Heterogeneity in Cancer
Simple Summary
Abstract
1. Introduction
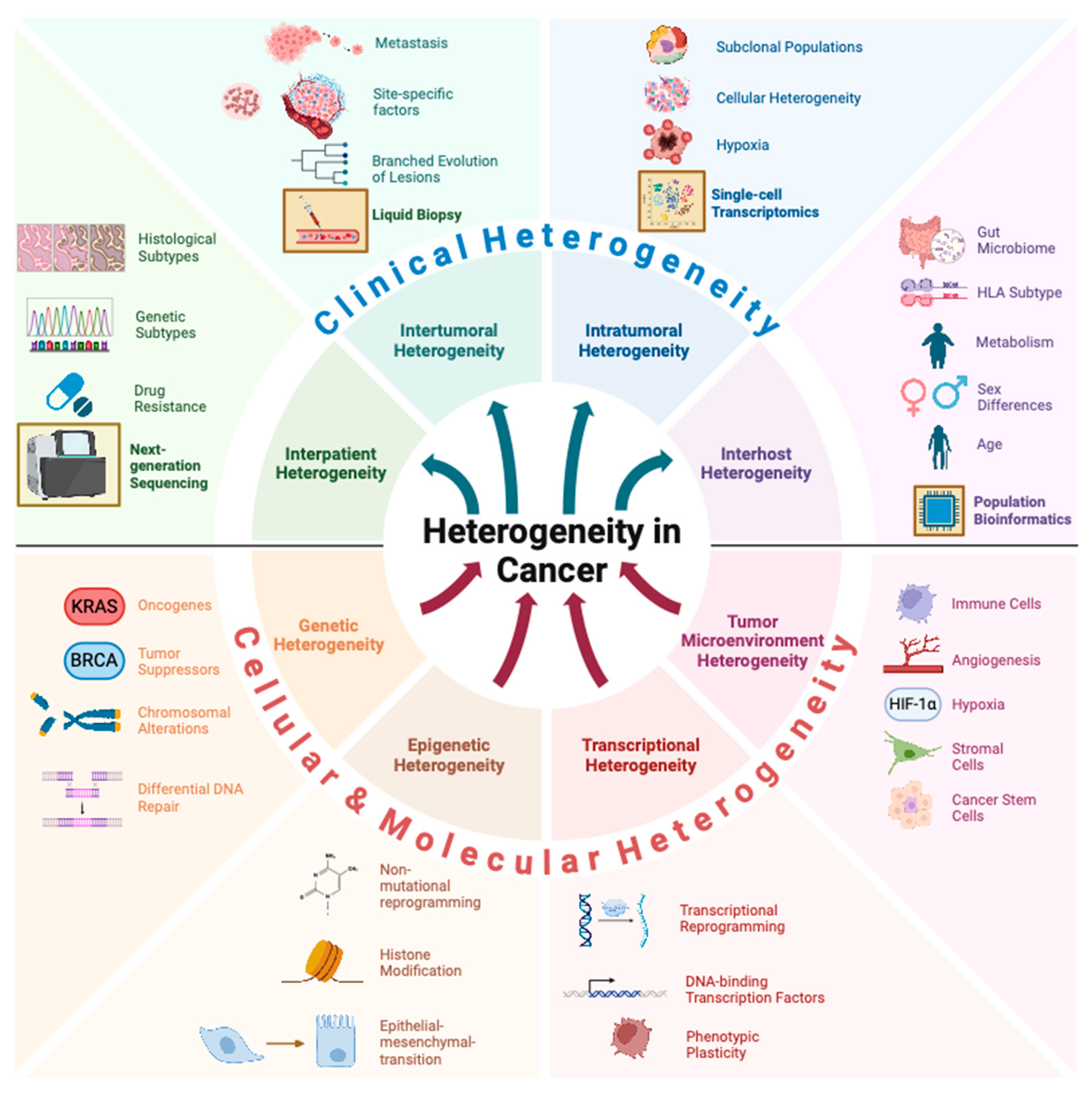
2. Cellular and Molecular Heterogeneity
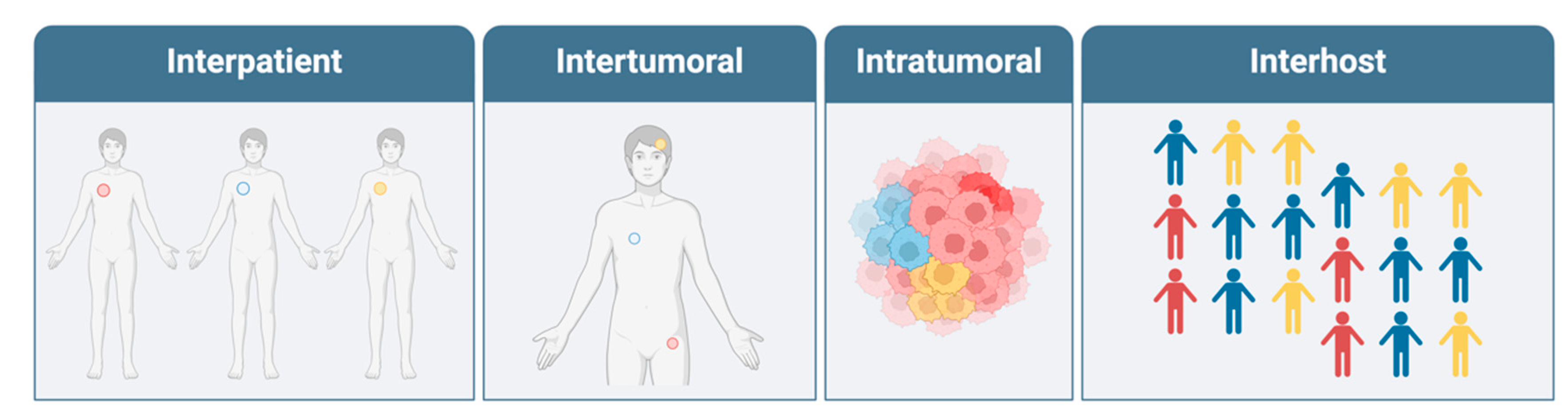
3. Interpatient Heterogeneity
4. Genetic Driver Mutations
| Technology | Principle | Application | Ref. |
|---|---|---|---|
| IHC | Chromogenically visualized antibody | Tissue level visualization of protein expression, including mutant proteins | [4] |
| In Situ Hybridization | Complementary DNA/RNA probes | Detection of genetic variants and copy number alterations at single-cell resolution | [22] |
| PCR | DNA amplification using primers | Highly sensitive detection of wide range of genetic variants including repeat expansions | [28] |
| Gene Panels | Hybridization probes arranged in array | Detection of select arrangement of genes with deep coverage | [29] |
| NGS | Amplification and massively parallel sequencing of sample DNA | Large scale sequencing for comprehensive genomic analysis and detection of novel genetic signatures | [5,24,25,26,27] |
5. Epigenetic Heterogeneity
| Technology | Principle | Application | Ref. |
|---|---|---|---|
| Methylation sensitive PCR | Amplification of sodium bisulfite treated DNA using primers | Highly sensitive and specific determination of the methylation status of CpG in particular genomic regions | [52] |
| Bisulfite Sequencing | Amplification and massively parallel sequencing of sodium bisulfite treated DNA | Genome-wide characterization of gene regulatory status through methylation assessment of promoter regions | [48] |
| ChIP-seq/PCR | Immunoprecipitation of histone modified DNA | Assessment of the genome-wide (seq) or single region (PCR) histone modification profile | [53,54,55] |
| ATAC-seq | Assessment of transposase- accessible genome regions | Genome-wide investigation of chromatin accessibility | [51,56] |
6. Transcriptional Regulation
7. Tumor Microenvironment Heterogeneity
8. Intra-Tumoral Heterogeneity
9. Spatial Omics
10. Multi-Omics
| Technology | Principle | Application | Ref. |
|---|---|---|---|
| Multi-site biopsies | Genomic or proteomic analysis of spatially distinct tumor samples | Uncovering heterogenous clonal populations spread out within a patient’s tumor | [101] |
| Single-cell RNA-seq | Captures gene expression at the single-cell level | Analyzing cellular diversity, identifying cell subtypes, and studying differential gene expression across cell types | [97,102] |
| Immuno- fluorescence | Antibodies conjugated to fluorescent dyes to detect protein expression | Localizing specific proteins within cells or tissues and studying protein–protein interactions | [104] |
| Spatial transcriptomics (e.g., Visium) | Barcoded RNA probes to map RNA-seq data to spatial locations in tissue | Visualizing gene expression in tissue sections, linking gene activity to tissue histology | [106] |
| Spatial epigenomics (e.g., epigenetic MERFISH) | Antibody-based probing of histone modifications with concomitant in situ transcriptions | Highly multiplexed spatial mapping of epigenetic markers and chromatin organization at high resolution in tissues | [107,108] |
| Spatial multi-omics (e.g., GeoMX) | Combines IF with spatial transcriptomics through UV cleavage of ROIs | High-throughput spatial profiling of both transcriptome and protein panel in a tissue sample | [113] |
11. Inter-Tumoral Heterogeneity
| Technology | Principle | Application | Ref. |
|---|---|---|---|
| Blood cytokine analysis | ELISA based quantification of blood cytokines | Characterization of the anti-tumor immune response and inflammation | [133] |
| cfDNA | Sequencing of DNA isolated from blood | Non-invasive measurement of tumor DNA signature for screening, diagnosis, monitoring, tracking tumor heterogeneity, and directing targeted therapy | [124,127] |
| cfDNA methylation | Analysis of methylation patterns of tumor cfDNA | Non-invasive characterization of cancer DNA methylation patterns and detection of resistance mechanisms | [130] |
| TCR sequencing | Sequencing of T-cell receptor genes to profile immune response | Analysis of the immune repertoire by tracking of the anti-tumor immune response through TIL expansion detection | [132] |
12. Host Heterogeneity
| Technology | Principle | Application | Ref. |
|---|---|---|---|
| HLA sequencing | DNA sequencing of the human leukocyte antigen (HLA) genes | Characterization, for prognostic and potentially therapeutic purposes, of the relationship between anti-tumor immune response and HLA type | [136] |
| Drug metabolism phenotyping | Genetic analysis of cytochrome variants affecting drug metabolism | Pharmacogenetic identification of poor and ultra-rapid metabolizers of chemotherapeutics for optimization of anti-neoplastic regimens | [148,149] |
| Population bioinformatics | Bioinformatic and AI driven synthesis of multi-omics and environmental factors in populations and individuals | Population-level identification of risk factors, prediction of individual disease risk, and prediction of therapy efficacy (e.g., immune checkpoint blockade response rate) | [150,151] |
13. Conclusions
| Topic | Author | Title | Year | Ref. |
|---|---|---|---|---|
| Genetic heterogeneity | Reiter et al. | An analysis of genetic heterogeneity in untreated cancers | 2019 | [94] |
| Genetic heterogeneity | Burrell et al. | The causes and consequences of genetic heterogeneity in cancer evolution | 2013 | [158] |
| Epigenetic heterogeneity | Sacco et al. | Epithelial–Mesenchymal Plasticity and Epigenetic Heterogeneity in Cancer | 2024 | [159] |
| Epigenetic heterogeneity | Carter et al. | The epigenetic basis of cellular heterogeneity | 2021 | [160] |
| Intratumoral heterogeneity | Ramon Y Cajal et al. | Clinical implications of intratumor heterogeneity: challenges and opportunities | 2020 | [161] |
| Intratumoral heterogeneity | Marusyk et al. | Intra-tumour heterogeneity: a looking glass for cancer? | 2012 | [162] |
| Intertumoral heterogeneity | Vogelstein et al. | Cancer Genome Landscapes | 2013 | [93] |
| Intertumoral heterogeneity | Buikhuisen et al. | Exploring and modelling colon cancer inter-tumour heterogeneity: opportunities and challenges | 2020 | [163] |
| Interhost heterogeneity | Zaal et al. | The Influence of Metabolism on Drug Response in Cancer | 2018 | [164] |
| Interhost heterogeneity | Hazini et al. | Deregulation of HLA-I in cancer and its central importance for immunotherapy | 2021 | [165] |
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Chaffer, C.L.; Weinberg, R.A. A perspective on cancer cell metastasis. Science 2011, 331, 1559–1564. [Google Scholar] [CrossRef] [PubMed]
- Marks, E.; Rizvi, S.M.; Sarwani, N.; Yang, Z.; El-Deiry, W.S. A case of heterogeneous sensitivity to panitumumab in cetuximab-refractory colorectal adenocarcinoma metastases. Cancer Biol. Ther. 2015, 16, 377–382. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Russo, M.; Siravegna, G.; Blaszkowsky, L.S.; Corti, G.; Crisafulli, G.; Ahronian, L.G.; Mussolin, B.; Kwak, E.L.; Buscarino, M.; Lazzari, L.; et al. Tumor Heterogeneity and Lesion-Specific Response to Targeted Therapy in Colorectal Cancer. Cancer Discov. 2016, 6, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Ortiz Hidalgo, C. Immunohistochemistry in Historical Perspective: Knowing the Past to Understand the Present. Methods Mol. Biol. 2022, 2422, 17–31. [Google Scholar] [CrossRef]
- Zhao, E.Y.; Jones, M.; Jones, S.J.M. Whole-Genome Sequencing in Cancer. Cold Spring Harb. Perspect. Med. 2019, 9, a034579. [Google Scholar] [CrossRef]
- Perou, C.M.; Sørlie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef]
- Harbeck, N.; Penault-Llorca, F.; Cortes, J.; Gnant, M.; Houssami, N.; Poortmans, P.; Ruddy, K.; Tsang, J.; Cardoso, F. Breast cancer. Nat. Rev. Dis. Primers 2019, 5, 66. [Google Scholar] [CrossRef]
- Holst, F.; Stahl, P.R.; Ruiz, C.; Hellwinkel, O.; Jehan, Z.; Wendland, M.; Lebeau, A.; Terracciano, L.; Al-Kuraya, K.; Jänicke, F.; et al. Estrogen receptor alpha (ESR1) gene amplification is frequent in breast cancer. Nat. Genet. 2007, 39, 655–660. [Google Scholar] [CrossRef]
- Dustin, D.; Gu, G.; Fuqua, S.A.W. ESR1 mutations in breast cancer. Cancer 2019, 125, 3714–3728. [Google Scholar] [CrossRef]
- Welboren, W.J.; Stunnenberg, H.G.; Sweep, F.C.; Span, P.N. Identifying estrogen receptor target genes. Mol. Oncol. 2007, 1, 138–143. [Google Scholar] [CrossRef]
- Lønning, P.E. The potency and clinical efficacy of aromatase inhibitors across the breast cancer continuum. Ann. Oncol. 2011, 22, 503–514. [Google Scholar] [CrossRef] [PubMed]
- Hackshaw, A.; Roughton, M.; Forsyth, S.; Monson, K.; Reczko, K.; Sainsbury, R.; Baum, M. Long-term benefits of 5 years of tamoxifen: 10-year follow-up of a large randomized trial in women at least 50 years of age with early breast cancer. J. Clin. Oncol. 2011, 29, 1657–1663. [Google Scholar] [CrossRef] [PubMed]
- Irani, S.; Tan, W.; Li, Q.; Toy, W.; Jones, C.; Gadiya, M.; Marra, A.; Katzenellenbogen, J.A.; Carlson, K.E.; Katzenellenbogen, B.S.; et al. Somatic estrogen receptor α mutations that induce dimerization promote receptor activity and breast cancer proliferation. J. Clin. Investig. 2024, 134, e163242. [Google Scholar] [CrossRef] [PubMed]
- Toy, W.; Weir, H.; Razavi, P.; Lawson, M.; Goeppert, A.U.; Mazzola, A.M.; Smith, A.; Wilson, J.; Morrow, C.; Wong, W.L.; et al. Activating ESR1 Mutations Differentially Affect the Efficacy of ER Antagonists. Cancer Discov. 2017, 7, 277–287. [Google Scholar] [CrossRef]
- Wang, H.; Miao, J.; Wen, Y.; Xia, X.; Chen, Y.; Huang, M.; Chen, S.; Zhao, Z.; Zhang, Y.; Chen, C.; et al. Molecular Landscape of ERBB2 Alterations in 14,956 Solid Tumors. Pathol. Oncol. Res. 2022, 28, 1610360. [Google Scholar] [CrossRef]
- Krishnamurti, U.; Silverman, J.F. HER2 in breast cancer: A review and update. Adv. Anat. Pathol. 2014, 21, 100–107. [Google Scholar] [CrossRef]
- Yarden, Y.; Sliwkowski, M.X. Untangling the ErbB signalling network. Nat. Rev. Mol. Cell Biol. 2001, 2, 127–137. [Google Scholar] [CrossRef]
- Diwanji, D.; Trenker, R.; Thaker, T.M.; Wang, F.; Agard, D.A.; Verba, K.A.; Jura, N. Structures of the HER2-HER3-NRG1β complex reveal a dynamic dimer interface. Nature 2021, 600, 339–343. [Google Scholar] [CrossRef]
- Ali, S.; Hendry, J.; Le, D.; Mondal, P.K.; Sami, A.; Chalchal, H.; Haider, K.; Ahmed, O.; El-Gayed, A.; Wright, P.; et al. Efficacy of adjuvant trastuzumab in women with HER2-positive T1a or bN0M0 breast cancer: A population-based cohort study. Sci. Rep. 2022, 12, 1068. [Google Scholar] [CrossRef]
- Swain, S.M.; Shastry, M.; Hamilton, E. Targeting HER2-positive breast cancer: Advances and future directions. Nat. Rev. Drug Discov. 2023, 22, 101–126. [Google Scholar] [CrossRef]
- Wu, X.; Yang, H.; Yu, X.; Qin, J.J. Drug-resistant HER2-positive breast cancer: Molecular mechanisms and overcoming strategies. Front. Pharmacol. 2022, 13, 1012552. [Google Scholar] [CrossRef] [PubMed]
- El-Deiry, W.S.; Goldberg, R.M.; Lenz, H.J.; Shields, A.F.; Gibney, G.T.; Tan, A.R.; Brown, J.; Eisenberg, B.; Heath, E.I.; Phuphanich, S.; et al. The current state of molecular testing in the treatment of patients with solid tumors, 2019. CA Cancer J. Clin. 2019, 69, 305–343. [Google Scholar] [CrossRef] [PubMed]
- Tung, N.; Garber, J.E. PARP inhibition in breast cancer: Progress made and future hopes. NPJ Breast Cancer 2022, 8, 47. [Google Scholar] [CrossRef] [PubMed]
- Griffith, M.; Miller, C.A.; Griffith, O.L.; Krysiak, K.; Skidmore, Z.L.; Ramu, A.; Walker, J.R.; Dang, H.X.; Trani, L.; Larson, D.E.; et al. Optimizing cancer genome sequencing and analysis. Cell Syst. 2015, 1, 210–223. [Google Scholar] [CrossRef] [PubMed]
- Frampton, G.M.; Fichtenholtz, A.; Otto, G.A.; Wang, K.; Downing, S.R.; He, J.; Schnall-Levin, M.; White, J.; Sanford, E.M.; An, P.; et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat. Biotechnol. 2013, 31, 1023–1031. [Google Scholar] [CrossRef] [PubMed]
- Sholl, L.M.; Do, K.; Shivdasani, P.; Cerami, E.; Dubuc, A.M.; Kuo, F.C.; Garcia, E.P.; Jia, Y.; Davineni, P.; Abo, R.P.; et al. Institutional implementation of clinical tumor profiling on an unselected cancer population. JCI Insight 2016, 1, e87062. [Google Scholar] [CrossRef]
- Kim, R.Y.; Xu, H.; Myllykangas, S.; Ji, H. Genetic-based biomarkers and next-generation sequencing: The future of personalized care in colorectal cancer. PerMed 2011, 8, 331–345. [Google Scholar] [CrossRef]
- Raj, G.V.; Moreno, J.G.; Gomella, L.G. Utilization of polymerase chain reaction technology in the detection of solid tumors. Cancer 1998, 82, 1419–1442. [Google Scholar] [CrossRef]
- Nagahashi, M.; Shimada, Y.; Ichikawa, H.; Kameyama, H.; Takabe, K.; Okuda, S.; Wakai, T. Next generation sequencing-based gene panel tests for the management of solid tumors. Cancer Sci. 2019, 110, 6–15. [Google Scholar] [CrossRef]
- Gopinathan, G.; Diekwisch, T.G.H. Epigenetics and Early Development. J. Dev. Biol. 2022, 10, 26. [Google Scholar] [CrossRef]
- Zovkic, I.B.; Guzman-Karlsson, M.C.; Sweatt, J.D. Epigenetic regulation of memory formation and maintenance. Learn Mem. 2013, 20, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Tilló, E.; Lázaro, A.; Torrent, R.; Cuatrecasas, M.; Vaquero, E.C.; Castells, A.; Engel, P.; Postigo, A. ZEB1 represses E-cadherin and induces an EMT by recruiting the SWI/SNF chromatin-remodeling protein BRG1. Oncogene 2010, 29, 3490–3500. [Google Scholar] [CrossRef]
- Lindner, P.; Paul, S.; Eckstein, M.; Hampel, C.; Muenzner, J.K.; Erlenbach-Wuensch, K.; Ahmed, H.P.; Mahadevan, V.; Brabletz, T.; Hartmann, A.; et al. EMT transcription factor ZEB1 alters the epigenetic landscape of colorectal cancer cells. Cell Death Dis. 2020, 11, 147. [Google Scholar] [CrossRef]
- Barrallo-Gimeno, A.; Nieto, M.A. The Snail genes as inducers of cell movement and survival: Implications in development and cancer. Development 2005, 132, 3151–3161. [Google Scholar] [CrossRef]
- Singh, A.; Settleman, J. EMT, cancer stem cells and drug resistance: An emerging axis of evil in the war on cancer. Oncogene 2010, 29, 4741–4751. [Google Scholar] [CrossRef]
- Guo, W.; Keckesova, Z.; Donaher, J.L.; Shibue, T.; Tischler, V.; Reinhardt, F.; Itzkovitz, S.; Noske, A.; Zürrer-Härdi, U.; Bell, G.; et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell 2012, 148, 1015–1028. [Google Scholar] [CrossRef]
- Lim, S.; Becker, A.; Zimmer, A.; Lu, J.; Buettner, R.; Kirfel, J. SNAI1-mediated epithelial-mesenchymal transition confers chemoresistance and cellular plasticity by regulating genes involved in cell death and stem cell maintenance. PLoS ONE 2013, 8, e66558. [Google Scholar] [CrossRef]
- Li, J.W.; Deng, Q.M.; Zhu, J.L.; Min, W.; Hu, X.Y.; Yu Chen, H.; Luo, Z.; Lin, L.L.; Wei, X.L.; Zhang, Y.Q.; et al. Methylation of ESR1 promoter induced by SNAI2-DNMT3B complex promotes epithelial-mesenchymal transition and correlates with poor prognosis in ERα-positive breast cancers. MedComm (2020) 2023, 4, e403. [Google Scholar] [CrossRef]
- Kirn, V.; Strake, L.; Thangarajah, F.; Richters, L.; Eischeid, H.; Koitzsch, U.; Odenthal, M.; Fries, J. ESR1-promoter-methylation status in primary breast cancer and its corresponding metastases. Clin. Exp. Metastasis 2018, 35, 707–712. [Google Scholar] [CrossRef] [PubMed]
- Butler, M.; Pongor, L.; Su, Y.T.; Xi, L.; Raffeld, M.; Quezado, M.; Trepel, J.; Aldape, K.; Pommier, Y.; Wu, J. MGMT Status as a Clinical Biomarker in Glioblastoma. Trends Cancer 2020, 6, 380–391. [Google Scholar] [CrossRef] [PubMed]
- Erasimus, H.; Gobin, M.; Niclou, S.; Van Dyck, E. DNA repair mechanisms and their clinical impact in glioblastoma. Mutat. Res. Rev. Mutat. Res. 2016, 769, 19–35. [Google Scholar] [CrossRef]
- Harris, L.C.; Potter, P.M.; Tano, K.; Shiota, S.; Mitra, S.; Brent, T.P. Characterization of the promoter region of the human O6-methylguanine-DNA methyltransferase gene. Nucleic Acids Res. 1991, 19, 6163–6167. [Google Scholar] [CrossRef]
- Esteller, M.; Hamilton, S.R.; Burger, P.C.; Baylin, S.B.; Herman, J.G. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999, 59, 793–797. [Google Scholar] [PubMed]
- Christmann, M.; Nagel, G.; Horn, S.; Krahn, U.; Wiewrodt, D.; Sommer, C.; Kaina, B. MGMT activity, promoter methylation and immunohistochemistry of pretreatment and recurrent malignant gliomas: A comparative study on astrocytoma and glioblastoma. Int. J. Cancer 2010, 127, 2106–2118. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.; Tanaka, M.; Trepel, J.; Reinhold, W.C.; Rajapakse, V.N.; Pommier, Y. Temozolomide in the Era of Precision Medicine. Cancer Res. 2017, 77, 823–826. [Google Scholar] [CrossRef]
- Heyn, H.; Vidal, E.; Ferreira, H.J.; Vizoso, M.; Sayols, S.; Gomez, A.; Moran, S.; Boque-Sastre, R.; Guil, S.; Martinez-Cardus, A.; et al. Epigenomic analysis detects aberrant super-enhancer DNA methylation in human cancer. Genome Biol. 2016, 17, 11. [Google Scholar] [CrossRef]
- Saghafinia, S.; Mina, M.; Riggi, N.; Hanahan, D.; Ciriello, G. Pan-Cancer Landscape of Aberrant DNA Methylation across Human Tumors. Cell Rep. 2018, 25, 1066–1080.e1068. [Google Scholar] [CrossRef]
- Audia, J.E.; Campbell, R.M. Histone Modifications and Cancer. Cold Spring Harb. Perspect Biol. 2016, 8, a019521. [Google Scholar] [CrossRef]
- Corces, M.R.; Granja, J.M.; Shams, S.; Louie, B.H.; Seoane, J.A.; Zhou, W.; Silva, T.C.; Groeneveld, C.; Wong, C.K.; Cho, S.W.; et al. The chromatin accessibility landscape of primary human cancers. Science 2018, 362, eaav1898. [Google Scholar] [CrossRef] [PubMed]
- Herman, J.G.; Graff, J.R.; Myöhänen, S.; Nelkin, B.D.; Baylin, S.B. Methylation-specific PCR: A novel PCR assay for methylation status of CpG islands. Proc. Natl. Acad. Sci. USA 1996, 93, 9821–9826. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.-L.; Lin, X.; Yu, Y.-L.; Chen, L.; Hu, Q.-X.; Chen, M.; Cao, N.; Zhao, C.; Wang, C.-Y.; Huang, C.-W.; et al. Genome-wide profiling in colorectal cancer identifies PHF19 and TBC1D16 as oncogenic super enhancers. Nat. Commun. 2021, 12, 6407. [Google Scholar] [CrossRef] [PubMed]
- Giannoni-Luza, S.; Acosta, O.; Murillo Carrasco, A.G.; Danos, P.; Cotrina Concha, J.M.; Guerra Miller, H.; Pinto, J.A.; Aguilar, A.; Araujo, J.M.; Fujita, R.; et al. Chip-based digital Polymerase Chain Reaction as quantitative technique for the detection of PIK3CA mutations in breast cancer patients. Heliyon 2022, 8, e11396. [Google Scholar] [CrossRef]
- Grosselin, K.; Durand, A.; Marsolier, J.; Poitou, A.; Marangoni, E.; Nemati, F.; Dahmani, A.; Lameiras, S.; Reyal, F.; Frenoy, O.; et al. High-throughput single-cell ChIP-seq identifies heterogeneity of chromatin states in breast cancer. Nat. Genet. 2019, 51, 1060–1066. [Google Scholar] [CrossRef]
- Grandi, F.C.; Modi, H.; Kampman, L.; Corces, M.R. Chromatin accessibility profiling by ATAC-seq. Nat. Protoc. 2022, 17, 1518–1552. [Google Scholar] [CrossRef]
- Patel, G.K.; Chugh, N.; Tripathi, M. Neuroendocrine Differentiation of Prostate Cancer-An Intriguing Example of Tumor Evolution at Play. Cancers 2019, 11, 1405. [Google Scholar] [CrossRef]
- Takuma, S.; Inoue, Y.; Karayama, M.; Tsuchiya, K.; Tsukui, H.; Hozumi, H.; Suzuki, Y.; Furuhashi, K.; Enomoto, N.; Fujisawa, T.; et al. EGFR-Mutated Lung Adenocarcinoma Successfully Treated With Osimertinib After Spontaneous Transformation to SCLC and Adenocarcinoma With Neuroendocrine Differentiation: Case Report. JTO Clin. Res. Rep. 2022, 3, 100264. [Google Scholar] [CrossRef]
- Davies, A.; Nouruzi, S.; Ganguli, D.; Namekawa, T.; Thaper, D.; Linder, S.; Karaoğlanoğlu, F.; Omur, M.E.; Kim, S.; Kobelev, M.; et al. An androgen receptor switch underlies lineage infidelity in treatment-resistant prostate cancer. Nat. Cell Biol. 2021, 23, 1023–1034. [Google Scholar] [CrossRef]
- Bluemn, E.G.; Coleman, I.M.; Lucas, J.M.; Coleman, R.T.; Hernandez-Lopez, S.; Tharakan, R.; Bianchi-Frias, D.; Dumpit, R.F.; Kaipainen, A.; Corella, A.N.; et al. Androgen Receptor Pathway-Independent Prostate Cancer Is Sustained through FGF Signaling. Cancer Cell 2017, 32, 474–489.e476. [Google Scholar] [CrossRef]
- Xie, Y.; Ning, S.; Hu, J. Molecular mechanisms of neuroendocrine differentiation in prostate cancer progression. J. Cancer Res. Clin. Oncol. 2022, 148, 1813–1823. [Google Scholar] [CrossRef] [PubMed]
- Rubin, M.A.; Bristow, R.G.; Thienger, P.D.; Dive, C.; Imielinski, M. Impact of Lineage Plasticity to and from a Neuroendocrine Phenotype on Progression and Response in Prostate and Lung Cancers. Mol Cell 2020, 80, 562–577. [Google Scholar] [CrossRef] [PubMed]
- Bhagirath, D.; Liston, M.; Patel, N.; Akoto, T.; Lui, B.; Yang, T.L.; To, D.M.; Majid, S.; Dahiya, R.; Tabatabai, Z.L.; et al. MicroRNA determinants of neuroendocrine differentiation in metastatic castration-resistant prostate cancer. Oncogene 2020, 39, 7209–7223. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.L.; Sood, A.; Rahimi, H.A.; Wang, W.; Gupta, N.; Hicks, J.; Mosier, S.; Gocke, C.D.; Epstein, J.I.; Netto, G.J.; et al. Rb loss is characteristic of prostatic small cell neuroendocrine carcinoma. Clin. Cancer Res. 2014, 20, 890–903. [Google Scholar] [CrossRef]
- Ku, S.Y.; Rosario, S.; Wang, Y.; Mu, P.; Seshadri, M.; Goodrich, Z.W.; Goodrich, M.M.; Labbé, D.P.; Gomez, E.C.; Wang, J.; et al. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science 2017, 355, 78–83. [Google Scholar] [CrossRef]
- Nava Rodrigues, D.; Casiraghi, N.; Romanel, A.; Crespo, M.; Miranda, S.; Rescigno, P.; Figueiredo, I.; Riisnaes, R.; Carreira, S.; Sumanasuriya, S.; et al. RB1 Heterogeneity in Advanced Metastatic Castration-Resistant Prostate Cancer. Clin. Cancer Res. 2019, 25, 687–697. [Google Scholar] [CrossRef]
- Cejas, P.; Xie, Y.; Font-Tello, A.; Lim, K.; Syamala, S.; Qiu, X.; Tewari, A.K.; Shah, N.; Nguyen, H.M.; Patel, R.A.; et al. Subtype heterogeneity and epigenetic convergence in neuroendocrine prostate cancer. Nat. Commun. 2021, 12, 5775. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, T.; Hong, D.; Dong, B.; Wang, Y.; Huang, H.; Zhang, W.; Lian, B.; Ji, B.; Shi, H.; et al. Single-cell transcriptional regulation and genetic evolution of neuroendocrine prostate cancer. iScience 2022, 25, 104576. [Google Scholar] [CrossRef]
- Drake, J.M.; Graham, N.A.; Lee, J.K.; Stoyanova, T.; Faltermeier, C.M.; Sud, S.; Titz, B.; Huang, J.; Pienta, K.J.; Graeber, T.G.; et al. Metastatic castration-resistant prostate cancer reveals intrapatient similarity and interpatient heterogeneity of therapeutic kinase targets. Proc. Natl. Acad. Sci. USA 2013, 110, E4762–E4769. [Google Scholar] [CrossRef]
- Beltran, H.; Oromendia, C.; Danila, D.C.; Montgomery, B.; Hoimes, C.; Szmulewitz, R.Z.; Vaishampayan, U.; Armstrong, A.J.; Stein, M.; Pinski, J.; et al. A Phase II Trial of the Aurora Kinase A Inhibitor Alisertib for Patients with Castration-resistant and Neuroendocrine Prostate Cancer: Efficacy and Biomarkers. Clin. Cancer Res. 2019, 25, 43–51. [Google Scholar] [CrossRef]
- VanDeusen, H.R.; Ramroop, J.R.; Morel, K.L.; Bae, S.Y.; Sheahan, A.V.; Sychev, Z.; Lau, N.A.; Cheng, L.C.; Tan, V.M.; Li, Z.; et al. Targeting RET Kinase in Neuroendocrine Prostate Cancer. Mol. Cancer Res. 2020, 18, 1176–1188. [Google Scholar] [CrossRef] [PubMed]
- Ge, R.; Wang, Z.; Cheng, L. Tumor microenvironment heterogeneity an important mediator of prostate cancer progression and therapeutic resistance. NPJ Precis. Oncol. 2022, 6, 31. [Google Scholar] [CrossRef] [PubMed]
- Hinshaw, D.C.; Shevde, L.A. The Tumor Microenvironment Innately Modulates Cancer Progression. Cancer Res. 2019, 79, 4557–4566. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Geng, H.; Liu, Y.; Liu, L.; Chen, Y.; Wu, F.; Liu, Z.; Ling, S.; Wang, Y.; Zhou, L. Hot and cold tumors: Immunological features and the therapeutic strategies. MedComm (2020) 2023, 4, e343. [Google Scholar] [CrossRef]
- Lippitz, B.E.; Harris, R.A. Cytokine patterns in cancer patients: A review of the correlation between interleukin 6 and prognosis. Oncoimmunology 2016, 5, e1093722. [Google Scholar] [CrossRef]
- van der Sijde, F.; Dik, W.A.; Mustafa, D.A.M.; Vietsch, E.E.; Besselink, M.G.; Debets, R.; Koerkamp, B.G.; Haberkorn, B.C.M.; Homs, M.Y.V.; Janssen, Q.P.; et al. Serum cytokine levels are associated with tumor progression during FOLFIRINOX chemotherapy and overall survival in pancreatic cancer patients. Front. Immunol. 2022, 13, 898498. [Google Scholar] [CrossRef]
- Xiao, Y.; Wang, Z.; Zhao, M.; Deng, Y.; Yang, M.; Su, G.; Yang, K.; Qian, C.; Hu, X.; Liu, Y.; et al. Single-Cell Transcriptomics Revealed Subtype-Specific Tumor Immune Microenvironments in Human Glioblastomas. Front. Immunol. 2022, 13, 914236. [Google Scholar] [CrossRef]
- Rooney, M.S.; Shukla, S.A.; Wu, C.J.; Getz, G.; Hacohen, N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 2015, 160, 48–61. [Google Scholar] [CrossRef]
- Ganesan, S.; Mehnert, J. Biomarkers for Response to Immune Checkpoint Blockade. Annu. Rev. Cancer Biol. 2020, 4, 331–351. [Google Scholar] [CrossRef]
- Costa, A.; Kieffer, Y.; Scholer-Dahirel, A.; Pelon, F.; Bourachot, B.; Cardon, M.; Sirven, P.; Magagna, I.; Fuhrmann, L.; Bernard, C.; et al. Fibroblast Heterogeneity and Immunosuppressive Environment in Human Breast Cancer. Cancer Cell 2018, 33, 463–479.e410. [Google Scholar] [CrossRef]
- Nassar, D.; Blanpain, C. Cancer Stem Cells: Basic Concepts and Therapeutic Implications. Annu. Rev. Pathol. 2016, 11, 47–76. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Ni, J.; Beretov, J.; Graham, P.; Li, Y. Cancer stem cell in breast cancer therapeutic resistance. Cancer Treat. Rev. 2018, 69, 152–163. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.Y.; Lin, Y.S.; Liang, C.W. Heterogeneity of cancer stem cell-related marker expression is associated with three-dimensional structures in malignant pleural effusion produced by lung adenocarcinoma. Cytopathology 2024, 35, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Rich, J.N. Cancer stem cells: Understanding tumor hierarchy and heterogeneity. Medicine 2016, 95, S2–S7. [Google Scholar] [CrossRef]
- Bocci, F.; Gearhart-Serna, L.; Boareto, M.; Ribeiro, M.; Ben-Jacob, E.; Devi, G.R.; Levine, H.; Onuchic, J.N.; Jolly, M.K. Toward understanding cancer stem cell heterogeneity in the tumor microenvironment. Proc. Natl. Acad. Sci. USA 2019, 116, 148–157. [Google Scholar] [CrossRef]
- Ye, Y.; Hu, Q.; Chen, H.; Liang, K.; Yuan, Y.; Xiang, Y.; Ruan, H.; Zhang, Z.; Song, A.; Zhang, H.; et al. Characterization of Hypoxia-associated Molecular Features to Aid Hypoxia-Targeted Therapy. Nat. Metab. 2019, 1, 431–444. [Google Scholar] [CrossRef]
- Yang, S.; Qian, L.; Li, Z.; Li, Y.; Bai, J.; Zheng, B.; Chen, K.; Qiu, X.; Cai, G.; Wang, S.; et al. Integrated Multi-Omics Landscape of Liver Metastases. Gastroenterology 2023, 164, 407–423.e417. [Google Scholar] [CrossRef]
- Höglund, M.; Gisselsson, D.; Säll, T.; Mitelman, F. Coping with complexity. multivariate analysis of tumor karyotypes. Cancer Genet. Cytogenet. 2002, 135, 103–109. [Google Scholar] [CrossRef]
- El-Deiry, W.S.; Taylor, B.; Neal, J.W. Tumor Evolution, Heterogeneity, and Therapy for Our Patients with Advanced Cancer: How Far Have We Come? Am. Soc. Clin. Oncol. Educ. Book 2017, 37, e8–e15. [Google Scholar] [CrossRef]
- Lyon, M.F. Sex chromatin and gene action in the mammalian X-chromosome. Am. J. Hum. Genet. 1962, 14, 135–148. [Google Scholar]
- Beutler, E.; Yeh, M.; Fairbanks, V.F. The normal human female as a mosaic of X-chromosome activity: Studies using the gene for C-6-PD-deficiency as a marker. Proc. Natl. Acad. Sci. USA 1962, 48, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Vogelstein, B.; Fearon, E.R.; Hamilton, S.R.; Feinberg, A.P. Use of restriction fragment length polymorphisms to determine the clonal origin of human tumors. Science 1985, 227, 642–645. [Google Scholar] [CrossRef] [PubMed]
- Vogelstein, B.; Papadopoulos, N.; Velculescu, V.E.; Zhou, S.; Diaz, L.A., Jr.; Kinzler, K.W. Cancer genome landscapes. Science 2013, 339, 1546–1558. [Google Scholar] [CrossRef] [PubMed]
- Reiter, J.G.; Baretti, M.; Gerold, J.M.; Makohon-Moore, A.P.; Daud, A.; Iacobuzio-Donahue, C.A.; Azad, N.S.; Kinzler, K.W.; Nowak, M.A.; Vogelstein, B. An analysis of genetic heterogeneity in untreated cancers. Nat. Rev. Cancer 2019, 19, 639–650. [Google Scholar] [CrossRef] [PubMed]
- Maley, C.C.; Galipeau, P.C.; Finley, J.C.; Wongsurawat, V.J.; Li, X.; Sanchez, C.A.; Paulson, T.G.; Blount, P.L.; Risques, R.-A.; Rabinovitch, P.S.; et al. Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat. Genet. 2006, 38, 468–473. [Google Scholar] [CrossRef] [PubMed]
- de Bruin, E.C.; McGranahan, N.; Mitter, R.; Salm, M.; Wedge, D.C.; Yates, L.; Jamal-Hanjani, M.; Shafi, S.; Murugaesu, N.; Rowan, A.J.; et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 2014, 346, 251–256. [Google Scholar] [CrossRef]
- Puram, S.V.; Tirosh, I.; Parikh, A.S.; Patel, A.P.; Yizhak, K.; Gillespie, S.; Rodman, C.; Luo, C.L.; Mroz, E.A.; Emerick, K.S.; et al. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell 2017, 171, 1611–1624.e1624. [Google Scholar] [CrossRef]
- Chen, C.C.; Tran, W.; Song, K.; Sugimoto, T.; Obusan, M.B.; Wang, L.; Sheu, K.M.; Cheng, D.; Ta, L.; Varuzhanyan, G.; et al. Temporal evolution reveals bifurcated lineages in aggressive neuroendocrine small cell prostate cancer trans-differentiation. Cancer Cell 2023, 41, 2066–2082.e2069. [Google Scholar] [CrossRef]
- Nguyen, H.M.; Vessella, R.L.; Morrissey, C.; Brown, L.G.; Coleman, I.M.; Higano, C.S.; Mostaghel, E.A.; Zhang, X.; True, L.D.; Lam, H.M.; et al. LuCaP Prostate Cancer Patient-Derived Xenografts Reflect the Molecular Heterogeneity of Advanced Disease an--d Serve as Models for Evaluating Cancer Therapeutics. Prostate 2017, 77, 654–671. [Google Scholar] [CrossRef]
- Bishop, J.L.; Thaper, D.; Vahid, S.; Davies, A.; Ketola, K.; Kuruma, H.; Jama, R.; Nip, K.M.; Angeles, A.; Johnson, F.; et al. The Master Neural Transcription Factor BRN2 Is an Androgen Receptor-Suppressed Driver of Neuroendocrine Differentiation in Prostate Cancer. Cancer Discov. 2017, 7, 54–71. [Google Scholar] [CrossRef]
- Gerlinger, M.; Rowan, A.J.; Horswell, S.; Math, M.; Larkin, J.; Endesfelder, D.; Gronroos, E.; Martinez, P.; Matthews, N.; Stewart, A.; et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012, 366, 883–892. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Love, J.C.; Navin, N.E.; Pachter, L.; Stubbington, M.J.; Svensson, V.; Sweedler, J.V.; Teichmann, S.A. Single-cell analysis at the threshold. Nat. Biotechnol. 2016, 34, 1111–1118. [Google Scholar] [CrossRef] [PubMed]
- Vandereyken, K.; Sifrim, A.; Thienpont, B.; Voet, T. Methods and applications for single-cell and spatial multi-omics. Nat. Rev. Genet. 2023, 24, 494–515. [Google Scholar] [CrossRef] [PubMed]
- Im, K.; Mareninov, S.; Diaz, M.F.P.; Yong, W.H. An Introduction to Performing Immunofluorescence Staining. Methods Mol. Biol. 2019, 1897, 299–311. [Google Scholar] [CrossRef]
- Jin, Y.; Zuo, Y.; Li, G.; Liu, W.; Pan, Y.; Fan, T.; Fu, X.; Yao, X.; Peng, Y. Advances in spatial transcriptomics and its applications in cancer research. Mol. Cancer 2024, 23, 129. [Google Scholar] [CrossRef]
- Marx, V. Method of the Year: Spatially resolved transcriptomics. Nat. Methods 2021, 18, 9–14. [Google Scholar] [CrossRef]
- Lu, T.; Ang, C.E.; Zhuang, X. Spatially resolved epigenomic profiling of single cells in complex tissues. Cell 2022, 185, 4448–4464.e4417. [Google Scholar] [CrossRef]
- Deng, Y.; Bartosovic, M.; Kukanja, P.; Zhang, D.; Liu, Y.; Su, G.; Enninful, A.; Bai, Z.; Castelo-Branco, G.; Fan, R. Spatial-CUT&Tag: Spatially resolved chromatin modification profiling at the cellular level. Science 2022, 375, 681–686. [Google Scholar] [CrossRef]
- Zhao, E.; Stone, M.R.; Ren, X.; Guenthoer, J.; Smythe, K.S.; Pulliam, T.; Williams, S.R.; Uytingco, C.R.; Taylor, S.E.B.; Nghiem, P.; et al. Spatial transcriptomics at subspot resolution with BayesSpace. Nat. Biotechnol. 2021, 39, 1375–1384. [Google Scholar] [CrossRef]
- Li, H.; Zhou, J.; Li, Z.; Chen, S.; Liao, X.; Zhang, B.; Zhang, R.; Wang, Y.; Sun, S.; Gao, X. A comprehensive benchmarking with practical guidelines for cellular deconvolution of spatial transcriptomics. Nat. Commun. 2023, 14, 1548. [Google Scholar] [CrossRef]
- Liu, Z.; Wu, D.; Zhai, W.; Ma, L. SONAR enables cell type deconvolution with spatially weighted Poisson-Gamma model for spatial transcriptomics. Nat. Commun. 2023, 14, 4727. [Google Scholar] [CrossRef] [PubMed]
- Vickovic, S.; Lötstedt, B.; Klughammer, J.; Mages, S.; Segerstolpe, Å.; Rozenblatt-Rosen, O.; Regev, A. SM-Omics is an automated platform for high-throughput spatial multi-omics. Nat. Commun. 2022, 13, 795. [Google Scholar] [CrossRef] [PubMed]
- Zollinger, D.R.; Lingle, S.E.; Sorg, K.; Beechem, J.M.; Merritt, C.R. GeoMx™ RNA Assay: High Multiplex, Digital, Spatial Analysis of RNA in FFPE Tissue. In In Situ Hybridization Protocols; Nielsen, B.S., Jones, J., Eds.; Springer: New York, NY, USA, 2020; pp. 331–345. [Google Scholar]
- Yu, X.; Zhao, H.; Wang, R.; Chen, Y.; Ouyang, X.; Li, W.; Sun, Y.; Peng, A. Cancer epigenetics: From laboratory studies and clinical trials to precision medicine. Cell Death Discov. 2024, 10, 28. [Google Scholar] [CrossRef] [PubMed]
- Rozenblatt-Rosen, O.; Regev, A.; Oberdoerffer, P.; Nawy, T.; Hupalowska, A.; Rood, J.E.; Ashenberg, O.; Cerami, E.; Coffey, R.J.; Demir, E.; et al. The Human Tumor Atlas Network: Charting Tumor Transitions across Space and Time at Single-Cell Resolution. Cell 2020, 181, 236–249. [Google Scholar] [CrossRef]
- Sammut, S.-J.; Crispin-Ortuzar, M.; Chin, S.-F.; Provenzano, E.; Bardwell, H.A.; Ma, W.; Cope, W.; Dariush, A.; Dawson, S.-J.; Abraham, J.E.; et al. Multi-omic machine learning predictor of breast cancer therapy response. Nature 2022, 601, 623–629. [Google Scholar] [CrossRef]
- Khadirnaikar, S.; Shukla, S.; Prasanna, S.R.M. Machine learning based combination of multi-omics data for subgroup identification in non-small cell lung cancer. Sci. Rep. 2023, 13, 4636. [Google Scholar] [CrossRef]
- Hong, M.K.; Macintyre, G.; Wedge, D.C.; Van Loo, P.; Patel, K.; Lunke, S.; Alexandrov, L.B.; Sloggett, C.; Cmero, M.; Marass, F.; et al. Tracking the origins and drivers of subclonal metastatic expansion in prostate cancer. Nat. Commun. 2015, 6, 6605. [Google Scholar] [CrossRef]
- Gundem, G.; Van Loo, P.; Kremeyer, B.; Alexandrov, L.B.; Tubio, J.M.C.; Papaemmanuil, E.; Brewer, D.S.; Kallio, H.M.L.; Högnäs, G.; Annala, M.; et al. The evolutionary history of lethal metastatic prostate cancer. Nature 2015, 520, 353–357. [Google Scholar] [CrossRef]
- Komarova, N.L.; Wodarz, D. Drug resistance in cancer: Principles of emergence and prevention. Proc. Natl. Acad. Sci. USA 2005, 102, 9714–9719. [Google Scholar] [CrossRef]
- Durrett, R.; Moseley, S. Evolution of resistance and progression to disease during clonal expansion of cancer. Theor. Popul. Biol. 2010, 77, 42–48. [Google Scholar] [CrossRef]
- Turke, A.B.; Zejnullahu, K.; Wu, Y.L.; Song, Y.; Dias-Santagata, D.; Lifshits, E.; Toschi, L.; Rogers, A.; Mok, T.; Sequist, L.; et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell 2010, 17, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Diaz, L.A., Jr.; Williams, R.T.; Wu, J.; Kinde, I.; Hecht, J.R.; Berlin, J.; Allen, B.; Bozic, I.; Reiter, J.G.; Nowak, M.A.; et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 2012, 486, 537–540. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.R.; Zhou, L.; El-Deiry, W.S. Circulating Tumor Cells Versus Circulating Tumor DNA in Colorectal Cancer: Pros and Cons. Curr. Colorectal. Cancer Rep. 2016, 12, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Telekes, A.; Horváth, A. The Role of Cell-Free DNA in Cancer Treatment Decision Making. Cancers 2022, 14, 6115. [Google Scholar] [CrossRef]
- Claus, J.; De Smet, D.; Breyne, J.; Wesolowski, J.; Himpe, U.; Demedts, I.; Martens, G.A. Patient-centric thresholding of Cobas® EGFR mutation Test v2 for surveillance of EGFR-mutated metastatic non-small cell lung cancer. Sci. Rep. 2024, 14, 18191. [Google Scholar] [CrossRef]
- Newman, A.M.; Lovejoy, A.F.; Klass, D.M.; Kurtz, D.M.; Chabon, J.J.; Scherer, F.; Stehr, H.; Liu, C.L.; Bratman, S.V.; Say, C.; et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat. Biotechnol. 2016, 34, 547–555. [Google Scholar] [CrossRef]
- Jamal-Hanjani, M.; Wilson, G.A.; Horswell, S.; Mitter, R.; Sakarya, O.; Constantin, T.; Salari, R.; Kirkizlar, E.; Sigurjonsson, S.; Pelham, R.; et al. Detection of ubiquitous and heterogeneous mutations in cell-free DNA from patients with early-stage non-small-cell lung cancer. Ann. Oncol. 2016, 27, 862–867. [Google Scholar] [CrossRef]
- De Mattos-Arruda, L.; Weigelt, B.; Cortes, J.; Won, H.H.; Ng, C.K.Y.; Nuciforo, P.; Bidard, F.C.; Aura, C.; Saura, C.; Peg, V.; et al. Capturing intra-tumor genetic heterogeneity by de novo mutation profiling of circulating cell-free tumor DNA: A proof-of-principle. Ann. Oncol. 2014, 25, 1729–1735. [Google Scholar] [CrossRef]
- Galardi, F.; Luca, F.; Romagnoli, D.; Biagioni, C.; Moretti, E.; Biganzoli, L.; Leo, A.D.; Migliaccio, I.; Malorni, L.; Benelli, M. Cell-Free DNA-Methylation-Based Methods and Applications in Oncology. Biomolecules 2020, 10, 1677. [Google Scholar] [CrossRef]
- Ortiz-Muñoz, G.; Brown, M.; Carbone, C.B.; Pechuan-Jorge, X.; Rouilly, V.; Lindberg, H.; Ritter, A.T.; Raghupathi, G.; Sun, Q.; Nicotra, T.; et al. In situ tumour arrays reveal early environmental control of cancer immunity. Nature 2023, 618, 827–833. [Google Scholar] [CrossRef]
- Han, J.; Duan, J.; Bai, H.; Wang, Y.; Wan, R.; Wang, X.; Chen, S.; Tian, Y.; Wang, D.; Fei, K.; et al. TCR Repertoire Diversity of Peripheral PD-1(+)CD8(+) T Cells Predicts Clinical Outcomes after Immunotherapy in Patients with Non-Small Cell Lung Cancer. Cancer Immunol. Res. 2020, 8, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Boutsikou, E.; Domvri, K.; Hardavella, G.; Tsiouda, D.; Zarogoulidis, K.; Kontakiotis, T. Tumour necrosis factor, interferon-gamma and interleukins as predictive markers of antiprogrammed cell-death protein-1 treatment in advanced non-small cell lung cancer: A pragmatic approach in clinical practice. Ther. Adv. Med. Oncol. 2018, 10, 1758835918768238. [Google Scholar] [CrossRef] [PubMed]
- Sivapalan, L.; Murray, J.C.; Canzoniero, J.V.; Landon, B.; Jackson, J.; Scott, S.; Lam, V.; Levy, B.P.; Sausen, M.; Anagnostou, V. Liquid biopsy approaches to capture tumor evolution and clinical outcomes during cancer immunotherapy. J. Immunother. Cancer 2023, 11, e005924. [Google Scholar] [CrossRef] [PubMed]
- Cogdill, A.P.; Andrews, M.C.; Wargo, J.A. Hallmarks of response to immune checkpoint blockade. Br. J. Cancer 2017, 117, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Chowell, D.; Morris, L.G.T.; Grigg, C.M.; Weber, J.K.; Samstein, R.M.; Makarov, V.; Kuo, F.; Kendall, S.M.; Requena, D.; Riaz, N.; et al. Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science 2018, 359, 582–587. [Google Scholar] [CrossRef]
- Routy, B.; Le Chatelier, E.; Derosa, L.; Duong, C.P.M.; Alou, M.T.; Daillère, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018, 359, 91–97. [Google Scholar] [CrossRef]
- Gopalakrishnan, V.; Spencer, C.N.; Nezi, L.; Reuben, A.; Andrews, M.C.; Karpinets, T.V.; Prieto, P.A.; Vicente, D.; Hoffman, K.; Wei, S.C.; et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018, 359, 97–103. [Google Scholar] [CrossRef]
- Kugel, C.H., 3rd; Douglass, S.M.; Webster, M.R.; Kaur, A.; Liu, Q.; Yin, X.; Weiss, S.A.; Darvishian, F.; Al-Rohil, R.N.; Ndoye, A.; et al. Age Correlates with Response to Anti-PD1, Reflecting Age-Related Differences in Intratumoral Effector and Regulatory T-Cell Populations. Clin. Cancer Res. 2018, 24, 5347–5356. [Google Scholar] [CrossRef]
- Shaikh, S.R.; Beck, M.A.; Alwarawrah, Y.; MacIver, N.J. Emerging mechanisms of obesity-associated immune dysfunction. Nat. Rev. Endocrinol. 2024, 20, 136–148. [Google Scholar] [CrossRef]
- McQuade, J.L.; Daniel, C.R.; Hess, K.R.; Mak, C.; Wang, D.Y.; Rai, R.R.; Park, J.J.; Haydu, L.E.; Spencer, C.; Wongchenko, M.; et al. Association of body-mass index and outcomes in patients with metastatic melanoma treated with targeted therapy, immunotherapy, or chemotherapy: A retrospective, multicohort analysis. Lancet Oncol. 2018, 19, 310–322. [Google Scholar] [CrossRef]
- Huang, S.Y.; Su, P.J.; Lin, C.T.; Kuo, M.C.; Chen, Y.H.; Wu, C.C.; Luo, H.L.; Chen, C.H.; Chou, C.C.; Huang, C.C.; et al. The impact of body mass index on survival endpoints among patients with metastatic urothelial carcinoma undergoing treatment with immune checkpoint inhibitors: A real-world multicenter analysis. Cancer Med. 2024, 13, e7008. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Aguilar, E.G.; Luna, J.I.; Dunai, C.; Khuat, L.T.; Le, C.T.; Mirsoian, A.; Minnar, C.M.; Stoffel, K.M.; Sturgill, I.R.; et al. Paradoxical effects of obesity on T cell function during tumor progression and PD-1 checkpoint blockade. Nat. Med. 2019, 25, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Botticelli, A.; Onesti, C.E.; Zizzari, I.; Cerbelli, B.; Sciattella, P.; Occhipinti, M.; Roberto, M.; Di Pietro, F.; Bonifacino, A.; Ghidini, M.; et al. The sexist behaviour of immune checkpoint inhibitors in cancer therapy? Oncotarget 2017, 8, 99336–99346. [Google Scholar] [CrossRef] [PubMed]
- Conforti, F.; Pala, L.; Bagnardi, V.; Viale, G.; De Pas, T.; Pagan, E.; Pennacchioli, E.; Cocorocchio, E.; Ferrucci, P.F.; De Marinis, F.; et al. Sex-Based Heterogeneity in Response to Lung Cancer Immunotherapy: A Systematic Review and Meta-Analysis. J. Natl. Cancer Inst. 2019, 111, 772–781. [Google Scholar] [CrossRef]
- Rubin, J.B. The spectrum of sex differences in cancer. Trends Cancer 2022, 8, 303–315. [Google Scholar] [CrossRef]
- Kammula, A.V.; Schäffer, A.A.; Rajagopal, P.S.; Kurzrock, R.; Ruppin, E. Outcome differences by sex in oncology clinical trials. Nat. Commun. 2024, 15, 2608. [Google Scholar] [CrossRef]
- Elfaki, I.; Mir, R.; Almutairi, F.M.; Duhier, F.M.A. Cytochrome P450: Polymorphisms and Roles in Cancer, Diabetes and Atherosclerosis. Asian Pac. J. Cancer Prev. 2018, 19, 2057–2070. [Google Scholar] [CrossRef]
- Edwardson, D.W.; Narendrula, R.; Chewchuk, S.; Mispel-Beyer, K.; Mapletoft, J.P.; Parissenti, A.M. Role of Drug Metabolism in the Cytotoxicity and Clinical Efficacy of Anthracyclines. Curr. Drug Metab. 2015, 16, 412–426. [Google Scholar] [CrossRef]
- Placido, D.; Yuan, B.; Hjaltelin, J.X.; Zheng, C.; Haue, A.D.; Chmura, P.J.; Yuan, C.; Kim, J.; Umeton, R.; Antell, G.; et al. A deep learning algorithm to predict risk of pancreatic cancer from disease trajectories. Nat. Med. 2023, 29, 1113–1122. [Google Scholar] [CrossRef]
- Chang, T.G.; Cao, Y.; Sfreddo, H.J.; Dhruba, S.R.; Lee, S.H.; Valero, C.; Yoo, S.K.; Chowell, D.; Morris, L.G.T.; Ruppin, E. LORIS robustly predicts patient outcomes with immune checkpoint blockade therapy using common clinical, pathologic and genomic features. Nat. Cancer 2024, 5, 1158–1175. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Fares, J.; Fares, M.Y.; Khachfe, H.H.; Salhab, H.A.; Fares, Y. Molecular principles of metastasis: A hallmark of cancer revisited. Signal Transduct. Target. Ther. 2020, 5, 28. [Google Scholar] [CrossRef] [PubMed]
- Sprouffske, K.; Kerr, G.; Li, C.; Prahallad, A.; Rebmann, R.; Waehle, V.; Naumann, U.; Bitter, H.; Jensen, M.R.; Hofmann, F.; et al. Genetic heterogeneity and clonal evolution during metastasis in breast cancer patient-derived tumor xenograft models. Comput. Struct. Biotechnol. J. 2020, 18, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.; Vegesna, R.; Mukherjee, S.; Kammula, A.V.; Dhruba, S.R.; Wu, W.; Kerr, D.L.; Nair, N.U.; Jones, M.G.; Yosef, N.; et al. PERCEPTION predicts patient response and resistance to treatment using single-cell transcriptomics of their tumors. Nat. Cancer 2024, 5, 938–952. [Google Scholar] [CrossRef]
- Zhang, T.; Zhang, L.; Payne, P.R.O.; Li, F. Synergistic Drug Combination Prediction by Integrating Multiomics Data in Deep Learning Models. Methods Mol. Biol. 2021, 2194, 223–238. [Google Scholar] [CrossRef]
- Feldt, S.L.; Bestvina, C.M. The Role of MET in Resistance to EGFR Inhibition in NSCLC: A Review of Mechanisms and Treatment Implications. Cancers 2023, 15, 2998. [Google Scholar] [CrossRef]
- Burrell, R.A.; McGranahan, N.; Bartek, J.; Swanton, C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature 2013, 501, 338–345. [Google Scholar] [CrossRef]
- Sacco, J.L.; Gomez, E.W. Epithelial-Mesenchymal Plasticity and Epigenetic Heterogeneity in Cancer. Cancers 2024, 16, 3289. [Google Scholar] [CrossRef]
- Carter, B.; Zhao, K. The epigenetic basis of cellular heterogeneity. Nat. Rev. Genet. 2021, 22, 235–250. [Google Scholar] [CrossRef]
- Ramón, Y.C.S.; Sesé, M.; Capdevila, C.; Aasen, T.; De Mattos-Arruda, L.; Diaz-Cano, S.J.; Hernández-Losa, J.; Castellví, J. Clinical implications of intratumor heterogeneity: Challenges and opportunities. J. Mol. Med. 2020, 98, 161–177. [Google Scholar] [CrossRef]
- Marusyk, A.; Almendro, V.; Polyak, K. Intra-tumour heterogeneity: A looking glass for cancer? Nat. Rev. Cancer 2012, 12, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Buikhuisen, J.Y.; Torang, A.; Medema, J.P. Exploring and modelling colon cancer inter-tumour heterogeneity: Opportunities and challenges. Oncogenesis 2020, 9, 66. [Google Scholar] [CrossRef] [PubMed]
- Zaal, E.A.; Berkers, C.R. The Influence of Metabolism on Drug Response in Cancer. Front. Oncol. 2018, 8, 500. [Google Scholar] [CrossRef] [PubMed]
- Hazini, A.; Fisher, K.; Seymour, L. Deregulation of HLA-I in cancer and its central importance for immunotherapy. J. Immunother. Cancer 2021, 9, e002899. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
MacDonald, W.J.; Purcell, C.; Pinho-Schwermann, M.; Stubbs, N.M.; Srinivasan, P.R.; El-Deiry, W.S. Heterogeneity in Cancer. Cancers 2025, 17, 441. https://doi.org/10.3390/cancers17030441
MacDonald WJ, Purcell C, Pinho-Schwermann M, Stubbs NM, Srinivasan PR, El-Deiry WS. Heterogeneity in Cancer. Cancers. 2025; 17(3):441. https://doi.org/10.3390/cancers17030441
Chicago/Turabian StyleMacDonald, William J., Connor Purcell, Maximilian Pinho-Schwermann, Nolan M. Stubbs, Praveen R. Srinivasan, and Wafik S. El-Deiry. 2025. "Heterogeneity in Cancer" Cancers 17, no. 3: 441. https://doi.org/10.3390/cancers17030441
APA StyleMacDonald, W. J., Purcell, C., Pinho-Schwermann, M., Stubbs, N. M., Srinivasan, P. R., & El-Deiry, W. S. (2025). Heterogeneity in Cancer. Cancers, 17(3), 441. https://doi.org/10.3390/cancers17030441







