] |
HER3 (Cell Signalling Technology) H score system | 44 Metastatic brain cancer HER3 (91%) | Tissue from breast cancer brain metastases had significantly higher levels of HER3 expression than primary tumours, supporting that HER3 is a potential target for BC brain metastases. |
| (Tomasich et al., 2023) [85] | HER3 (Cell Signalling Technology, 12708, RRID: AB_2721919 >10% expression was considered positive | Metastatic brain cancer HER3 180 (75%) | HER3 expression did not correlate with overall survival from Brain metastasis diagnosis. |
| Mulliqi et al., 2025 Current study | Rabbit anti-HER3 mAb Clone: SP71 Isotype: IgG Mouse anti-human HER4 mAb Clone: HFR1 Isotype: IgG2b, kappa ≥5% expression was considered positive | 80 Glioblastoma 19% HER3 (≥5%) 18%HER3(≥10%) 15% HER3 (≥20%) 4% HER3(≥50%) 71%HER4 (≥5%) 68%HER4(≥10%) 56%HER4(≥20%) 25% HER4(≥50%) | HER3 expression was found not to be of prognostic significance. There was found to be a prognostic significance in years of HER2 and its co expression with HER4 on the overall survival. |
| Study | Antibody Used/Scoring System | Number of Specimens Examined (% of Positive Cases) | Summary |
|---|---|---|---|
| (Shinojima et al., 2003) [100] | EGFRvIII DH8.3 no staining), 1(light or focal), 2 (moderate), and 3 (strong). | 87 supratentorial glioblastoma (45%) | EGFRvIII overexpression, was not predictive for OS. However, in patients with EGFR amplification, multivariate analysis revealed that EGFRvIII overexpression was an independent, significant, poor prognostic factor for OS (p = 0.0044, HR = 2.71). |
| (Aldape et al., 2004) [101] | Rabbit anti EGFRvIII Zymed, San Francisco, CA, USA >10% expression was considered positive | 168 Glioblastoma (41.3%) Anaplastic astrocytoma (21.4%) | EGFRvIII had positivity but no association with overall survival among GBM patients (p = 0.84) but being highly associated with reduced survival among Anaplastic astrocytoma (AA) patients (p = 0.0016). The antibody recognizes both wtEGFR and EGFRvIII. |
| (Heimberger et al., 2005) [65] | Rabbit anti-EGFRvIII polyclonal antibody (Zymed, San Francisco, CA, USA). >10% expression was considered positive | 196 Glioblastoma multiforme (31%) | Neither the overexpressed wild-type EGFR nor EGFRvIII was an independent predictor of median overall survival. The antibody recognizes both wtEGFR and EGFRvIII. |
| (Pelloski et al., 2007) [96] | EGFRvIII Zymed, Carlsbad, CA, USA) >10% expression was considered positive | 649 Glioblastoma (31%) | No significant association between shorter overall survival time and positivity for EGFRvIII (p 0.056) and p-Akt (p 0.095) and extent of resection (p 0.091). |
| (Felsberg et al., 2017) [15] | EGFRvIII monoclonal mouse antibody E30 (Dako) Semiquantitative scored | 106 Glioblastoma (56.6%) | EGFRvIII positivity was not associated with different progression-free or overall survival |
| (Nozawa et al., 2019) [102] | EGFRvIII (clone L8A4; Absolute Antibody, Oxford, UK. >30% expression was considered positive | 67 Glioblastoma (19.4%) | EGFRvIII-expression in patients with glioblastoma was not significantly associated with a favorable outcome. |
| Mulliqi et al., 2025 Current study | Mouse anti-EGFRvIII mAb. Clone: DH8.5 (IgG1) ≥5% expression was considered positive | 80 Glioblastoma 85% (≥5%) 59% (≥10%) 41% (≥20%) 4% (≥50%) | EGFRvIII had no prognostic significance. |
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| EGFR | Epidermal growth factor receptor |
| HER | Human Epidermal Growth Factor Receptor |
| EGFRvIII | Mutated version of Epidermal growth factor receptor |
| CD44 | Cluster differentiation 44 |
| CD109 | Cluster differentiation 109 |
| GBM | Glioblastoma |
References
- Hynes, N.E.; MacDonald, G. ErbB receptors and signaling pathways in cancer. Curr. Opin. Cell Biol. 2009, 21, 177–184. [Google Scholar] [CrossRef]
- Pan, P.C.; Magge, R.S. Mechanisms of EGFR Resistance in Glioblastoma. Int. J. Mol. Sci. 2020, 21, 8471. [Google Scholar] [CrossRef]
- Bai, X.; Sun, P.; Wang, X.; Long, C.; Liao, S.; Dang, S.; Zhuang, S.; Du, Y.; Zhang, X.; Li, N.; et al. Structure and dynamics of the EGFR/HER2 heterodimer. Cell Discov. 2023, 9, 18. [Google Scholar] [CrossRef]
- Lee, A.; Arasaratnam, M.; Chan, D.L.H.; Khasraw, M.; Howell, V.M.; Wheeler, H. Anti-epidermal growth factor receptor therapy for glioblastoma in adults. Cochrane Database Syst. Rev. 2020, 5, CD013238. [Google Scholar] [CrossRef]
- Ezzati, S.; Salib, S.; Balasubramaniam, M.; Aboud, O. Epidermal Growth Factor Receptor Inhibitors in Glioblastoma: Current Status and Future Possibilities. Int. J. Mol. Sci. 2024, 25, 2316. [Google Scholar] [CrossRef] [PubMed]
- Velpula, K.K.; Dasari, V.R.; Asuthkar, S.; Gorantla, B.; Tsung, A.J. EGFR and c-Met Cross Talk in Glioblastoma and Its Regulation by Human Cord Blood Stem Cells. Transl. Oncol. 2012, 5, 379–392. [Google Scholar] [CrossRef] [PubMed]
- Kalman, B.; Szep, E.; Garzuly, F.; Post, D.E. Epidermal growth factor receptor as a therapeutic target in glioblastoma. Neuromol. Med. 2013, 15, 420–434. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Chang, S.S.; Hsu, J.L.; Hung, M.C. Signaling cross-talk in the resistance to HER family receptor targeted therapy. Oncogene 2014, 33, 1073–1081. [Google Scholar] [CrossRef]
- Paolillo, M.; Boselli, C.; Schinelli, S. Glioblastoma under Siege: An Overview of Current Therapeutic Strategies. Brain Sci. 2018, 8, 15. [Google Scholar] [CrossRef]
- Narita, Y.; Arakawa, Y.; Yamasaki, F.; Nishikawa, R.; Aoki, T.; Kanamori, M.; Nagane, M.; Kumabe, T.; Hirose, Y.; Ichikawa, T. A randomized, double-blind, phase III trial of personalized peptide vaccination for recurrent glioblastoma. Neuro-Oncology 2019, 21, 348–359. [Google Scholar]
- Zanders, E.D.; Svensson, F.; Bailey, D.S. Therapy for glioblastoma: Is it working? Drug Discov. Today 2019, 24, 1193–1201. [Google Scholar] [CrossRef] [PubMed]
- Kizilbash, S.H. Why has targeting EGFR aberrations in glioblastoma therapy had limited success? Expert Rev. Anticancer Ther. 2022, 22, 1261–1263. [Google Scholar] [CrossRef] [PubMed]
- Haynik, D.M.; Roma, A.A.; Prayson, R.A. HER-2/neu expression in glioblastoma multiforme. Appl. Immunohistochem. Mol. Morphol. 2007, 15, 56–58. [Google Scholar] [CrossRef]
- Zadeh, G.; Bhat, K.P.; Aldape, K. EGFR and EGFRvIII in glioblastoma: Partners in crime. Cancer Cell 2013, 24, 403–404. [Google Scholar] [CrossRef]
- Felsberg, J.; Hentschel, B.; Kaulich, K.; Gramatzki, D.; Zacher, A.; Malzkorn, B.; Kamp, M.; Sabel, M.; Simon, M.; Westphal, M.; et al. Epidermal Growth Factor Receptor Variant III (EGFRvIII) Positivity in EGFR-Amplified Glioblastomas: Prognostic Role and Comparison between Primary and Recurrent Tumors. Clin. Cancer Res. 2017, 23, 6846–6855. [Google Scholar] [CrossRef] [PubMed]
- An, Z.; Aksoy, O.; Zheng, T.; Fan, Q.W.; Weiss, W.A. Epidermal growth factor receptor and EGFRvIII in glioblastoma: Signaling pathways and targeted therapies. Oncogene 2018, 37, 1561–1575. [Google Scholar] [CrossRef]
- Mulliqi, E.; Khelwatty, S.; Morgan, A.; Ashkan, K.; Modjtahedi, H. Synergistic Effects of Neratinib in Combination With Palbociclib or Miransertib in Brain Cancer Cells. World J. Oncol. 2024, 15, 492–505. [Google Scholar] [CrossRef]
- Pointer, K.B.; Clark, P.A.; Zorniak, M.; Alrfaei, B.M.; Kuo, J.S. Glioblastoma cancer stem cells: Biomarker and therapeutic advances. Neurochem. Int. 2014, 71, 1–7. [Google Scholar] [CrossRef]
- Gimple, R.C.; Yang, K.; Halbert, M.E.; Agnihotri, S.; Rich, J.N. Brain cancer stem cells: Resilience through adaptive plasticity and hierarchical heterogeneity. Nat. Rev. Cancer 2022, 22, 497–514. [Google Scholar] [CrossRef]
- Lin, S.; Li, K.; Qi, L. Cancer stem cells in brain tumors: From origin to clinical implications. MedComm 2023, 4, e341. [Google Scholar] [CrossRef]
- Jijiwa, M.; Demir, H.; Gupta, S.; Leung, C.; Joshi, K.; Orozco, N.; Huang, T.; Yildiz, V.O.; Shibahara, I.; de Jesus, J.A.; et al. CD44v6 regulates growth of brain tumor stem cells partially through the AKT-mediated pathway. PLoS ONE 2011, 6, e24217. [Google Scholar] [CrossRef]
- Inoue, A.; Ohnishi, T.; Nishikawa, M.; Ohtsuka, Y.; Kusakabe, K.; Yano, H.; Tanaka, J.; Kunieda, T. A Narrative Review on CD44’s Role in Glioblastoma Invasion, Proliferation, and Tumor Recurrence. Cancers 2023, 15, 4898. [Google Scholar] [CrossRef]
- Yin, J.; Zhang, H.; Wu, X.; Zhang, Y.; Li, J.; Shen, J.; Zhao, Y.; Xiao, Z.; Lu, L.; Huang, C.; et al. CD44 inhibition attenuates EGFR signaling and enhances cisplatin sensitivity in human EGFR wild-type non-small-cell lung cancer cells. Int. J. Mol. Med. 2020, 45, 1783–1792. [Google Scholar] [CrossRef] [PubMed]
- Filppu, P.; Tanjore Ramanathan, J.; Granberg, K.J.; Gucciardo, E.; Haapasalo, H.; Lehti, K.; Nykter, M.; Le Joncour, V.; Laakkonen, P. CD109-GP130 interaction drives glioblastoma stem cell plasticity and chemoresistance through STAT3 activity. JCI Insight 2021, 6, e141486. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.M.; Murakumo, Y.; Hagiwara, S.; Jiang, P.; Mii, S.; Kalyoncu, E.; Saito, S.; Suzuki, C.; Sakurai, Y.; Numata, Y.; et al. CD109 attenuates TGF-beta1 signaling and enhances EGF signaling in SK-MG-1 human glioblastoma cells. Biochem. Biophys. Res. Commun. 2015, 459, 252–258. [Google Scholar] [CrossRef]
- Shiraki, Y.; Mii, S.; Enomoto, A.; Momota, H.; Han, Y.P.; Kato, T.; Ushida, K.; Kato, A.; Asai, N.; Murakumo, Y.; et al. Significance of perivascular tumour cells defined by CD109 expression in progression of glioma. J. Pathol. 2017, 243, 468–480. [Google Scholar] [CrossRef]
- Mo, X.T.; Leung, T.H.; Tang, H.W.; Siu, M.K.; Wan, P.K.; Chan, K.K.; Cheung, A.N.; Ngan, H.Y. CD109 mediates tumorigenicity and cancer aggressiveness via regulation of EGFR and STAT3 signalling in cervical squamous cell carcinoma. Br. J. Cancer 2020, 123, 833–843. [Google Scholar] [CrossRef]
- Zhou, S.; Hassan, A.; Kungyal, T.; Tabaries, S.; Luna, J.; Siegel, P.M.; Philip, A. CD109 Is a Critical Determinant of EGFR Expression and Signaling, and Tumorigenicity in Squamous Cell Carcinoma Cells. Cancers 2022, 14, 3672. [Google Scholar] [CrossRef]
- Khan, T.; Seddon, A.M.; Khelwatty, S.A.; Dalgleish, A.; Bagwan, I.; Mudan, S.; Modjtahedi, H. The Co-expression of HER Family Members and CD109 is common in Pancreatic Cancer. Med. Res. Arch. 2023, 11. [Google Scholar] [CrossRef]
- Arias-Pinilla, G.A.; Dalgleish, A.G.; Mudan, S.; Bagwan, I.; Walker, A.J.; Modjtahedi, H. Development of novel monoclonal antibodies against CD109 overexpressed in human pancreatic cancer. Oncotarget 2018, 9, 19994–20007. [Google Scholar] [CrossRef]
- Khelwatty, S.A.; Puvanenthiran, S.; Essapen, S.; Bagwan, I.; Seddon, A.M.; Modjtahedi, H. HER2 Expression Is Predictive of Survival in Cetuximab Treated Patients with RAS Wild Type Metastatic Colorectal Cancer. Cancers 2021, 13, 638. [Google Scholar] [CrossRef] [PubMed]
- Puvanenthiran, S.; Essapen, S.; Haagsma, B.; Bagwan, I.; Green, M.; Khelwatty, S.A.; Seddon, A.; Modjtahedi, H. Co-expression and prognostic significance of the HER family members, EGFRvIII, c-MET, CD44 in patients with ovarian cancer. Oncotarget 2018, 9, 19662–19674. [Google Scholar] [CrossRef]
- Sergina, N.V.; Moasser, M.M. The HER family and cancer: Emerging molecular mechanisms and therapeutic targets. Trends Mol. Med. 2007, 13, 527–534. [Google Scholar] [CrossRef]
- Roskoski, R., Jr. The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol. Res. 2014, 79, 34–74. [Google Scholar] [CrossRef]
- Connell, C.M.; Doherty, G.J. Activating HER2 mutations as emerging targets in multiple solid cancers. ESMO Open 2017, 2, e000279. [Google Scholar] [CrossRef] [PubMed]
- Pitcher, J.L.; Alexander, N.; Miranda, P.J.; Johns, T.G. ErbB4 in the brain: Focus on high grade glioma. Front. Oncol. 2022, 12, 983514. [Google Scholar] [CrossRef]
- Gao, L.; Zhang, Y.; Feng, M.; Shen, M.; Yang, L.; Wei, B.; Zhou, Y.; Zhang, Z. HER3: Updates and current biology function, targeted therapy and pathologic detecting methods. Life Sci. 2024, 357, 123087. [Google Scholar] [CrossRef] [PubMed]
- Modjtahedi, H.; Ali, S.; Essapen, S. Therapeutic application of monoclonal antibodies in cancer: Advances and challenges. Br. Med. Bull. 2012, 104, 41–59. [Google Scholar] [CrossRef]
- Hsu, J.L.; Hung, M.C. The role of HER2, EGFR, and other receptor tyrosine kinases in breast cancer. Cancer Metastasis Rev. 2016, 35, 575–588. [Google Scholar] [CrossRef]
- Keating, G.M. Afatinib: A Review in Advanced Non-Small Cell Lung Cancer. Target. Oncol. 2016, 11, 825–835. [Google Scholar] [CrossRef]
- Levantini, E.; Maroni, G.; Del Re, M.; Tenen, D.G. EGFR signaling pathway as therapeutic target in human cancers. Semin. Cancer Biol. 2022, 85, 253–275. [Google Scholar] [CrossRef] [PubMed]
- Swain, S.M.; Shastry, M.; Hamilton, E. Targeting HER2-positive breast cancer: Advances and future directions. Nat. Rev. Drug Discov. 2023, 22, 101–126. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.; Yang, X.; Tai, H.; Zhong, X.; Luo, T.; Zheng, H. HER2-targeted therapies in cancer: A systematic review. Biomark. Res. 2024, 12, 16. [Google Scholar] [CrossRef]
- Dunn, G.P.; Rinne, M.L.; Wykosky, J.; Genovese, G.; Quayle, S.N.; Dunn, I.F.; Agarwalla, P.K.; Chheda, M.G.; Campos, B.; Wang, A.; et al. Emerging insights into the molecular and cellular basis of glioblastoma. Genes. Dev. 2012, 26, 756–784. [Google Scholar] [CrossRef] [PubMed]
- Vanderbeek, A.M.; Rahman, R.; Fell, G.; Ventz, S.; Chen, T.; Redd, R.; Parmigiani, G.; Cloughesy, T.F.; Wen, P.Y.; Trippa, L.; et al. The clinical trials landscape for glioblastoma: Is it adequate to develop new treatments? Neuro Oncol. 2018, 20, 1034–1043. [Google Scholar] [CrossRef]
- Harrison, R.A.; Anderson, M.D.; Cachia, D.; Kamiya-Matsuoka, C.; Weathers, S.S.; O’Brien, B.J.; Penas-Prado, M.; Yung, W.K.A.; Wu, J.; Yuan, Y.; et al. Clinical trial participation of patients with glioblastoma at The University of Texas MD Anderson Cancer Center. Eur. J. Cancer 2019, 112, 83–93. [Google Scholar] [CrossRef]
- Aldaz, P.; Arozarena, I. Tyrosine kinase inhibitors in adult glioblastoma: An (un) closed chapter? Cancers 2021, 13, 5799. [Google Scholar] [CrossRef]
- Bagley, S.J.; Kothari, S.; Rahman, R.; Lee, E.Q.; Dunn, G.P.; Galanis, E.; Chang, S.M.; Nabors, L.B.; Ahluwalia, M.S.; Stupp, R.; et al. Glioblastoma Clinical Trials: Current Landscape and Opportunities for Improvement. Clin. Cancer Res. 2022, 28, 594–602. [Google Scholar] [CrossRef]
- Lauko, A.; Lo, A.; Ahluwalia, M.S.; Lathia, J.D. Cancer cell heterogeneity & plasticity in glioblastoma and brain tumors. Semin. Cancer Biol. 2022, 82, 162–175. [Google Scholar] [CrossRef]
- Shah, H.A.; Mishra, A.; Gouzoulis, M.J.; Ben-Shalom, N.; D’Amico, R.S. Analysis of factors leading to early termination in glioblastoma-related clinical trials. J. Neurooncol. 2022, 158, 489–495. [Google Scholar] [CrossRef]
- Orian-Rousseau, V. CD44 Acts as a Signaling Platform Controlling Tumor Progression and Metastasis. Front. Immunol. 2015, 6, 154. [Google Scholar] [CrossRef]
- Hassn Mesrati, M.; Syafruddin, S.E.; Mohtar, M.A.; Syahir, A. CD44: A Multifunctional Mediator of Cancer Progression. Biomolecules 2021, 11, 1850. [Google Scholar] [CrossRef]
- Varela, M.; Ranuncolo, S.M.; Morand, A.; Lastiri, J.; De Kier Joffe, E.B.; Puricelli, L.I.; Pallotta, M.G. EGF-R and PDGF-R, but not bcl-2, overexpression predict overall survival in patients with low-grade astrocytomas. J. Surg. Oncol. 2004, 86, 34–40. [Google Scholar] [CrossRef]
- Chakravarti, A.; Seiferheld, W.; Tu, X.; Wang, H.; Zhang, H.Z.; Ang, K.K.; Hammond, E.; Curran, W., Jr.; Mehta, M. Immunohistochemically determined total epidermal growth factor receptor levels not of prognostic value in newly diagnosed glioblastoma multiforme: Report from the Radiation Therapy Oncology Group. Int. J. Radiat. Oncol. Biol. Phys. 2005, 62, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Song, Y.J.; Lee, H.S.; Hur, W.J.; Sung, K.H.; Kim, K.U.; Choi, S.S.; Kim, S.J.; Kim, D.C. Epidermal growth factor receptor is related to poor survival in glioblastomas: Single-institution experience. Yonsei Med. J. 2013, 54, 101–107. [Google Scholar] [CrossRef]
- Hobbs, J.; Nikiforova, M.N.; Fardo, D.W.; Bortoluzzi, S.; Cieply, K.; Hamilton, R.L.; Horbinski, C. Paradoxical relationship between the degree of EGFR amplification and outcome in glioblastomas. Am. J. Surg. Pathol. 2012, 36, 1186–1193. [Google Scholar] [CrossRef] [PubMed]
- Abdulghani, M.M.; Abbas, M.N.; Mohammed, W.R. Immunohistochemical Expression of Epidermal Growth Factor Receptor in Astrocytic Tumors in Iraqi Patients. Open Access Maced. J. Med. Sci. 2019, 7, 3514–3520. [Google Scholar] [CrossRef] [PubMed]
- Han, W.; Lo, H.W. Landscape of EGFR signaling network in human cancers: Biology and therapeutic response in relation to receptor subcellular locations. Cancer Lett. 2012, 318, 124–134. [Google Scholar] [CrossRef]
- Brand, T.M.; Iida, M.; Luthar, N.; Starr, M.M.; Huppert, E.J.; Wheeler, D.L. Nuclear EGFR as a molecular target in cancer. Radiother. Oncol. 2013, 108, 370–377. [Google Scholar] [CrossRef]
- Sgosti, R.M.; Leuthold, M.; Gullick, W.J.; Yasargil, M.G.; Wiestler, O.D. Expression of the epidermal growth factor receptor in astrocytic tumours is specifically associated with glioblastoma multiforme. Virchows Arch. A Pathol. Anat. Histopathol. 1992, 420, 321–325. [Google Scholar]
- Zhu, A.; Shaeffer, J.; Leslie, S.; Kolm, P.; El-Mahdi, A.M. Epidermal growth factor receptor: An independent predictor of survival in astrocytic tumors given definitive irradiation. Int. J. Radiat. Oncol. Biol. Phys. 1996, 34, 809–815. [Google Scholar] [PubMed]
- Korshunov, A.; Golanov, A.; Timirgaz, V. Immunohistochemical markers for intracranial ependymoma recurrence. An analysis of 88 cases. J. Neurol. Sci. 2000, 177, 72–82. [Google Scholar]
- Andersson, U.; Guo, D.; Malmer, B.; Bergenheim, A.T.; Brännström, T.; Hedman, H.; Henriksson, R. Epidermal growth factor receptor family (EGFR, ErbB2-4) in gliomas and meningiomas. Acta Neuropathol. 2004, 108, 135–142. [Google Scholar] [PubMed]
- Nishikawa, R.; Sugiyama, T.; Narita, Y.; Furnari, F.; Cavenee, W.K.; Matsutani, M. Immunohistochemical analysis of the mutant epidermal growth factor, ΔEGFR, in glioblastoma. Brain Tumor Pathol. 2004, 21, 53–56. [Google Scholar] [PubMed]
- Heimberger, A.B.; Hlatky, R.; Suki, D.; Yang, D.; Weinberg, J.; Gilbert, M.; Sawaya, R.; Aldape, K. Prognostic effect of epidermal growth factor receptor and EGFRvIII in glioblastoma multiforme patients. Clin. Cancer Res. 2005, 11, 1462–1466. [Google Scholar]
- Mendrzyk, F.; Korshunov, A.; Benner, A.; Toedt, G.; Pfister, S.; Radlwimmer, B.; Lichter, P. Identification of gains on 1q and epidermal growth factor receptor overexpression as independent prognostic markers in intracranial ependymoma. Clin. Cancer Res. 2006, 12 Pt 1, 2070–2079. [Google Scholar]
- Umesh, S.; Tandon, A.; Santosh, V.; Anandh, B.; Sampath, S.; Chandramouli, B.A.; Sastry Kolluri, V.R. Clinical and immunohistochemical prognostic factors in adult glioblastoma patients. Clin. Neuropathol. 2009, 28, 362–372. [Google Scholar]
- Nabika, S.; Kiya, K.; Satoh, H.; Mizoue, T.; Kondo, H.; Katagiri, M.; Nishisaka, T.; Sugiyama, K.; Kurisu, K. Prognostic significance of expression patterns of EGFR family, p21 and p27 in high-grade astrocytoma. Hiroshima J. Med. Sci. 2010, 59, 65–70. [Google Scholar]
- Senetta, R.; Miracco, C.; Lanzafame, S.; Chiusa, L.; Caltabiano, R.; Galia, A.; Stella, G.; Cassoni, P. Epidermal growth factor receptor and caveolin-1 coexpression identifies adult supratentorial ependymomas with rapid unfavorable outcomes. Neuro Oncol. 2011, 13, 176–183. [Google Scholar]
- Lee, K.S.; Choe, G.; Nam, K.H.; Seo, A.N.; Yun, S.; Kim, K.J.; Cho, H.J.; Park, S.H. Immunohistochemical classification of primary and secondary glioblastomas. Korean J. Pathol. 2013, 47, 541–548. [Google Scholar]
- Michaelsen, S.R.; Christensen, I.J.; Grunnet, K.; Stockhausen, M.T.; Broholm, H.; Kosteljanetz, M.; Poulsen, H.S. Clinical variables serve as prognostic factors in a model for survival from glioblastoma multiforme: An observational study of a cohort of consecutive non-selected patients from a single institution. BMC Cancer 2013, 13, 402. [Google Scholar] [CrossRef]
- Saha, R.; Chatterjee, U.; Mandal, S.; Saha, K.; Chatterjee, S.; Ghosh, S.N. Expression of phosphatase and tensin homolog, epidermal growth factor receptor, and Ki-67 in astrocytoma: A prospective study in a tertiary care hospital. Indian. J. Med. Paediatr. Oncol. 2014, 35, 149–155. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Montgomery, R.M.; Queiroz Lde, S.; Rogerio, F. EGFR, p53, IDH-1 and MDM2 immunohistochemical analysis in glioblastoma: Therapeutic and prognostic correlation. Arq. Neuropsiquiatr. 2015, 73, 561–568. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tini, P.; Cerase, A.; Cevenini, G.; Carbone, S.F.; Miracco, C.; Pirtoli, L. Epidermal Growth Factor Receptor Expression May Correlate with Survival Through Clinical and Radiological Features of Aggressiveness in Glioblastoma Treated with Radiochemotherapy. Anticancer. Res. 2015, 35, 4117–4124. [Google Scholar]
- Tripathy, K.; Das, B.; Singh, A.K.; Misra, A.; Misra, S.; Misra, S.S. Prognostic Significance of Epidermal Growth Factor Receptor in Patients of Glioblastoma Multiforme. J. Clin. Diagn. Res. 2017, 11, EC05–EC8. [Google Scholar] [CrossRef]
- Amirpour, Z.; Bahari, A.; Nafisi, B.; Rahmani, K.; Taghipour Zahir, S. Prognosis and survival study in patients with glioblastoma multiform and its relationship with EGFR expression. Iran. J. Neurosurg. 2020, 6, 113–120. [Google Scholar] [CrossRef]
- Miratashi Yazdi, S.A.; Bakhshi, N.; Nazar, E.; Tabriz, H.M.; Gorji, R. Epidermal growth factor receptor (EGFR) expression in high grade glioma and relationship with histopathologic findings, a cross sectional study. Int. J. Surg. Open 2022, 46, 100527. [Google Scholar] [CrossRef]
- Shimozaki, K.; Fukuoka, S.; Ooki, A.; Yamaguchi, K. HER2-low gastric cancer: Is the subgroup targetable? ESMO Open 2024, 9, 103679. [Google Scholar] [CrossRef]
- Ramezani, M.; Siami, S.; Rezaei, M.; Khazaei, S.; Sadeghi, M. An immunohistochemical study of HER2 expression in primary brain tumors. Biomedicine 2020, 10, 21–27. [Google Scholar] [CrossRef]
- Meurer, R.T.; Martins, D.T.; Hilbig, A.; Ribeiro Mde, C.; Roehe, A.V.; Barbosa-Coutinho, L.M.; Fernandes Mda, C. Immunohistochemical expression of markers Ki-67, neun, synaptophysin, p53 and HER2 in medulloblastoma and its correlation with clinicopathological parameters. Arq. Neuropsiquiatr. 2008, 66, 385–390. [Google Scholar] [CrossRef]
- Mineo, J.F.; Bordron, A.; Baroncini, M.; Maurage, C.A.; Ramirez, C.; Siminski, R.M.; Berthou, C.; Dam Hieu, P. Low HER2-expressing glioblastomas are more often secondary to anaplastic transformation of low-grade glioma. J. Neurooncol. 2007, 85, 281–287. [Google Scholar] [CrossRef] [PubMed]
- Haapasalo, H.; Hyytinen, E.; Sallinen, P.; Helin, H.; Kallioniemi, O.P.; Isola, J. c-erbB-2 in astrocytomas: Infrequent overexpression by immunohistochemistry and absence of gene amplification by fluorescence in situ hybridization. Br. J. Cancer 1996, 73, 620–623. [Google Scholar]
- Koka, V.; Potti, A.; Forseen, S.E.; Pervez, H.; Fraiman, G.N.; Koch, M.; Levitt, R. Role of Her-2/neu overexpression and clinical determinants of early mortality in glioblastoma multiforme. Am. J. Clin. Oncol. 2003, 26, 332–335. [Google Scholar] [PubMed]
- Donoghue, J.F.; Kerr, L.T.; Alexander, N.W.; Greenall, S.A.; Longano, A.B.; Gottardo, N.G.; Wang, R.; Tabar, V.; Adams, T.E.; Mischel, P.S.; et al. Activation of ERBB4 in Glioblastoma Can Contribute to Increased Tumorigenicity and Influence Therapeutic Response. Cancers 2018, 10, 243. [Google Scholar] [CrossRef]
- Arnli, M.B.; Meta, R.; Lydersen, S.; Torp, S.H. HER3 and HER4 are highly expressed in human meningiomas. Pathol. Res. Pract. 2019, 215, 152551. [Google Scholar]
- Kusuhara, S.; Kogawa, T.; Shimokawa, M.; Funasaka, C.; Kondoh, C.; Harano, K.; Matsubara, N.; Naito, Y.; Hosono, A.; Satomi, K. 264P Increased membrane HER3 expression in brain metastases compared to primary tumors in breast cancer. Ann. Oncol. 2022, 33, S658. [Google Scholar]
- Tomasich, E.; Steindl, A.; Paiato, C.; Hatziioannou, T.; Kleinberger, M.; Berchtold, L.; Puhr, R.; Hainfellner, J.A.; Mullauer, L.; Widhalm, G.; et al. Frequent Overexpression of HER3 in Brain Metastases from Breast and Lung Cancer. Clin. Cancer Res. 2023, 29, 3225–3236. [Google Scholar] [CrossRef]
- Kabraji, S.; Lin, N.U. Keeping It in the Family: HER3 as a Target in Brain Metastases. Clin. Cancer Res. 2023, 29, 2961–2963. [Google Scholar] [CrossRef] [PubMed]
- Majumder, A. HER3: Toward the Prognostic Significance, Therapeutic Potential, Current Challenges, and Future Therapeutics in Different Types of Cancer. Cells 2023, 12, 2517. [Google Scholar] [CrossRef]
- Uliano, J.; Corvaja, C.; Curigliano, G.; Tarantino, P. Targeting HER3 for cancer treatment: A new horizon for an old target. ESMO Open 2023, 8, 100790. [Google Scholar] [CrossRef]
- Moscatello, D.K.; Montgomery, R.B.; Sundareshan, P.; McDanel, H.; Wong, M.Y.; Wong, A.J. Transformational and altered signal transduction by a naturally occurring mutant EGF receptor. Oncogene 1996, 13, 85–96. [Google Scholar] [PubMed]
- Gan, H.K.; Cvrljevic, A.N.; Johns, T.G. The epidermal growth factor receptor variant III (EGFRvIII): Where wild things are altered. FEBS J. 2013, 280, 5350–5370. [Google Scholar] [CrossRef] [PubMed]
- Binder, D.C.; Ladomersky, E.; Lenzen, A.; Zhai, L.; Lauing, K.L.; Otto-Meyer, S.D.; Lukas, R.V.; Wainwright, D.A. Lessons learned from rindopepimut treatment in patients with EGFRvIII-expressing glioblastoma. Transl. Cancer Res. 2018, 7, S510–S513. [Google Scholar] [CrossRef]
- Iurlaro, R.; Waldhauer, I.; Planas-Rigol, E.; Bonfill-Teixidor, E.; Arias, A.; Nicolini, V.; Freimoser-Grundschober, A.; Cuartas, I.; Martinez-Moreno, A.; Martinez-Ricarte, F.; et al. A Novel EGFRvIII T-Cell Bispecific Antibody for the Treatment of Glioblastoma. Mol. Cancer Ther. 2022, 21, 1499–1509. [Google Scholar] [CrossRef]
- Rutkowska, A.; Strozik, T.; Jedrychowska-Danska, K.; Zamerska, A.; Jesionek-Kupnicka, D.; Kowalczyk, T.; Och, W.; Szostak, B.; Treda, C.; Wlodarczyk, A.; et al. Immunohistochemical detection of EGFRvIII in glioblastoma—Anti-EGFRvIII antibody validation for diagnostic and CAR-T purposes. Biochem. Biophys. Res. Commun. 2023, 685, 149133. [Google Scholar] [CrossRef] [PubMed]
- Pelloski, C.E.; Ballman, K.V.; Furth, A.F.; Zhang, L.; Lin, E.; Sulman, E.P.; Bhat, K.; McDonald, J.M.; Yung, W.K.; Colman, H.; et al. Epidermal growth factor receptor variant III status defines clinically distinct subtypes of glioblastoma. J. Clin. Oncol. 2007, 25, 2288–2294. [Google Scholar] [CrossRef]
- Rosenthal, M.; Curry, R.; Reardon, D.A.; Rasmussen, E.; Upreti, V.V.; Damore, M.A.; Henary, H.A.; Hill, J.S.; Cloughesy, T. Safety, tolerability, and pharmacokinetics of anti-EGFRvIII antibody-drug conjugate AMG 595 in patients with recurrent malignant glioma expressing EGFRvIII. Cancer Chemother. Pharmacol. 2019, 84, 327–336. [Google Scholar] [CrossRef]
- Schmidts, A.; Srivastava, A.A.; Ramapriyan, R.; Bailey, S.R.; Bouffard, A.A.; Cahill, D.P.; Carter, B.S.; Curry, W.T.; Dunn, G.P.; Frigault, M.J.; et al. Tandem chimeric antigen receptor (CAR) T cells targeting EGFRvIII and IL-13Ralpha2 are effective against heterogeneous glioblastoma. Neurooncol. Adv. 2023, 5, vdac185. [Google Scholar] [CrossRef]
- Bagley, S.J.; Binder, Z.A.; Lamrani, L.; Marinari, E.; Desai, A.S.; Nasrallah, M.P.; Maloney, E.; Brem, S.; Lustig, R.A.; Kurtz, G.; et al. Repeated peripheral infusions of anti-EGFRvIII CAR T cells in combination with pembrolizumab show no efficacy in glioblastoma: A phase 1 trial. Nat. Cancer 2024, 5, 517–531. [Google Scholar] [CrossRef]
- Shinojima, N.; Tada, K.; Shiraishi, S.; Kamiryo, T.; Kochi, M.; Nakamura, H.; Makino, K.; Saya, H.; Hirano, H.; Kuratsu, J.I.; et al. Prognostic value of epidermal growth factor receptor in patients with glioblastoma multiforme. Cancer Res. 2003, 63, 6962–6970. [Google Scholar]
- Aldape, K.D.; Ballman, K.; Furth, A.; Buckner, J.C.; Giannini, C.; Burger, P.C.; Scheithauer, B.W.; Jenkins, R.B.; James, C.D. Immunohistochemical detection of EGFRvIII in high malignancy grade astrocytomas and evaluation of prognostic significance. J. Neuropathol. Exp. Neurol. 2004, 63, 700–707. [Google Scholar]
- Nozawa, T.; Okada, M.; Natsumeda, M.; Eda, T.; Abe, H.; Tsukamoto, Y.; Okamoto, K.; Oishi, M.; Takahashi, H.; Fujii, Y.; et al. EGFRvIII Is Expressed in Cellular Areas of Tumor in a Subset of Glioblastoma. Neurol. Med. Chir. 2019, 59, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Bastola, S.; Pavlyukov, M.S.; Yamashita, D.; Ghosh, S.; Cho, H.; Kagaya, N.; Zhang, Z.; Minata, M.; Lee, Y.; Sadahiro, H.; et al. Glioma-initiating cells at tumor edge gain signals from tumor core cells to promote their malignancy. Nat. Commun. 2020, 11, 4660. [Google Scholar] [CrossRef] [PubMed]
- Thapa, R.; Wilson, G.D. The Importance of CD44 as a Stem Cell Biomarker and Therapeutic Target in Cancer. Stem Cells Int. 2016, 2016, 2087204. [Google Scholar] [CrossRef] [PubMed]
- Si, D.; Yin, F.; Peng, J.; Zhang, G. High Expression of CD44 Predicts a Poor Prognosis in Glioblastomas. Cancer Manag. Res. 2020, 12, 769–775. [Google Scholar] [CrossRef]
- Huang, H.H.; Wang, Y.C.; Chou, Y.C.; Yu, M.H.; Chao, T.K. The combination of aldehyde dehydrogenase 1 (ALDH1) and CD44 is associated with poor outcomes in endometrial cancer. PLoS ONE 2018, 13, e0206685. [Google Scholar] [CrossRef]
- Karan Krizanac, D.; Krasic Arapovic, A.; Skocibusic, S.; Pintaric, I.; Trgo, G.; Tomic, S. CD44 Immunoexpression is Unfavorable Predictor in Ovarian Serous Cancer. Appl. Immunohistochem. Mol. Morphol. 2018, 26, 398–402. [Google Scholar] [CrossRef]
- Sherif, O.; Khelwatty, S.A.; Bagwan, I.; Seddon, A.M.; Dalgleish, A.; Mudan, S.; Modjtahedi, H. Expression of EGFRvIII and its co-expression with wild-type EGFR, or putative cancer stem cell biomarkers CD44 or EpCAM are associated with poorer prognosis in patients with hepatocellular carcinoma. Oncol. Rep. 2024, 52, 172. [Google Scholar] [CrossRef]
- Zhou, S.; da Silva, S.D.; Siegel, P.M.; Philip, A. CD109 acts as a gatekeeper of the epithelial trait by suppressing epithelial to mesenchymal transition in squamous cell carcinoma cells in vitro. Sci. Rep. 2019, 9, 16317. [Google Scholar] [CrossRef]
- Kim, J.S.; Shin, M.J.; Lee, S.Y.; Kim, D.K.; Choi, K.U.; Suh, D.S.; Kim, D.; Kim, J.H. CD109 Promotes Drug Resistance in A2780 Ovarian Cancer Cells by Regulating the STAT3-NOTCH1 Signaling Axis. Int. J. Mol. Sci. 2023, 24, 10306. [Google Scholar] [CrossRef]
- Minata, M.; Audia, A.; Shi, J.; Lu, S.; Bernstock, J.; Pavlyukov, M.S.; Das, A.; Kim, S.H.; Shin, Y.J.; Lee, Y.; et al. Phenotypic Plasticity of Invasive Edge Glioma Stem-like Cells in Response to Ionizing Radiation. Cell Rep. 2019, 26, 1893–1905.e7. [Google Scholar] [CrossRef] [PubMed]
- Liffers, K.; Lamszus, K.; Schulte, A. EGFR Amplification and Glioblastoma Stem-Like Cells. Stem Cells Int. 2015, 2015, 427518. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.H.; Liao, C.C.; Chow, N.H.; Huang, L.L.; Chuang, J.I.; Wei, K.C.; Shin, J.W. Whether CD44 is an applicable marker for glioma stem cells. Am. J. Transl. Res. 2017, 9, 4785–4806. [Google Scholar]
- Bodey, B.; Kaiser, H.E.; Siegel, S.E. Epidermal growth factor receptor (EGFR) expression in childhood brain tumors. In Vivo 2005, 19, 931–941. [Google Scholar]
- Al-Janaby, T.; Khelwatty SBagwan, I.; Abbassi-Ghandi Modjahedi, H. The co-expression and prognostic significance of the HER-family members, EGFRvIII, CD44, CD109 and Claudin 18.2 in patients with adenocarcinoma. In Proceedings of the AACR, Chicago, IL, USA, 25–30 April 2025; p. 3341. [Google Scholar]
- Westphal, M.; Maire, C.L.; Lamszus, K. EGFR as a Target for Glioblastoma Treatment: An Unfulfilled Promise. CNS Drugs 2017, 31, 723–735. [Google Scholar] [CrossRef] [PubMed]
- Rabah, N.; Ait Mohand, F.E.; Kravchenko-Balasha, N. Understanding Glioblastoma Signaling, Heterogeneity, Invasiveness, and Drug Delivery Barriers. Int. J. Mol. Sci. 2023, 24, 14256. [Google Scholar] [CrossRef]
- Ge, Z.; Xu, M.; Ge, Y.; Huang, G.; Chen, D.; Ye, X.; Xiao, Y.; Zhu, H.; Yin, R.; Shen, H.; et al. Inhibiting G6PD by quercetin promotes degradation of EGFR T790M mutation. Cell Rep. 2023, 42, 113417. [Google Scholar] [CrossRef]
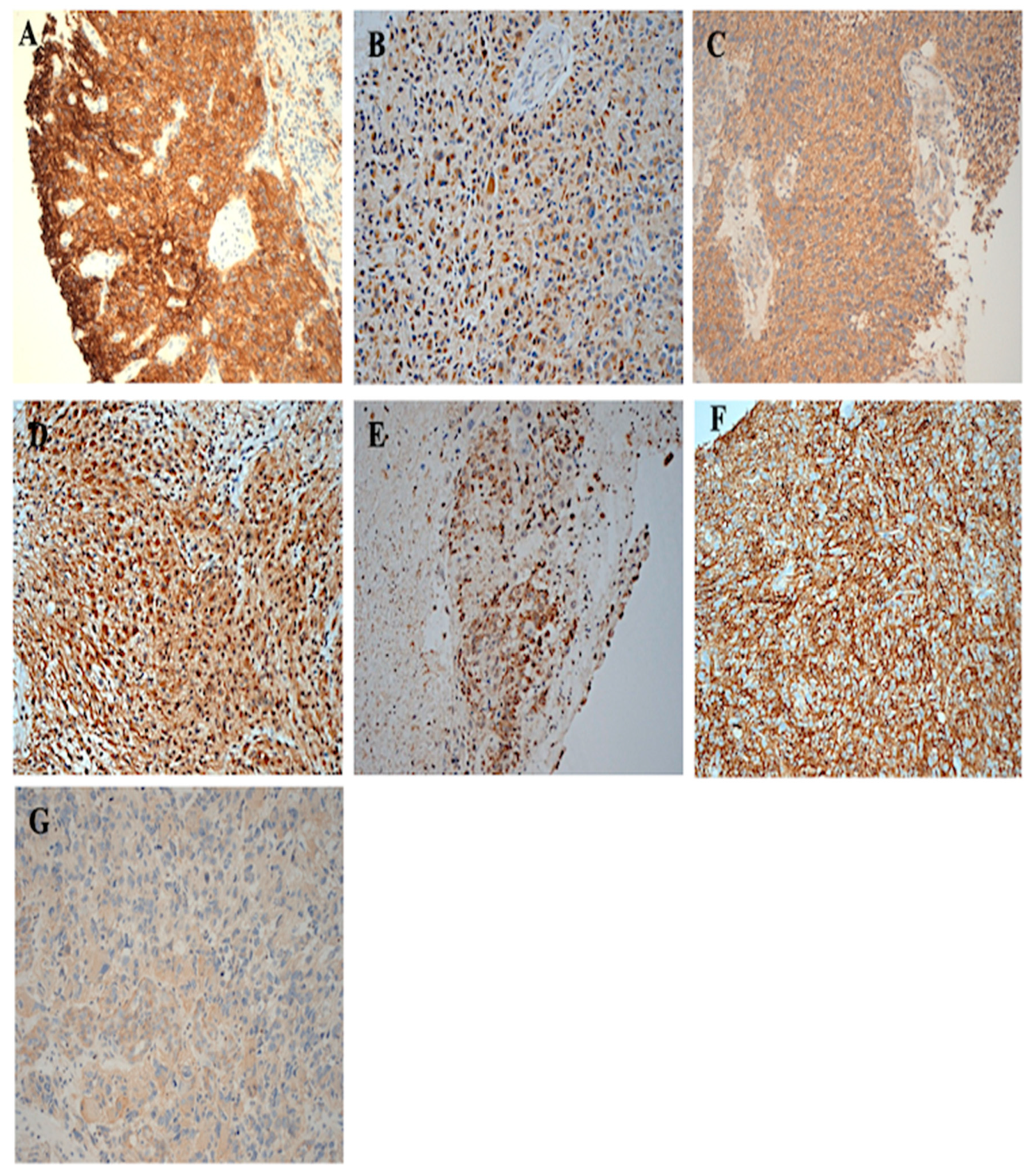
| Characteristics | Number of Patients (%) | Overall Survival in Years (Mean ± SE) | 95% CI | p-Value |
|---|---|---|---|---|
| Age | ||||
| <50 | 16 (20) | 3.43 ± 0.72 | 2.02–4.85 | <0.1 |
| ≥50 | 64 (80) | 1.67 ± 0.15 | 0.89–1.45 | |
| Gender | ||||
| Male | 49 (61.3) | 1.36 ± 0.29 | 1.06–2.21 | 0.848 |
| Female | 31 (38.8) | 1.61 ± 0.31 | 1.02–2.21 | |
| Location | ||||
| Frontal | 17 (21.3) | 1.18 ± 0.32 | 0.54–1.81 | 0.068 |
| Parietal | 19 (23.8) | 2.70 ± 0.65 | 1.42–3.96 | |
| Other | 44 (55) | 1.30 ± 0.92 | 0.92–1.67 | |
| Hemisphere | ||||
| Left | 39 (48.8) | 1.54 ± 0.30 | 0.96–2.12 | 0.191 |
| Right | 37 (46.3) | 1.83 ± 0.30 | 1.19–2.48 | |
| Both | 4 (5.0) | 0.50 ± 0.50 | 0–1.48 | |
| O6-methylguanine-DNA methyl-transferase (MGMT) | ||||
| Present | 41 (51.2) | 1.94 ± 0.32 | 1.33–2.56 | 0.066 |
| Absent | 39 (48.8) | 1.31 ± 0.28 | 0.76–1.85 | |
| Isocitrate dehydrogenase (IDH) | ||||
| Present | 2 (2.5) | 3.00 ± 0.72 | 1.16–4.83 | 0.091 |
| Absent | 78 (97.5) | 1.153 ± 0.208 | 0.208–1.14 | |
| Scoring Criteria | Wt-EGFR | HER2 | HER3 | HER4 | EGFRvIII | CD44 | CD109 |
|---|---|---|---|---|---|---|---|
| Cut-off value Number of positive (%) | |||||||
| ≥5% | 37 (46) | 60 (75) | 15 (19) | 57 (71) | 68 (85) | 76 (95) | 13 (16) |
| ≥10% | 32 (40) | 44 (55) | 14 (18) | 54 (68) | 47 (59) | 72 (90) | 13 (16) |
| ≥20% | 27 (34) | 35 (44) | 12 (15) | 45 (56) | 33 (41) | 66 (83) | 12 (15) |
| ≥50% | 19 (24) | 16 (20) | 3 (4) | 20 (25) | 3 (4) | 33 (41) | 6 (8) |
| Intensity | |||||||
| 1+ | 12 (15) | 58 (41) | 15 (19) | 43 (54) | 62 (78) | 53 (66) | 13 (16) |
| 2+ | 22 (27.5) | 13 (9) | 1 (1) | 18 (23) | 8 (10) | 45 (56) | 0 (0) |
| 3+ | 11 (8) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 13 (16) | 0 (0) |
| Sub-cellular localization | |||||||
| Membranous | 23 (29) | 4 (5) | 0 (0) | 16 (20) | 0 (0) | 76 (95) | 0 (0) |
| Cytoplasmic | 33 (41) | 60 (75) | 15 (18) | 40 (50) | 60 (75) | 1 (1) | 13 (16) |
| Nuclear | 1 (1) | 0 (0) | 0 (0) | 4 (15) | 10 (13) | 0 (0) | 1 (1.3) |
| Markers | Number of Positive Tumours (%) | |||
|---|---|---|---|---|
| ≥5% Cut Off | ≥10% Cut Off | ≥20% Cut Off | ≥50% Cut Off | |
| wtEGFR/HER2 | 28 (35) | 18 (23) | 10 (13) | 5 (6) |
| wtEGFR/HER3 | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| wtEGFR/HER4 | 24 (30) | 24 (30) | 14 (18) | 4 (5) |
| WtEGFR/EGFRvIII | 29 (36) | 14 (18) | 12 (15) | 0 (0) |
| wtEGFR/CD44 | 35 (44) | 30 (38) | 25 (31) | 10 (13) |
| wtEGFR/CD109 | 3 (4) | 2 (3) | 1 (1) | 1 (1) |
| wtEGFR/HER2/HER3 | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| wtEGFR/HER2/HER4 | 23 (29) | 16 (20) | 8 (10) | 2 (3) |
| WtEGFR/HER3/HER4 | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| wtEGFR/HER2/EGFRvIII | 22 (28) | 11 (14) | 5 (6) | 0 (0) |
| wtEGFR/HER2/CD44 | 25 (31) | 17 (21) | 10 (13) | 2 (3) |
| wtEGFR/HER2/CD109 | 3 (4) | 0 (0) | 0 (0) | 0 (0) |
| wtEGFR/HER3/EGFRvIII | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| wtEGFR/HER3/CD44 | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| wtEGFR/HER3/CD109 | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| wtEGFR/HER4/EGFRvIII | 19 (24) | 11 (14) | 5 (6) | 0 (0) |
| wtEGFR/EGFRvIII/CD44 | 29 (36) | 16 (20) | 12 (15) | 0 (0) |
| wtEGFR/EGFRvIII/CD109 | 4 (5) | 2 (3) | 0 (0) | 0 (0) |
| wtEGFR/HER2/HER3/HER4 | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| HER2/HER3 | 13 (16) | 11 (14) | 7 (9) | 1 (1) |
| HER2/HER4 | 50 (63) | 35 (44) | 24 (30) | 6 (8) |
| HER2/EGFRvIII | 52 (65) | 29 (36) | 17 (21) | 0 (0) |
| HER2/CD44 | 57 (71) | 41 (51) | 29 (36) | 6 (8) |
| HER2/CD109 | 11 (14) | 9 (11) | 6 (8) | 2 (3) |
| HER2/HER3/HER4 | 12 (15) | 10 (13) | 5 (6) | 0 (0) |
| HER2/HER3/EGFRvIII | 11 (14) | 9 (11) | 4 (5) | 0 (0) |
| HER2/HER3/CD44 | 14 (85) | 11 (14) | 7 (9) | 0 (0) |
| HER2/HER3/CD109 | 5 (6) | 4 (5) | 3 (4) | 0 (0) |
| HER2/HER4/EGFRvIII | 45 (56) | 21 (26) | 11 (14) | 0 (0) |
| HER2/HER4/CD44 | 48 (60) | 33 (41) | 19 (24) | 3 (4) |
| HER2/EGFRvIII/CD44 | 52 (65) | 27 (34) | 15 (19) | 0 (0) |
| HER2/HER4/CD109 | 11 (14) | 8 (10) | 2 (3) | 0 (0) |
| HER2/EGFRvIII/CD109 | 12 (15) | 7 (9) | 5 (6) | 0 (0) |
| HER2/CD44/CD109 | 12 (15) | 10 (13) | 8 (10) | 0 (0) |
| HER2/HER4/EGFRVIII/CD44 | 44 (55) | 19 (24) | 8 (10) | 0 (0) |
| HER3/HER4 | 13 (16) | 12 (15) | 9 (1) | 0 (0) |
| HER3/EGFRvIII | 12 (15) | 11 (14) | 6 (8) | 0 (0) |
| HER3/CD44 | 13 (16) | 14 (18) | 12 (15) | 1 (1) |
| HER3/CD109 | 4 (5) | 4 (5) | 3 (4) | 0 (0) |
| HER3/HER4/EGFRVIII | 12 (15) | 10 (13) | 6 (8) | 0 (0) |
| HER3/HER4/CD109 | 4 (5) | 4 (5) | 2 (2) | 0 (0) |
| HER3/HER4/CD44 | 13 (16) | 12 (15) | 9 (11) | 0 (0) |
| HER3/EGFRvIII/CD44 | 13 (16) | 11 (14) | 9 (11) | 0 (0) |
| HER3/EGFRVIII/CD109 | 5 (6) | 5 (6) | 3 (4) | 0 (0) |
| HER3/CD44/CD109 | 5 (63) | 5 (6) | 3 (4) | 0 (0) |
| HER4/EGFRvIII | 50 (63) | 31 (39) | 17 (21) | 1 (1) |
| HER4/CD44 | 54 (68) | 49 (61) | 35 (44) | 8 (10) |
| HER4/CD109 | 12 (15) | 8 (10) | 8 (10) | 1 (1) |
| HER4/EGFRvIII/CD44 | 48 (60) | 29 (36) | 14 (18) | 0 (0) |
| HER4/EGFRvIII/CD109 | 11 (14) | 8 (10) | 4 (5) | 0 (0) |
| HER4/CD44/CD109 | 12 (15) | 10 (13) | 7 (9) | 0 (0) |
| EGFRvIII/CD44 | 66 (83) | 44 (55) | 29 (36) | 1 (1) |
| EGFRvIII/CD109 | 13 (16) | 11 (14) | 5 (6) | 0 (0) |
| EGFRvIII/CD44/CD109 | 13 (16) | 11 (14) | 4 (5) | 0 (0) |
| CD44/CD109 | 13 (16) | 13 (16) | 11 (14) | 0 (0) |
| Overall Survival (Years) | |||
|---|---|---|---|
| Receptor Expression | Negative | Positive | p-Value |
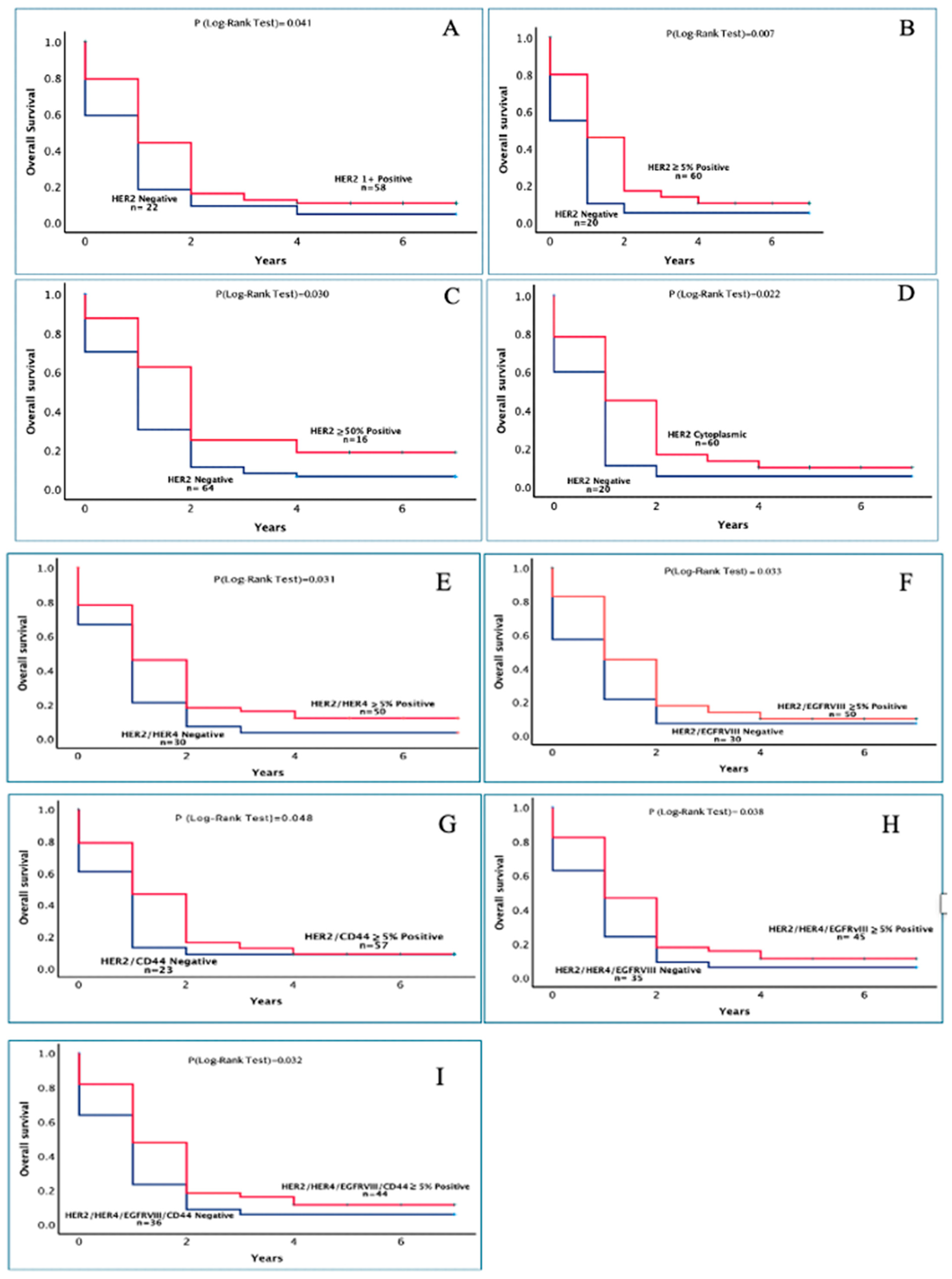
| HER2 Intensity 1+ | 1.09 ± 0.34 | 1.83 ± 0.26 | 0.041 |
| HER2 ≥ 5% | 0.90 ± 0.33 | 1.87 ± 0.25 | 0.7 |
| HER2 ≥ 50% | 1.39 ± 0.21 | 2.56 ± 0.57 | 0.030 |
| HER2 Cytoplasmic | 0.98 ± 0.35 | 1.83 ± 0.25 | 0.022 |
| HER2/HER4 ≥ 5% | 1.08 ± 0.25 | 1.94 ± 0.29 | 0.031 |
| HER2/EGFRVIII ≥ 5% | 1.43 ± 0.33 | 1.89 + 0.27 | 0.033 |
| HER2/CD44 ≥ 5% | 1.17 ± 0.39 | 1.81 ± 0.25 | 0.048 |
| HER2/HER4/EGFRVIII ≥ 5% | 1.19 ± 0.28 | 1.95 ± 0.30 | 0.038 |
| HER2/HER4/EGFRVIII/CD44 ≥ 5% | 1.19 ± 0.27 | 1.97 ± 0.27 | 0.032 |
| Expression in Years | Univariate | Multivariate | ||||
|---|---|---|---|---|---|---|
| Hazard Ratio | 95% CI | p-Value | Hazard Ratio | 95% CI | p-Value | |
| HER2 Intensity 1+ | 0.665 | 0.398–1.111 | 0.119 | 0.677 | 0.403–1.137 | 0.140 |
| HER2 ≥ 5% | 0.567 | 0.331–0.970 | 0.038 | 0.556 | 0.323–0.958 | 0.034 |
| HER2 ≥ 50% | 0.608 | 0.332–1.114 | 0.107 | 1.37 | 0.910–2.081 | 0.130 |
| HER2 Cytoplasmic | 0.612 | 0.354–1.056 | 0.078 | 0.650 | 0.373–1.132 | 0.128 |
| HER2/HER4 ≥ 5% | 0.669 | 0.414–1.083 | 0.102 | 0.706 | 0.433–1.151 | 0.162 |
| HER2/EGFRVIII ≥ 5% | 0.670 | 0.413–1.089 | 0.106 | 0.710 | 0.435–1.158 | 0.170 |
| HER2/CD44 ≥ 5% | 0.669 | 0.398–1.122 | 0.128 | 0.687 | 0.405–1.164 | 0.163 |
| HER2/HER4/EGFRVIII ≥ 5% | 0.686 | 0.686–1.098 | 0.116 | 0.718 | 0.445–1.156 | 0.173 |
| HER2/HER4/EGFRVIII/CD44 ≥ 5% | 0.678 | 0.424–1.085 | 0.105 | 0.702 | 0.436–1.133 | 0.147 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mulliqi, E.; Khelwatty, S.; Bagwan, I.; Kamaludin, A.; Morgan, A.; Long, N.; Ashkan, K.; Modjtahedi, H. The Co-Expression and Cellular Location of HER Family Members, EGFRvIII, Putative Cancer Stem Cell Biomarkers CD44 and CD109 in Patients with Glioblastoma, and Their Impacts on Prognosis. Cancers 2025, 17, 1221. https://doi.org/10.3390/cancers17071221
Mulliqi E, Khelwatty S, Bagwan I, Kamaludin A, Morgan A, Long N, Ashkan K, Modjtahedi H. The Co-Expression and Cellular Location of HER Family Members, EGFRvIII, Putative Cancer Stem Cell Biomarkers CD44 and CD109 in Patients with Glioblastoma, and Their Impacts on Prognosis. Cancers. 2025; 17(7):1221. https://doi.org/10.3390/cancers17071221
Chicago/Turabian StyleMulliqi, Ermira, Said Khelwatty, Izhar Bagwan, Ahmad Kamaludin, Anna Morgan, Natalie Long, Keyoumars Ashkan, and Helmout Modjtahedi. 2025. "The Co-Expression and Cellular Location of HER Family Members, EGFRvIII, Putative Cancer Stem Cell Biomarkers CD44 and CD109 in Patients with Glioblastoma, and Their Impacts on Prognosis" Cancers 17, no. 7: 1221. https://doi.org/10.3390/cancers17071221
APA StyleMulliqi, E., Khelwatty, S., Bagwan, I., Kamaludin, A., Morgan, A., Long, N., Ashkan, K., & Modjtahedi, H. (2025). The Co-Expression and Cellular Location of HER Family Members, EGFRvIII, Putative Cancer Stem Cell Biomarkers CD44 and CD109 in Patients with Glioblastoma, and Their Impacts on Prognosis. Cancers, 17(7), 1221. https://doi.org/10.3390/cancers17071221








