Comparing 68Ga-Pentixafor,18F-FDG PET/CT and Chemokine Receptor 4 Immunohistochemistry Staining in Breast Cancer: A Prospective Cross Sectional Study †
Simple Summary
Abstract
1. Introduction
2. Methods
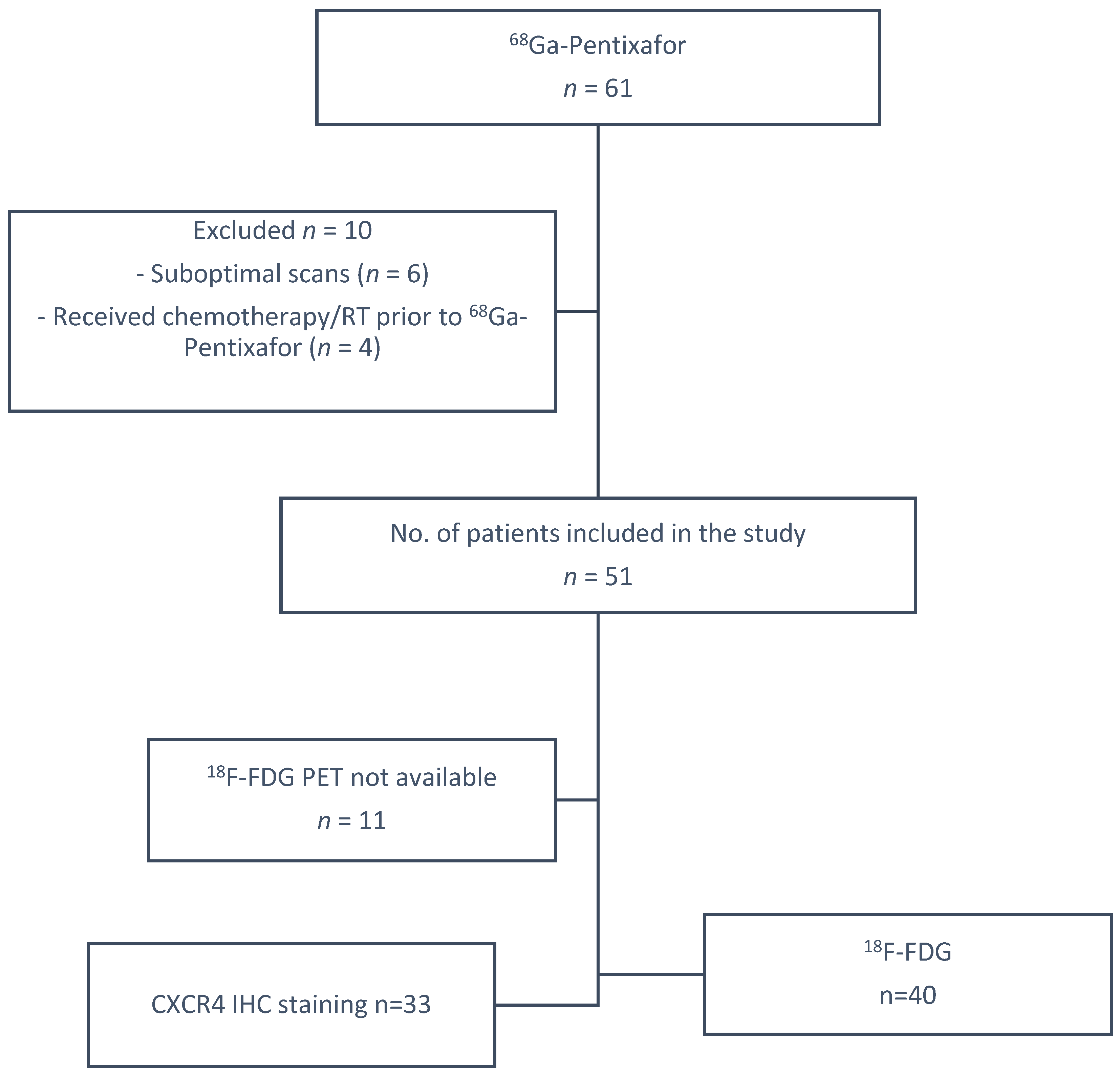
2.1. Patients
2.2. Radiochemistry of 68Ga-Pentixafor
2.3. 68Ga-Pentixafor PET/CT Imaging Procedure
2.4. [18F]FDG PET/CT Imaging Procedure
2.5. Image Analysis
2.6. Semi-Quantitative Analysis
2.7. Pathological Analysis
2.8. Immunohistochemistry
2.9. Follow-Up
2.10. Statistical Analysis
3. Results
3.1. Patient Clinical Characteristics
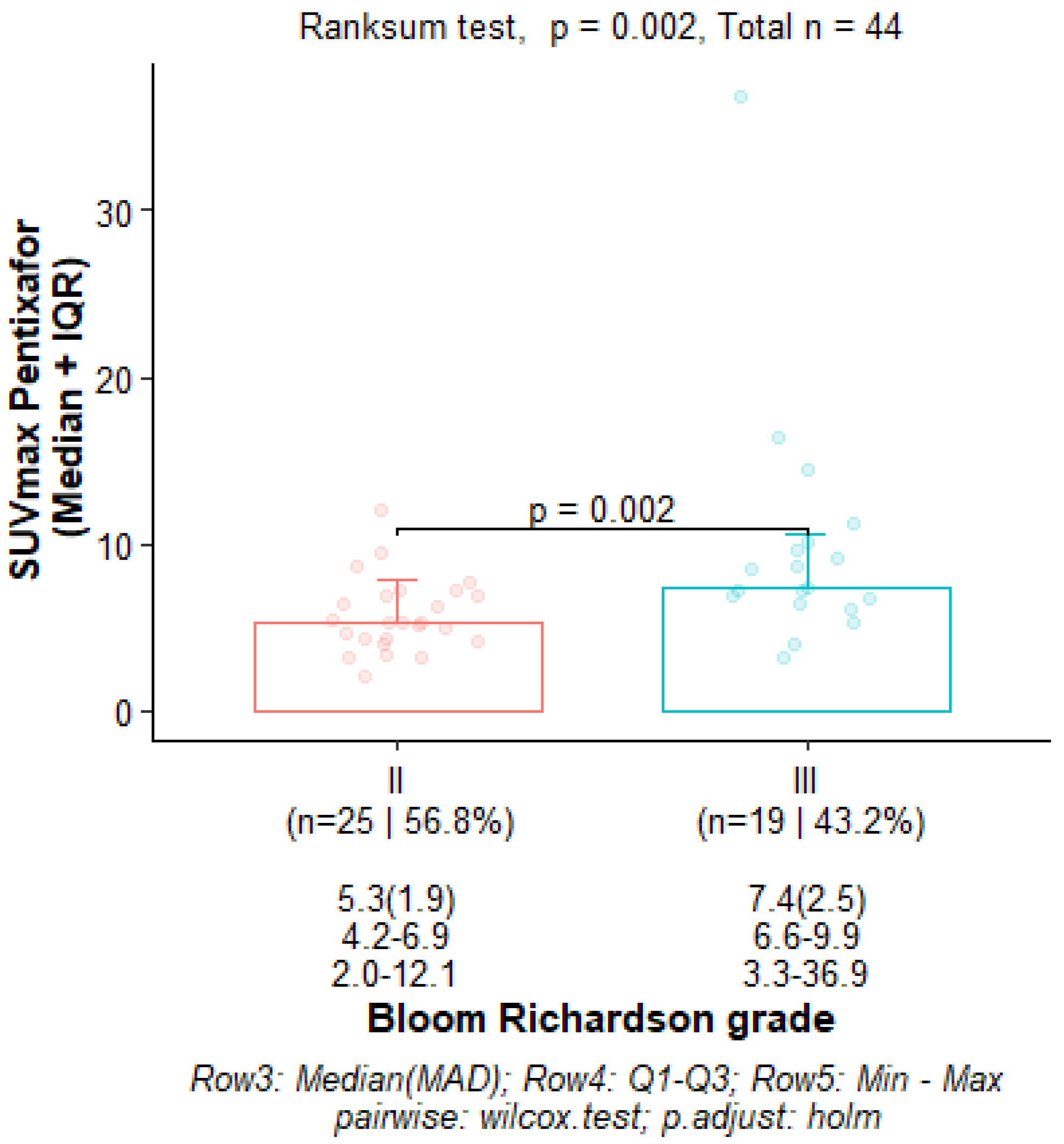
3.2. Correlation Between 68Ga-Pentixafor PET/CT Metrics and Prognostic Factors
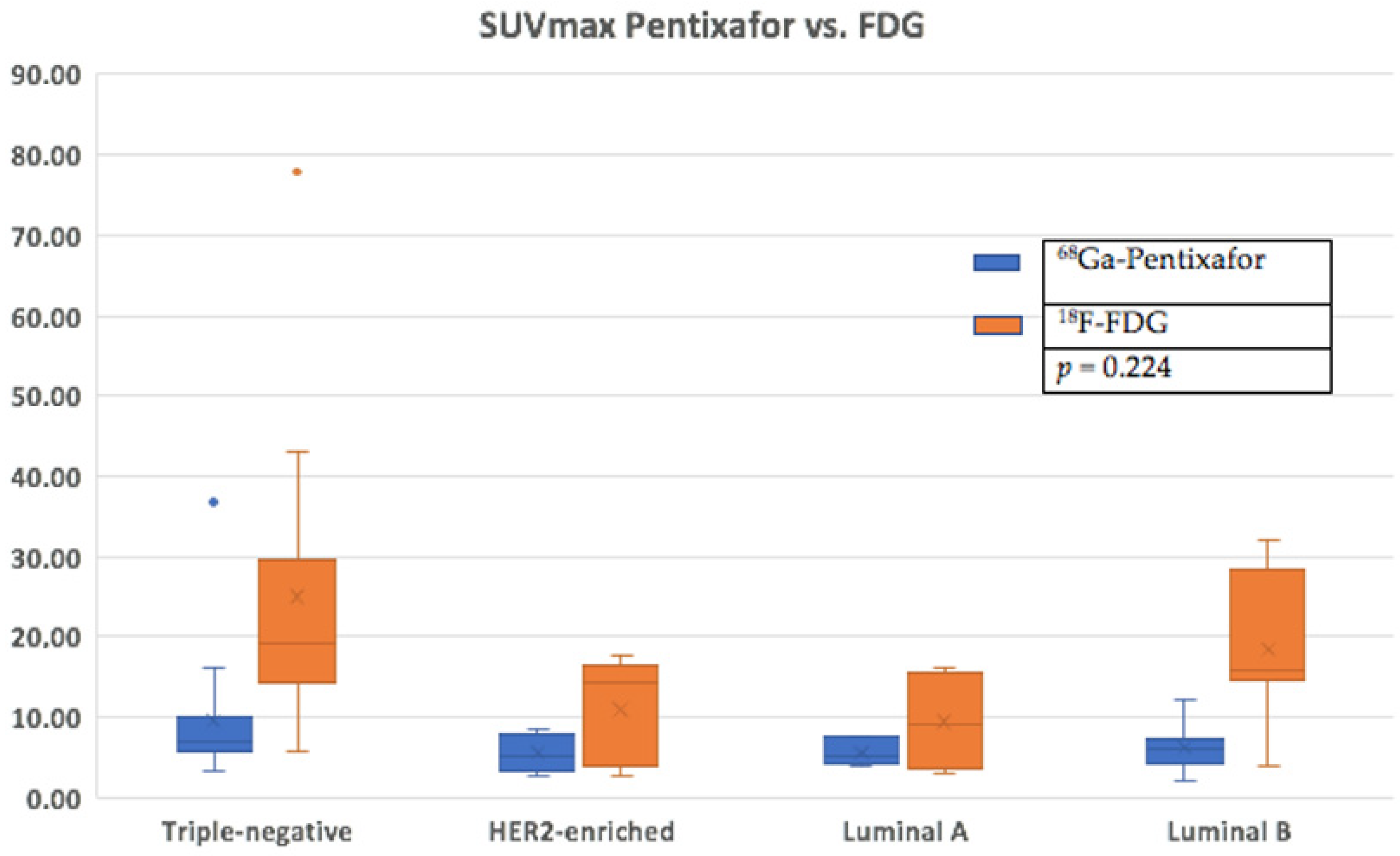
3.3. Comparison of FDG and Pentixafor
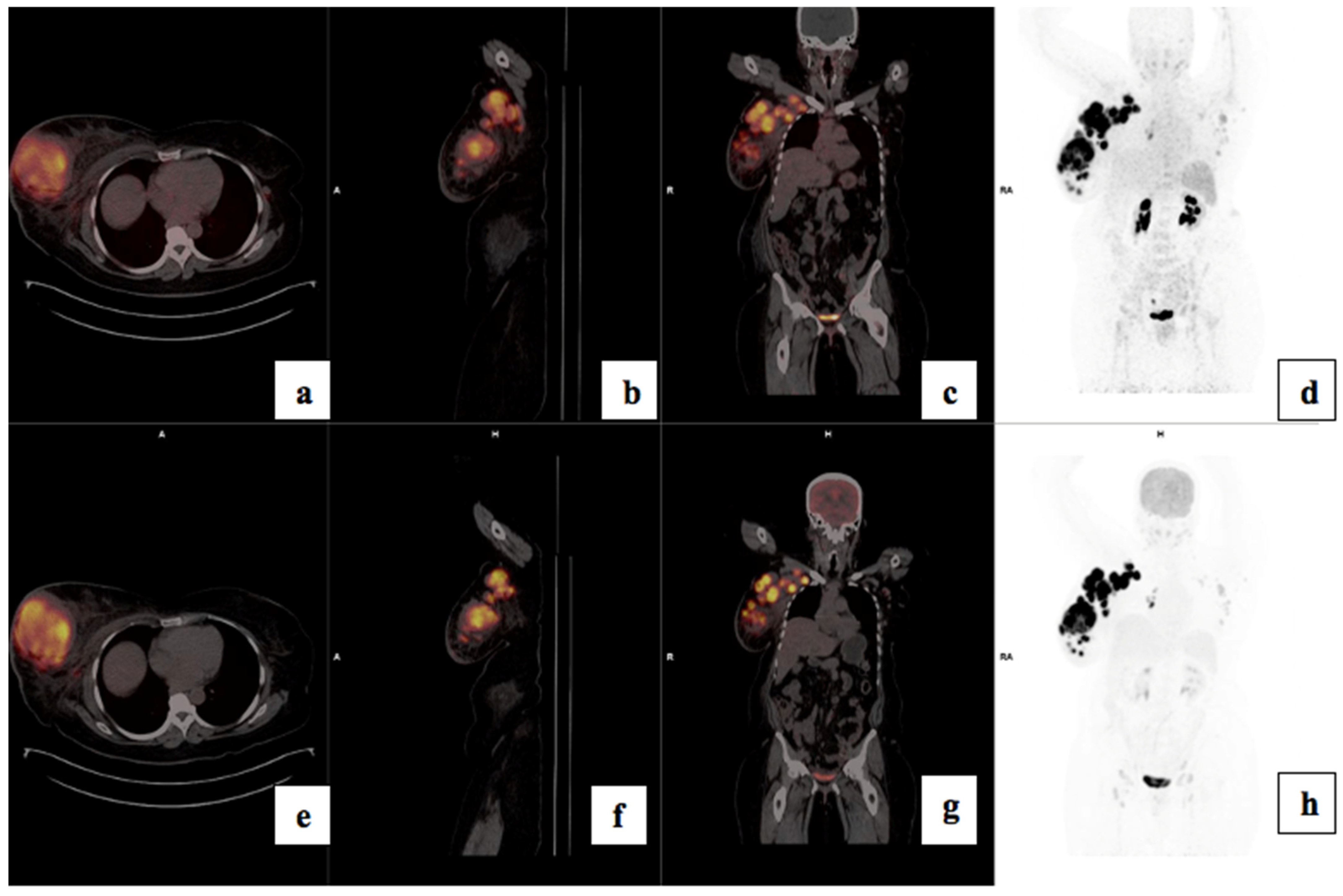
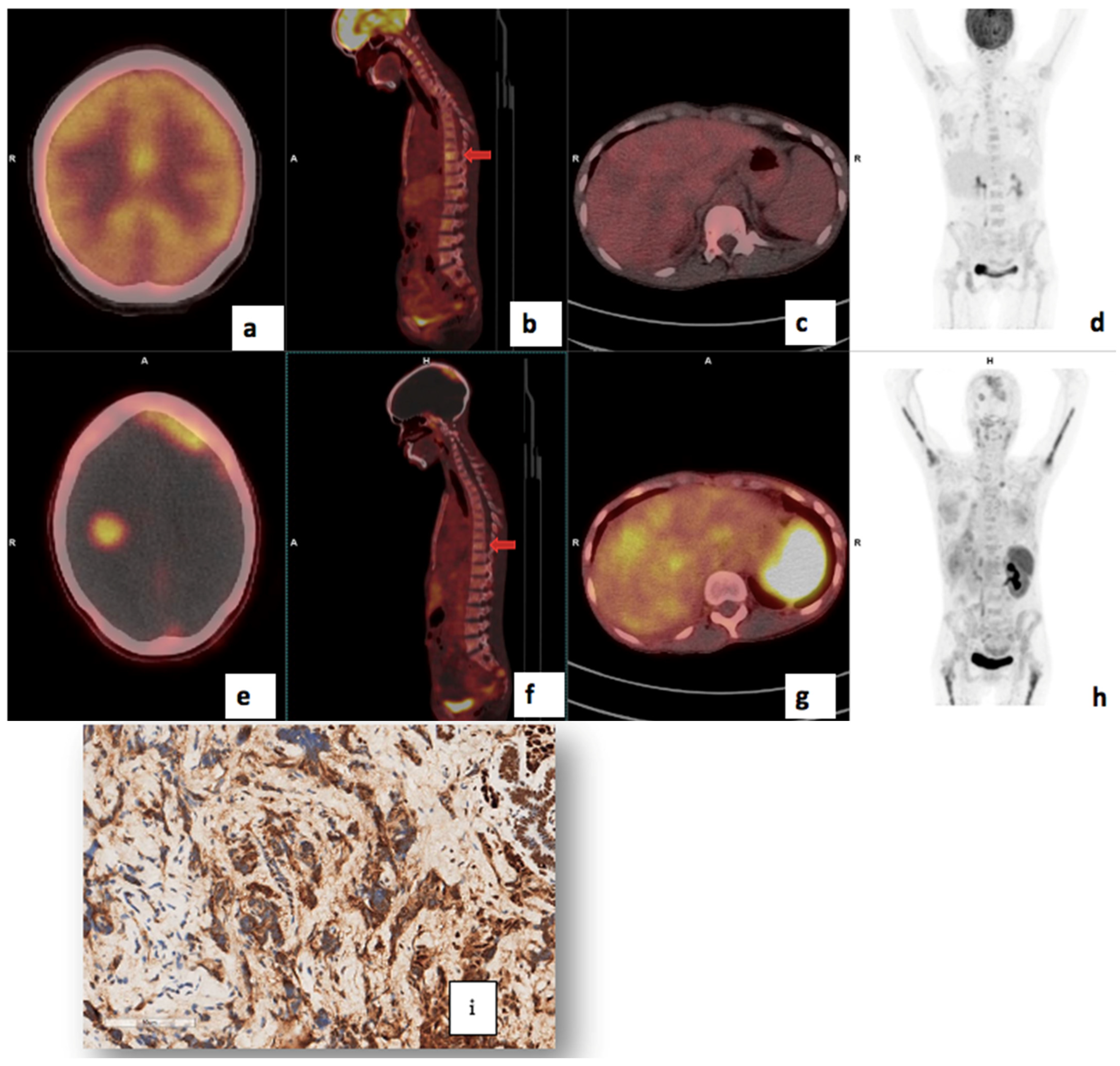
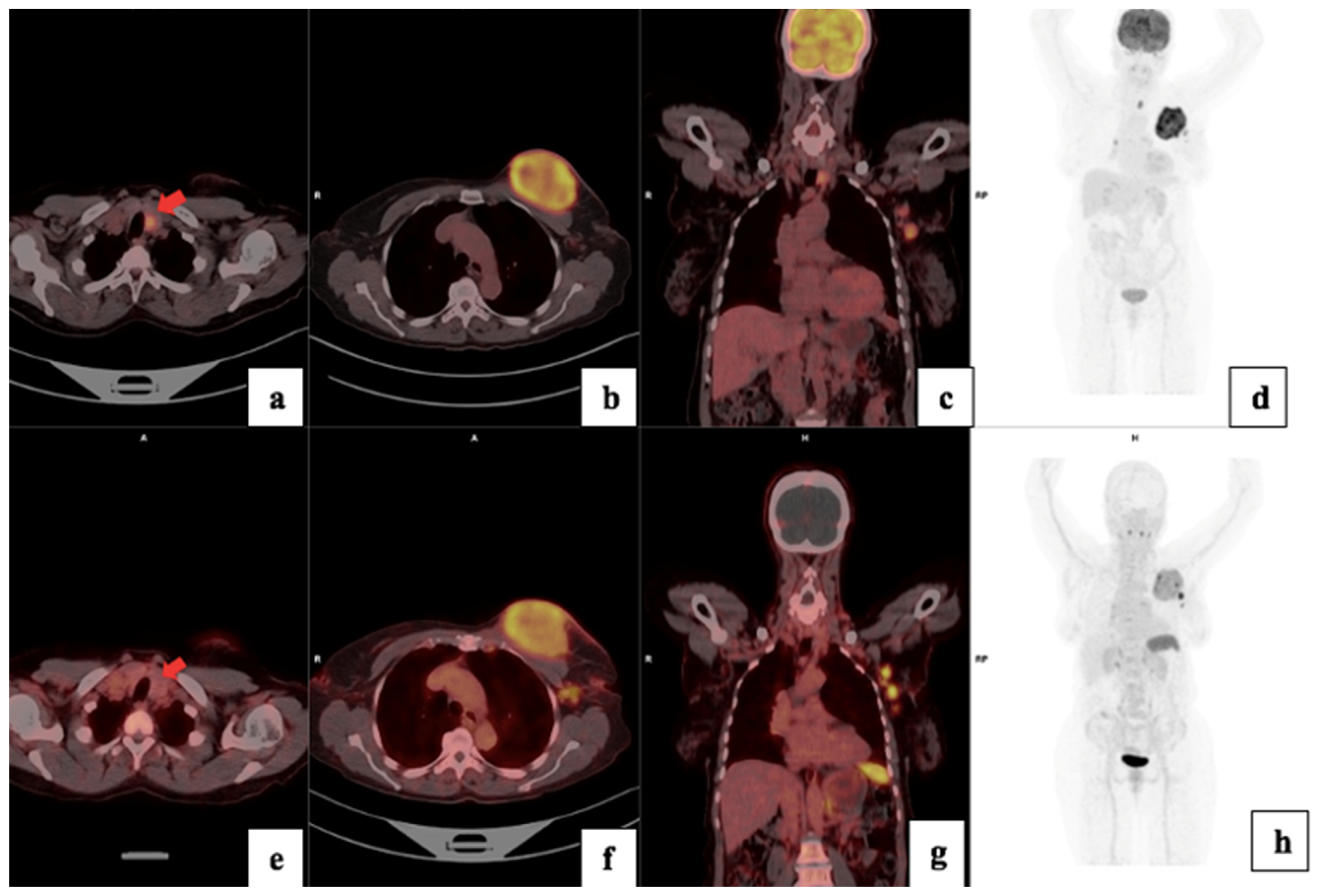
3.4. Visual Analysis
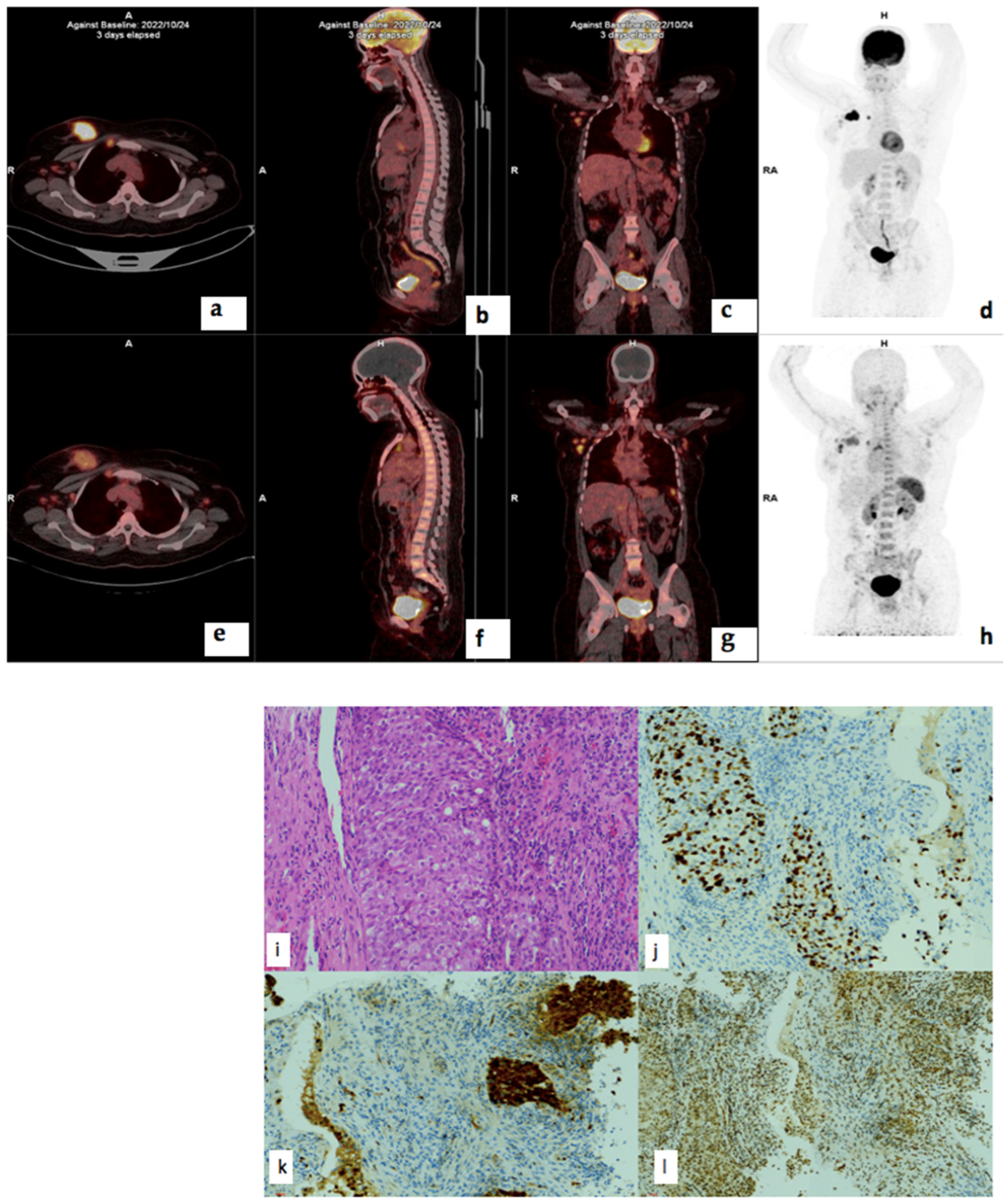
3.5. Treatment Response
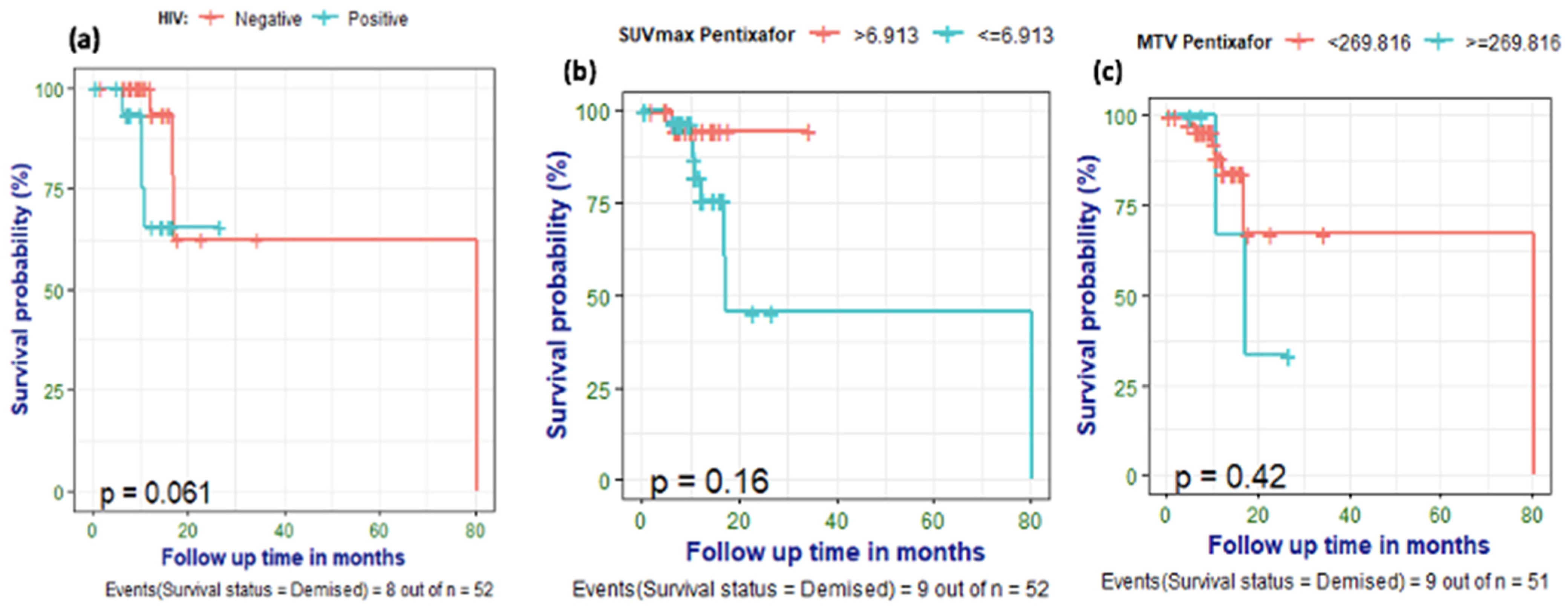
3.6. Survival Analysis
3.7. Correlation of PET/CT Metrics with CXCR4 Immunohistochemistry Staining
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CXCR4 | Chemokine receptor 4 |
| FDG | Fluorodeoxyglucose |
| HIV | Human Immunodeficiency Virus |
| IHC | Immunohistochemistry |
| PET | Positron Emission Tomography |
| RT | Radiation therapy |
| SUV | Standardised Uptake Value |
| TBR | Tumor-to-breast background ratio |
| TNBC | Triple negative breast cancer |
| TLR | Tumor-to-liver background ratio |
| MTV | Metabolic tumor volume |
| TLU | Total lesion uptake |
| TLG | Total lesion glycolysis |
References
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer statistics. CA Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Hon, J.D.C.; Singh, B.; Sahin, A.; Du, G.; Wang, J.; Wang, V.Y.; Deng, F.M.; Zhang, D.Y.; Monaco, M.E.; Lee, P. Breast cancer molecular subtypes: From TNBC to QNBC. J. Clin. Oncol. 2016, 6, 1864–1872. [Google Scholar]
- Joko-Fru, W.Y.; Miranda-Filho, A.; Soerjomataram, I.; Egue, M.; Akele-Akpo, M.T.; N’da, G.; Assefa, M.; Buziba, N.; Korir, A.; Kamate, B.; et al. Breast cancer survival in sub-saharan africa by age, stage at diagnosis, and human development index: A population-based registry study. Int. J. Cancer 2020, 146, 1208–1218. [Google Scholar] [CrossRef] [PubMed]
- Gradishar, W.J.; Moran, M.S.; Abraham, J.; Aft, R.; Agnese, D.; Allison, K.H.; Blair, S.L.; Burstein, H.J.; Dang, C.; Elias, A.D.; et al. NCCN Guidelines® Insights: Breast Cancer, Version 4. J. Natl. Compr. Cancer Netw. 2021, 19, 484–493. [Google Scholar] [CrossRef]
- Esteva, F.J.; Hubbard-Lucey, V.M.; Tang, J.; Pusztai, L. Immunotherapy and targeted therapy combinations in metastatic breast cancer. Lancet Oncol. 2019, 20, e175–e186. [Google Scholar] [CrossRef]
- Bao, S.; Darvishi, M.; Amin, A.H.; Al-Haideri, M.T.; Patra, I.; Kashikova, K.; Ahmad, I.; Alsaikhan, F.; Al-Qaim, Z.H.; Al-Gazally, M.E.; et al. CXC chemokine receptor 4 (CXCR4) blockade in cancer treatment. Z. Krebsforsch. Klin. Onkol. 2023, 149, 7945–7968. [Google Scholar] [CrossRef]
- Zhou, W.; Guo, S.; Liu, M.; Burow, M.E.; Wang, G. Targeting CXCL12/CXCR4 axis in tumor immunotherapy. Curr. Med. Chem. 2019, 26, 3026–3041. [Google Scholar] [CrossRef]
- Edmonds, C.E.; O’Brien, S.R.; McDonald, E.S.; Mankoff, D.A.; Pantel, A.R. PET imaging of breast cancer: Current applications and future directions. J. Breast Imaging 2024, 6, 586–600. [Google Scholar] [CrossRef]
- Sonkin, D.; Thomas, A.; Teicher, B.A. Cancer treatments: Past, present, and future. Cancer Genet. 2024, 286–287, 18–24. [Google Scholar] [CrossRef]
- Müller, A.; Homey, B.; Soto, H.; Ge, N.; Catron, D.; Buchanan, M.E.; McClanahan, T.; Murphy, E.; Yuan, W.; Wagner, S.N.; et al. Involvement of chemokine receptors in breast cancer metastasis. Nature 2001, 410, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Vag, T.; Steiger, K.; Rossmann, A.; Keller, U.; Noske, A.; Herhaus, P.; Ettl, J.; Niemeyer, M.; Wester, H.-J.; Schwaiger, M. PET imaging of chemokine receptor CXCR4 in patients with primary and recurrent breast carcinoma. EJNMMI Res. 2018, 8, 90. [Google Scholar] [CrossRef] [PubMed]
- Akonnor, A.; Makise, M.; Kuniyasu, A. CXCR4-targeted necrosis-inducing peptidomimetic for treating breast cancer. ACS Omega 2023, 8, 24467–24476. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Riese, D.J., 2nd; Shen, J. The role of the CXCL12/CXCR4/CXCR7 chemokine axis in cancer. Front. Pharmacol. 2020, 11, 574667. [Google Scholar] [CrossRef]
- Guo, K.; Feng, G.; Yan, Q.; Sun, L.; Zhang, K.; Shen, F.; Shen, M.; Ruan, S. CXCR4 and CXCR3 are two distinct prognostic biomarkers in breast cancer: Database mining for CXCR family members. Mol. Med. Rep. 2019, 20, 4791–4802. [Google Scholar] [CrossRef]
- Demmer, O.; Gourni, E.; Schumacher, U.; Kessler, H.; Wester, H.J. PET imaging of CXCR4 receptors in cancer by a new optimized ligand. ChemMedChem 2011, 6, 1789. [Google Scholar] [CrossRef]
- Ben-Batalla, I.; Seoane, S.; Garcia-Caballero, T.; Gallego, R.; Macia, M.; Gonzalez, L.O.; Vizoso, F.; Perez-Fernandez, R. Deregulation of the Pit-1 transcription factor in human breast cancer cells promotes tumor growth and metastasis. J. Clin. Investig. 2010, 120, 4289–4302. [Google Scholar] [CrossRef]
- Zhou, J.; Le, K.; Xu, M.; Ming, J.; Yang, W.; Zhang, Q.; Lu, L.; Xi, Z.; Ruan, S.; Huang, T. CXCR4 antagonist AMD3100 reverses the resistance to tamoxifen in breast cancer via inhibiting AKT phosphorylation. Mol. Ther. Oncolytics 2020, 18, 161–170. [Google Scholar] [CrossRef]
- Li, C.; Lang, J.; Wang, Y.; Cheng, Z.; Zu, M.; Li, F.; Sun, J.; Deng, Y.; Ji, T.; Nie, G.; et al. Self-assembly of CXCR4 antagonist peptide–docetaxel conjugates for breast tumor multi-organ metastasis inhibition. Acta Pharm. Sin. B 2023, 13, 3849–3861. [Google Scholar] [CrossRef]
- Yao, Q.; Xu, C.; Zhao, H.; Chen, H. CXCR4 in breast cancer: Oncogenic role and therapeutic targeting. Drug Des. Dev. Ther. 2015, 9, 4953–4964. [Google Scholar] [CrossRef]
- Chonco, B.; Nxasana, T.; Harry, L.; Buthelezi-Zulu, T.; Gabela, L.; Sathekge, M.; Vorster, M. [68Ga]Pentixafor-PET/CT for imaging of chemokine receptor 4 expression in patients with locally advanced breast cancer—initial experience. In Proceedings of the SNMMI Annual Meeting, Chicago, IL, USA, 24–27 June 2023; Volume 64, p. 1150. [Google Scholar]
- Xiang, J.; Hurchla, M.A.; Fontana, F.; Su, X.; Amend, S.R.; Esser, A.K.; Douglas, G.J.; Mudalagiriyappa, C.; Luker, K.E.; Pluard, T.; et al. CXCR4 protein epitope mimetic antagonist POL5551 disrupts metastasis and enhances chemotherapy effect in triple-negative breast cancer. Mol. Cancer Ther. 2015, 14, 2473–2485. [Google Scholar] [CrossRef] [PubMed]
- Regan, M.M.; Aebi, S.; André, F.; Barrios, C.; Bergh, J.; Bonnefoi, H.; Morales, D.B.; Brucker, S.; Burstein, H.; Cameron, D.; et al. Customizing local and systemic therapies for women with early breast cancer: The st. Gallen International Consensus Guidelines for treatment of early breast cancer. Ann. Oncol. 2021, 32, 1216–1235. [Google Scholar] [CrossRef]
- Remmele, W.; Stegner, H.E. Recommendation for uniform definition of an immunoreactive score (IRS) for immunohistochemical estrogen receptor detection (ER-ICA) in breast cancer tissue. Pathol. Res. Pract. 1987, 8, 138–140. [Google Scholar]
- Buck, A.K.; Haug, A.; Dreher, N.; Lambertini, A.; Higuchi, T.; Lapa, C.; Weich, A.; Pomper, M.G.; Wester, H.J.; Zehndner, A.; et al. Imaging of C-X-C motif chemokine receptor 4 expression in 690 patients with solid or hematologic neoplasms using [(68)Ga]-Pentixafor PET. J. Nucl. Med. 2022, 63, 1687–1692. [Google Scholar] [CrossRef]
- Chiodoni, C.; Cancila, V.; Renzi, T.A.; Perrone, M.; Tomirotti, A.M.; Sangaletti, S.; Botti, L.; Dugo, M.; Milani, M.; Bongiovanni, L.; et al. Transcriptional profiles and stromal changes reveal bone marrow adaptation to early breast cancer in association with deregulated circulating microRNAs. Cancer Res. 2020, 80, 484–498. [Google Scholar] [CrossRef]
- Yuan, J.; Yang, L.; Zhang, H.; Beeraka, N.M.; Zhang, D.; Wang, Q.; Wang, M.; Vikram, P.R.H.; Sethi, G.; Wang, G. Decoding tumor microenvironment: EMT modulation in breast cancer metastasis and therapeutic resistance, and implications of novel immune checkpoint blockers. Biomed. Pharmacother. 2024, 181, 117714. [Google Scholar] [CrossRef]
- Liu, H.; Dong, A.; Rasteh, A.M.; Wang, P.; Weng, J. Identification of the novel exhausted T cell CD8+ markers in breast cancer. Sci. Rep. 2024, 14, 19142. [Google Scholar] [CrossRef]
- Tan, K.; Naylor, M.J. Tumor microenvironment-immune cell interactions influencing breast cancer heterogeneity and disease progression. Front. Oncol. 2022, 12, 876451. [Google Scholar] [CrossRef]
- Mukherjee, D.; Zhao, J. The role of chemokine receptor CXCR4 in breast cancer metastasis. Am. J. Cancer Res. 2013, 3, 46. [Google Scholar]
- Chen, H.-W.; Du, C.-W.; Wei, X.-L.; Khoo, U.-S.; Zhang, G.-J. Cytoplasmic CXCR4 high-expression exhibits distinct poor clinicopathological characteristics and predicts poor prognosis in triple-negative breast cancer. Clin. Oncol. 2013, 13, 410–416. [Google Scholar]
- Darwish, M.M.; Riad, A.Y.; Salem, D.A.; Essa, A.E.; Shakweer, M.M.; Sherif, D.E.M. Prognostic implication of PD-L1 expression and associated tumor-infiltrating lymphocytes in metastatic breast cancer. Immunopathol. Persa 2022, 8, e18. [Google Scholar] [CrossRef]
- Vazquez, E.D.; Fang, X.; Levesque, L.A.; Huynh, M.; Venegas, C.; Lu, N.; Salazar, N. Chemokine receptors differentially expressed by race category and molecular subtype in the breast cancer TCGA cohort. Sci. Rep. 2022, 12, 10825. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Ordoñez, A.; Seoane, S.; Cabezas, P.; Eiro, N.; Sendon-Lago, J.; Macia, M.; Garcia-Caballero, T.; Gonzalez, L.O.; Sanchez, L.; Vizoso, F.; et al. Breast cancer metastasis to liver and lung is facilitated by Pit-1-CXCL12-CXCR4 axis. Oncogene 2018, 37, 1430–1444. [Google Scholar] [CrossRef] [PubMed]
- Caccuri, F.; Giordano, F.; Barone, I.; Mazzuca, P.; Giagulli, C.; Andò, S.; Caruso, A.; Marsico, S. HIV-1 matrix protein p17 and its variants promote human triple-negative breast cancer cell aggressiveness. Infect. Agents Cancer 2017, 12, 49. [Google Scholar] [CrossRef][Green Version]
- Vicenzi, E.; Liò, P.; Poli, G. The puzzling role of CXCR4 in human immunodeficiency virus infection. Theranostics 2013, 3, 18–25. [Google Scholar] [CrossRef]
- Dreher, N.; Hahner, S.; Fuß, C.T.; Schlötelburg, W.; Hartrampf, P.E.; Serfling, S.E.; Schirbel, A.; Samnick, S.; Higuchi, T.; Weich, A.; et al. CXCR4-directed PET/CT with [(68)Ga]-pentixafor in solid tumors: A comprehensive analysis of imaging findings and comparison with histopathology. Eur. J. Nucl. Med. Mol. Imaging 2024, 51, 1383–1394. [Google Scholar] [CrossRef]
- Buck, A.K.; Serfling, S.E.; Kraus, S.; Samnick, S.; Dreher, N.; Higuchi, T.; Rasche, L.; Einsele, H.; Werner, R.A. Theranostics in hematooncology. J. Nucl. Med. 2023, 64, 1009–1016. [Google Scholar] [CrossRef]
- Maurer, S.; Herhaus, P.; Lippenmeyer, R.; Hänscheid, H.; Kircher, M.; Schirbel, A.; Maurer, H.C.; Buck, A.K.; Wester, H.-J.; Einsele, H.; et al. Side effects of CXC-chemokine receptor 4–directed endoradiotherapy with pentixather before hematopoietic stem cell transplantation. J. Nucl. Med. 2019, 60, 1399–1405. [Google Scholar] [CrossRef]
- Bao, G.; Wang, Z.; Liu, L.; Zhang, B.; Song, S.; Wang, D.; Cheng, S.; Moon, E.S.; Roesch, F.; Zhao, J.; et al. Targeting CXCR4/CXCL12 axis via [(177)Lu]Lu-DOTAGA.(SA.FAPi)(2) with CXCR4 antagonist in triple-negative breast cancer. Eur. J. Nucl. Med. Mol. Imaging 2024, 51, 2744–2757. [Google Scholar] [CrossRef]
- Yang, J.; Tian, E.; Chen, L.; Liu, Z.; Ren, Y.; Mao, W.; Zhang, Y.; Zhang, J. Development and therapeutic perspectives of CXCR4 antagonists for disease therapy. Eur. J. Med. Chem. 2024, 275, 116594. [Google Scholar] [CrossRef]
- Wong, R.S.; Ong, R.J.; Lim, J.S. Immune checkpoint inhibitors in breast cancer: Development, mechanisms of resistance and potential management strategies. Cancer Drug Resist. 2023, 6, 768. [Google Scholar] [CrossRef] [PubMed]
- Chen, I.X.; Chauhan, V.P.; Posada, J.; Ng, M.R.; Wu, M.W.; Adstamongkonkul, P.; Huang, P.; Lindeman, N.; Langer, R.; Jain, R.K. Blocking CXCR4 alleviates desmoplasia, increases T-lymphocyte infiltration, and improves immunotherapy in metastatic breast cancer. Proc. Natl. Acad. Sci. USA 2019, 116, 4558–4566. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Guo, L.; Zhao, J.; Weng, H.; Zhao, B. CXCR4 over-expression and survival in cancer: A system review and meta-analysis. Oncotarget 2014, 6, 5022–5040. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhang, F.; Liu, Z.; Fan, Z. Immunotherapy for triple-negative breast cancer: Combination strategies to improve outcome. Cancers 2023, 15, 321. [Google Scholar] [CrossRef]
- Liu, S.; Xie, S.M.; Liu, W.; Gagea, M.; Hanker, A.B.; Nguyen, N.; Raghavendra, A.S.; Yang-Kolodji, G.; Chu, F.; Neelapu, S.S.; et al. Targeting CXCR4 abrogates resistance to trastuzumab by blocking cell cycle progression and synergizes with docetaxel in breast cancer treatment. Breast Cancer Res. 2023, 25, 62. [Google Scholar] [CrossRef]
- Kotb, R.M.; Ibrahim, S.S.; Mostafa, O.M.; Shahin, N.N. Potential role of CXCR4 in trastuzumab resistance in breast cancer patients. Biochim. Biophys. Acta Mol. Basis Dis. 2022, 1868, 166520. [Google Scholar] [CrossRef]
- Buck, A.K.; Serfling, S.E.; Lindner, T.; Hänscheid, H.; Schirbel, A.; Hahner, S.; Fassnacht, M.; Einsele, H.; Werner, R.A. CXCR4-targeted theranostics in oncology. Eur. J. Nucl. Med. Mol. Imaging 2022, 49, 4133–4144. [Google Scholar] [CrossRef]
- Garg, P.; Jallepalli, V.R.; Verma, S. Unravelling the CXCL12/CXCR4 axis in breast cancer: Insights into metastasis, microenvironment interactions, and therapeutic opportunities. Hum. Gene 2024, 40, 201272. [Google Scholar] [CrossRef]
- Lapa, C.; Hänscheid, H.; Kircher, M.; Schirbel, A.; Wunderlich, G.; Werner, R.A.; Samnick, S.; Kotzerke, J.; Einsele, H.; Buck, A.K.; et al. Feasibility of CXCR4-directed radioligand therapy in advanced diffuse large B-cell lymphoma. J. Nucl. Med. 2018, 60, 60–64. [Google Scholar] [CrossRef]
- Osl, T.; Schmidt, A.; Schwaiger, M.; Schottelius, M.; Wester, H.J. A new class of pentixafor-and pentixather-based theranostic agents with enhanced CXCR4-targeting efficiency. Theranostics 2020, 10, 8264. [Google Scholar] [CrossRef]
- Spahn, M.A.; Loy, T.V.; Celen, S.; Koole, M.; Deroose, C.M.; Cawthorne, C.; Vanduffel, W.; Schols, D.; Bormans, G.; Cleeren, F. Selective PET imaging of CXCR4 using the Al18F-labeled antagonist LY. Eur. J. Nucl. Med. Mol. Imaging 2024, 11, 1–6. [Google Scholar]
- Blot, E.; Couteulx, S.L.; Jamali, H.; Cornic, M.; Guillemet, C.; Duval, C.; Hellot, M.F.; Pille, J.Y.; Picquenot, J.M.; Veyret, C. CxCR4 membrane expression in node-negative breast cancer. Breast J. 2008, 14, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Salvucci, O.; Bouchard, A.; Baccarelli, A.; Deschenes, J.; Sauter, G.; Simon, R.; Bianchi, R.; Basik, M. The role of CXCR4 receptor expression in breast cancer: A large tissue microarray study. Breast Cancer Res. Treat. 2006, 97, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Cabioglu, N.; Onder, S.; Karatay, H.; Bayram, A.; Oner, G.; Tukenmez, M.; Muslumanoglu, M.; Igci, A.; Dinccag, A.; Ozmen, V.; et al. New emerging chemokine receptors: CCR5 or CXCR5 on tumor is associated with poor response to chemotherapy and poor prognosis in locally advanced triple-negative breast cancer. Cancers 2024, 16, 2388. [Google Scholar] [CrossRef]
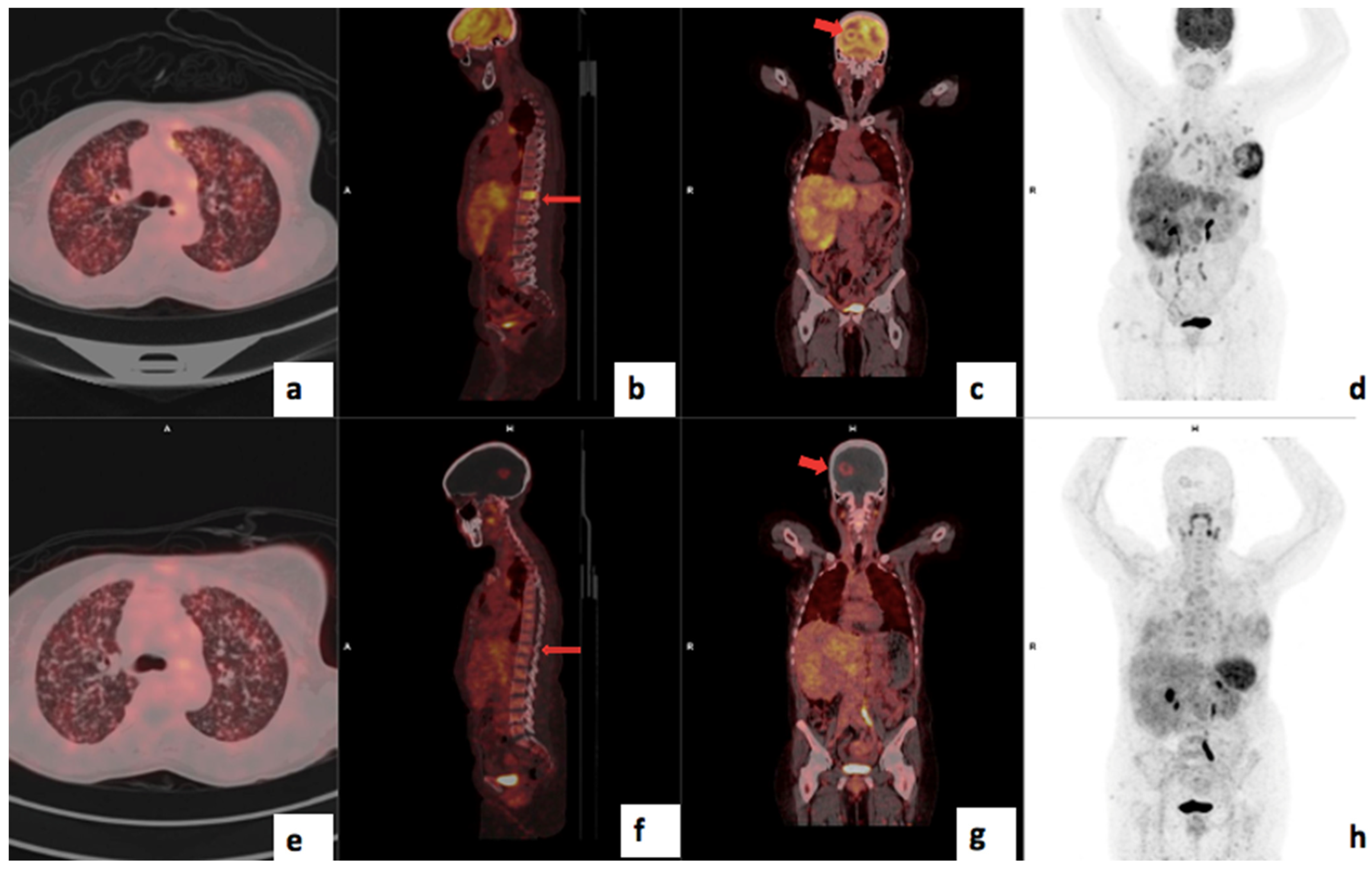
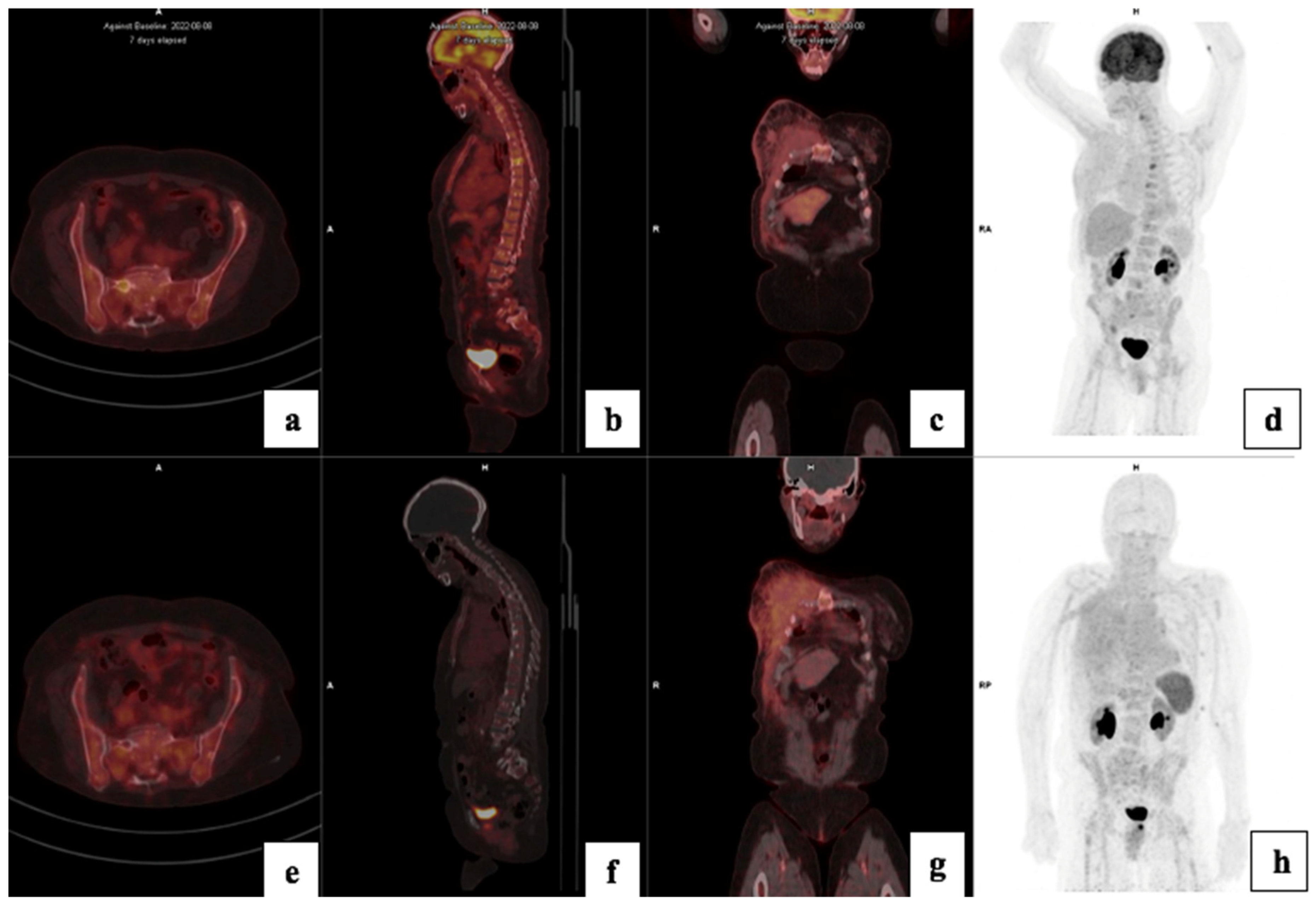
| Clinical Details | |
|---|---|
| Age median (IQR) | 51 (36–81) |
| Ki67 | 50 (10–90%) |
| T stage | N = 51 (%) |
| 1 | 1 (2.0%) |
| 2 | 5 (10.0%) |
| 3 | 12 (24.0%) |
| 4 | 33 (64.0%) |
| N stage | |
| N1 | 26 (52%) |
| N2 | 19 (38%) |
| Metastasis | |
| Internal mammary | 1 |
| Bone | 12 |
| Liver | 6 |
| Lung | 8 |
| Brain | 2 |
| Bloom Richardson grade | |
| II | 24 (55%) |
| III | 19 (45%) |
| Histology | |
| IDC | 45 (90%) |
| IDC (mucinous component) | 2 (4%) |
| ILC | 1 (2%) |
| Metaplastic | 1 (2%) |
| Molecular subtypes | |
| Luminal A | 5 (10%) |
| Luminal B | 19 (40%) |
| HER2-enriched | 5 (10%) |
| Triple-negative | 21 (40%) |
| Atypical | 1 (2%) |
| Receptor expression | |
| ER+/− | 24/26 |
| PR+/− | 31/19 |
| HER2+/− | 40/9 |
| HIV infection | |
| HIV-positive | 18 (37%) |
| HIV-negative | 33 (63%) |
| Race | |
| Black | 36 (69%) |
| Indian | 11 (21%) |
| White | 3 (8%) |
| Mixed-race | 1 (2%) |
| HIV | Negative (N = 33) | Positive (N = 18) | p-Value | Overall (N = 51) |
|---|---|---|---|---|
| TLU Pentixafor | Ranksum | |||
| Median(Q1–Q3) | 174 (105–557) | 376 (219–881) | 0.038 | 239 (112–677) |
| n(Min–Max) | 32 (6.21–1150) | 19 (60.7–5950) | 51 (6.21–5950) |
| Median SUVmax | Median MTV | Median TLU | Median TBR (Liver) | Median TBR (Breast) | Sensitivity (%) | Specificity (%) | |
|---|---|---|---|---|---|---|---|
| 68Ga-Pentixafor | 6.35 | 89 | 242 | 2.2 | 1.7 | 96 | 100 |
| 18F-FDG | 16.9 | 54 | 416 | 9.8 | 4.8 | 100 | 100 |
| Variable 1 | Variable 2 | Correlation Coefficient | p-Value |
|---|---|---|---|
| 68Ga-Pentixafor MTV | 18F-FDG MTV FDG | 0.523 | <0.001 |
| Ki67 | 18F-FDG SUVmean | 0.362 | 0.022 |
| CA 153 | 68Ga-Pentixafor TBR (liver) | 0.589 | 0.034 |
| 68Ga-Pentixafor TLU | 18F-MTV FDG | 0.477 | 0.002 |
| 68Ga-Pentixafor TLU | 18F-FDG TBR | 0.418 | 0.002 |
| 68Ga-Pentixafor TLU | 18F-FDG SUVmean | 0.426 | 0.005 |
| 68Ga-Pentixafor TBR(breast) | 18F-FDG SUVmax | 0.667 | <0.001 |
| 68Ga-Pentixafor TBR (breast) | 18F-FDG TLG | 0.583 | <0.001 |
| 68Ga-Pentixafor TBR (breast) | 18F-FDG SUVmean | 0.553 | <0.001 |
| 68Ga-Pentixafor TBR (breast) | 18F-FDG TBR max | 0.530 | <0.001 |
| Predictor | Direction | Optimal_Cutpoint | Youden | Accuracy | Sensitivity | Specificity | AUC | Prevalence |
|---|---|---|---|---|---|---|---|---|
| SUVmax Pentixafor | <= | 6.913 | 0.286 | 0.490 | 0.857 | 0.429 | 0.592 | 0.143 |
| SUVmean Pentixafor | <= | 3.291 | 0.348 | 0.646 | 0.714 | 0.634 | 0.662 | 0.146 |
| TLU_Pentixafor | >= | 3676.708 | −0.049 | 0.812 | 0.000 | 0.951 | 0.603 | 0.146 |
| MTV_Pentixafor | >= | 269.816 | 0.213 | 0.833 | 0.286 | 0.927 | 0.631 | 0.146 |
| TBR(liver) Pentixafor | <= | 4.486 | 0.095 | 0.224 | 1.000 | 0.095 | 0.558 | 0.143 |
| TBR(breast) Pentixafor_ | <= | 6.342 | 0.048 | 0.184 | 1.000 | 0.048 | 0.507 | 0.143 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hadebe, B.; Harry, L.; Gabela, L.; Nxasana, T.; Ndlovu, N.; Pillay, V.; Masikane, S.; Patel, M.; Mpanya, D.; Buccimaza, I.; et al. Comparing 68Ga-Pentixafor,18F-FDG PET/CT and Chemokine Receptor 4 Immunohistochemistry Staining in Breast Cancer: A Prospective Cross Sectional Study. Cancers 2025, 17, 763. https://doi.org/10.3390/cancers17050763
Hadebe B, Harry L, Gabela L, Nxasana T, Ndlovu N, Pillay V, Masikane S, Patel M, Mpanya D, Buccimaza I, et al. Comparing 68Ga-Pentixafor,18F-FDG PET/CT and Chemokine Receptor 4 Immunohistochemistry Staining in Breast Cancer: A Prospective Cross Sectional Study. Cancers. 2025; 17(5):763. https://doi.org/10.3390/cancers17050763
Chicago/Turabian StyleHadebe, Bawinile, Lerwine Harry, Lerato Gabela, Thembelihle Nxasana, Nontobeko Ndlovu, Venesen Pillay, Siphelele Masikane, Maryam Patel, Dineo Mpanya, Ines Buccimaza, and et al. 2025. "Comparing 68Ga-Pentixafor,18F-FDG PET/CT and Chemokine Receptor 4 Immunohistochemistry Staining in Breast Cancer: A Prospective Cross Sectional Study" Cancers 17, no. 5: 763. https://doi.org/10.3390/cancers17050763
APA StyleHadebe, B., Harry, L., Gabela, L., Nxasana, T., Ndlovu, N., Pillay, V., Masikane, S., Patel, M., Mpanya, D., Buccimaza, I., Msimang, M., Aldous, C., Sathekge, M., & Vorster, M. (2025). Comparing 68Ga-Pentixafor,18F-FDG PET/CT and Chemokine Receptor 4 Immunohistochemistry Staining in Breast Cancer: A Prospective Cross Sectional Study. Cancers, 17(5), 763. https://doi.org/10.3390/cancers17050763








